Transcriptome Reveals Granulosa Cells Coping through Redox, Inflammatory and Metabolic Mechanisms under Acute Heat Stress
Abstract
:1. Introduction
2. Methods
2.1. Granulosa Cell Culture, Heat Treatment and Cell Proliferation Assay
2.2. Reactive Oxygen Species, Apoptosis and Hormone Measurements
2.3. Statistical Analysis of Physiological Parameters
2.4. RNA Sequencing
2.5. Differential Expression, Validation and Functional Analyses of Differentially Expressed Genes
3. Results
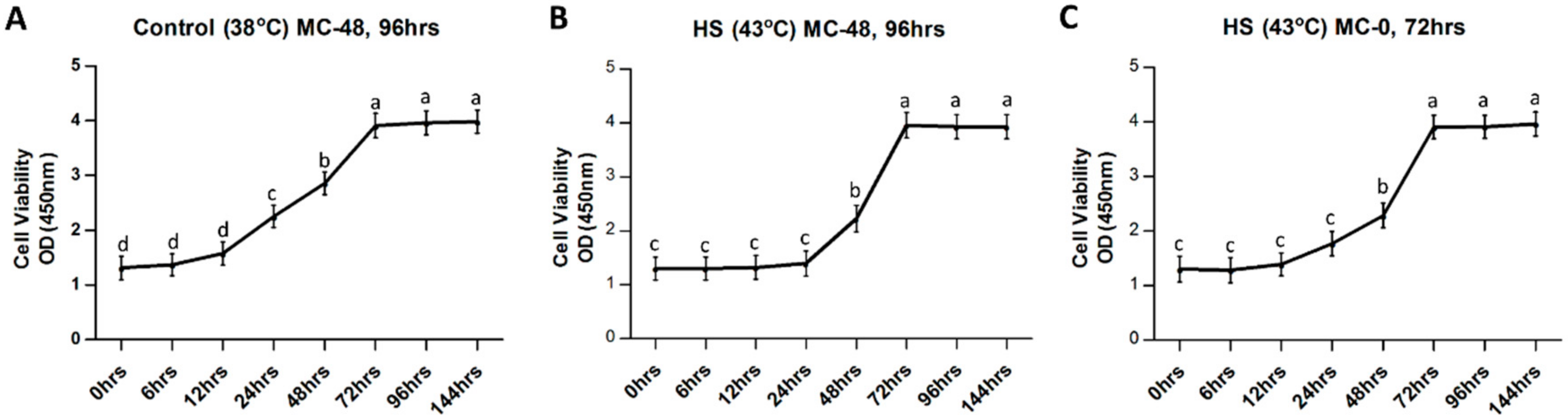
3.1. Influence of Heat Stress on Physical Parameters of Bovine Granulosa Cells
3.2. Role of Acute Heat Stress on GCs through Deep Sequencing
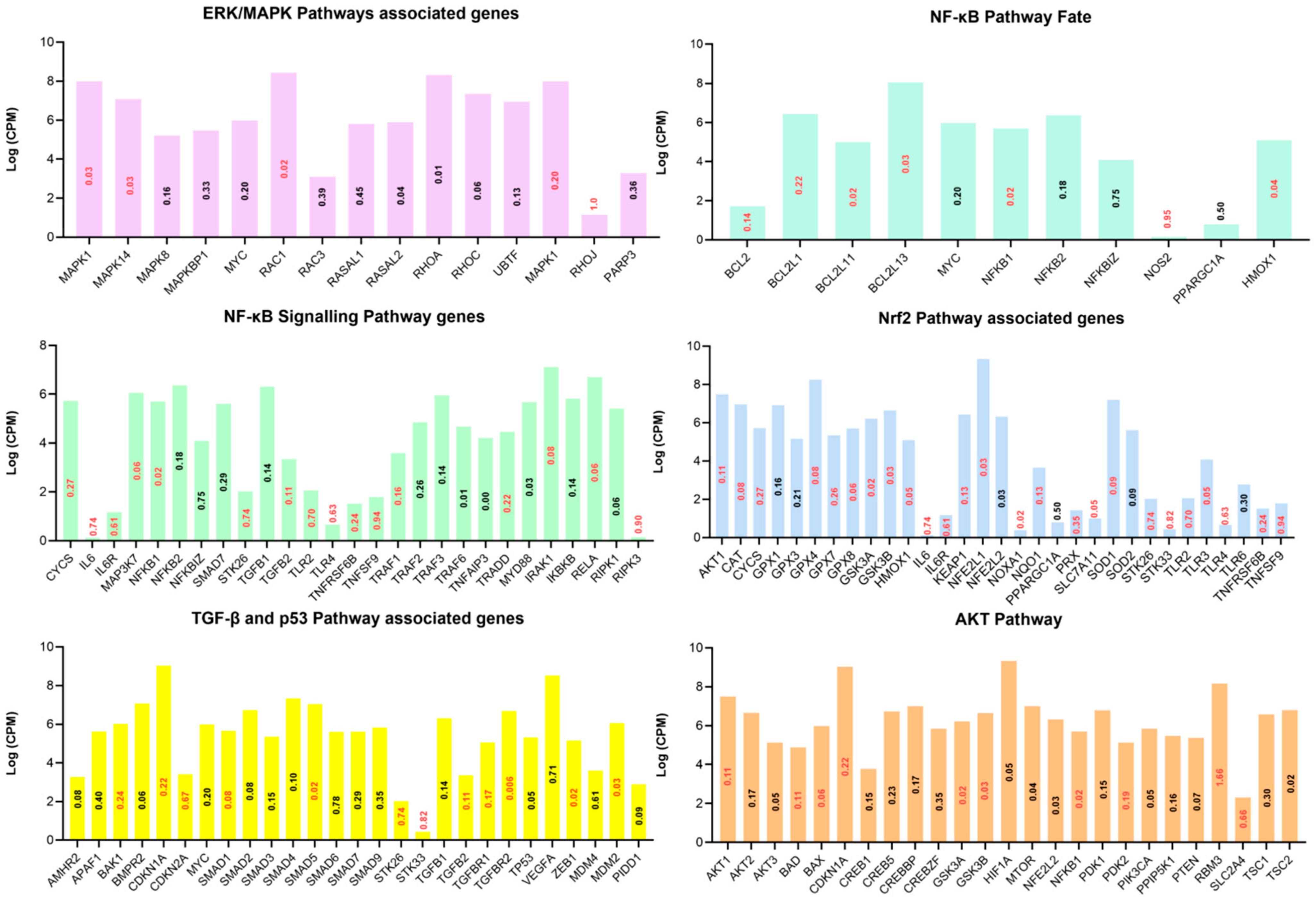
3.3. Pathway Analysis of Differentially Expressed Genes
3.4. Functional Annotation of Differentially Expressed Genes
3.5. Protein–Protein Interaction (PPI) Networks of DEGs under Acute Heat Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sherwood, S.C.; Huber, M. An adaptability limit to climate change due to heat stress. Proc. Natl. Acad. Sci. USA 2010, 107, 9552–9555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jyoteeshkumar Reddy, P.; Perkins-Kirkpatrick, S.E.; Sharples, J.J. Intensifying Australian Heatwave Trends and Their Sensitivity to Observational Data. Earth’s Future 2021, 9, e2020EF001924. [Google Scholar] [CrossRef]
- Li, X.X. Heat wave trends in Southeast Asia during 1979–2018: The impact of humidity. Sci. Total Environ. 2020, 721, 137664. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobell, D.B.; Gourdji, S.M. The Influence of Climate Change on Global Crop Productivity. Plant Physiol. 2012, 160, 1686–1697. [Google Scholar] [CrossRef] [Green Version]
- Rahimi, J.; Mutua, J.Y.; Notenbaert, A.M.O.; Marshall, K.; Butterbach-Bahl, K. Heat stress will detrimentally impact future livestock production in East Africa. Nat. Food 2021, 2, 88–96. [Google Scholar] [CrossRef]
- Kovats, R.S.; Hajat, S. Heat stress and public health: A critical review. Annu. Rev. Public Health 2008, 29, 41–55. [Google Scholar] [CrossRef]
- Kenney, W.L.; Craighead, D.H.; Alexander, L.M. Heat waves aging and human cardiovascular health. Med. Sci. Sports Exerc. 2014, 46, 1891–1899. [Google Scholar] [CrossRef] [Green Version]
- Das, R.; Sailo, L.; Verma, N.; Bharti, P.; Saikia, J.; Imtiwati; Kumar, R. Impact of heat stress on health and performance of dairy animals: A review. Vet. World 2016, 9, 260–268. [Google Scholar] [CrossRef] [Green Version]
- Collier, R.J.; Collier, J.L. Environmental Physiology of Livestock; Wiley: Hoboken, NJ, USA, 2012. [Google Scholar]
- Sammad, A.; Luo, H.; Qiu, W.; Galindez, J.M.; Wang, Y.; Guo, G.; Huang, X.; Wang, Y. Automated monitoring of seasonal and diurnal variation of rumination behaviour: Insights into thermotolerance management of Holstein cows. Biosyst. Eng. 2021. [Google Scholar] [CrossRef]
- Ray, D.; Correa-Calderon, A.; Armstrong, D.; Enns, M.; DeNise, S.; Howison, C. Thermoregulatory responses of Holstein and Brown Swiss Heat-Stressed dairy cows to two different cooling systems. Int. J. Biometeorol. 2004, 48, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Asseng, S.; Spänkuch, D.; Hernandez-Ochoa, I.M.; Laporta, J. The upper temperature thresholds of life. Lancet Planet. Health 2021, 5, e378–e385. [Google Scholar] [CrossRef]
- Pinto, S.; Hoffmann, G.; Ammon, C.; Amon, T. Critical THI thresholds based on the physiological parameters of lactating dairy cows. J. Therm. Biol. 2020, 88, 102523. [Google Scholar] [CrossRef] [PubMed]
- Sammad, A.; Wang, Y.J.; Umer, S.; Lirong, H.; Khan, I.; Khan, A.; Ahmad, B.; Wang, Y. Nutritional physiology and biochemistry of dairy cattle under the influence of heat stress: Consequences and opportunities. Animals 2020, 10, 793. [Google Scholar] [CrossRef]
- Sammad, A.; Khan, M.Z.; Abbas, Z.; Hu, L.; Ullah, Q.; Wang, Y.; Zhu, H.; Wang, Y. Major Nutritional Metabolic Alterations Influencing the Reproductive System of Postpartum Dairy Cows. Metabolites 2022, 12, 60. [Google Scholar] [CrossRef]
- Hansen, P.J. Embryonic mortality in cattle from the embryo’s perspective. J. Anim. Sci. 2002, 80, E33–E44. [Google Scholar] [CrossRef]
- Sammad, A.; Umer, S.; Shi, R.; Zhu, H.; Zhao, X.; Wang, Y. Dairy cow reproduction under the influence of heat stress. J. Anim. Physiol. Anim. Nutr. 2020, 104, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Destaw, W.M.; Kefyalew, A.W. Evaluation of the reproductive performance of Holstein Friesian dairy cows in Alage ATVET college, Ethiopia. Int. J. Livest. Prod. 2018, 9, 131–139. [Google Scholar] [CrossRef]
- Jahromi, B.N.; Mosallanezhad, Z.; Matloob, N.; Davari, M.; Ghobadifar, M.A. The potential role of granulosa cells in the maturation rate of immature human oocytes and embryo development: A co-culture study. Clin. Exp. Reprod. Med. 2015, 42, 111–117. [Google Scholar] [CrossRef]
- Diaz, F.J.; Wigglesworth, K.; Eppig, J.J. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J. Cell Sci. 2007, 120, 1009–1016. [Google Scholar] [CrossRef] [Green Version]
- Alemu, T.W.; Pandey, H.O.; Salilew Wondim, D.; Gebremedhn, S.; Neuhof, C.; Tholen, E.; Holker, M.; Schellander, K.; Tesfaye, D. Oxidative and endoplasmic reticulum stress defense mechanisms of bovine granulosa cells exposed to heat stress. Theriogenology 2018, 110, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Dou, J.; Wang, Y.; Jiang, X.; Khan, M.Z.; Luo, H.; Usman, T.; Zhu, H. Evaluation of heat stress effects on cellular and transcriptional adaptation of bovine granulosa cells. J. Anim. Sci. Biotechnol. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.L.; Liu, D.; Ding, G.R.; Liao, P.F.; Zhang, J.W. Hypoxia-inducible factor-1α and Wnt/β-catenin signaling pathways promote the invasion of hypoxic gastric cancer cells. Mol. Med. Rep. 2015, 12, 3365–3373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, Z. Heat stress reduces maturation and developmental capacity in bovine oocytes. Reprod. Fertil. Dev. 2021, 33, 66–75. [Google Scholar] [CrossRef]
- Roth, Z.; Meiden, R.; Braw-Tal, R.; Wolfenson, D. Immediate and delayed effects of heat stress on follicular development and its association with plasma FSH and inhibin concentration in cows. J. Reprod. Fertil. 2000, 120, 66–75. [Google Scholar] [CrossRef]
- Gaskins, A.J.; Mínguez-Alarcón, L.; VoPham, T.; Hart, J.E.; Chavarro, J.E.; Schwartz, J.; Souter, I.; Laden, F. Impact of ambient temperature on ovarian reserve. Fertil. Steril. 2021, 116, 1052–1060. [Google Scholar] [CrossRef]
- Wolfenson, D.; Roth, Z. Impact of heat stress on cow reproduction and fertility. Anim. Front. 2019, 9, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Tseng, J.K.; Chen, C.H.; Chou, P.C.; Yeh, S.P.; Ju, J.C. Influences of follicular size on parthenogenetic activation and in vitro heat shock on the cytoskeleton in cattle oocytes. Reprod. Domest. Anim. 2004, 39, 146–153. [Google Scholar] [CrossRef]
- Shehab-El-Deen, M.A.M.M.; Leroy, J.L.M.R.; Fadel, M.S.; Saleh, S.Y.A.; Maes, D.; Van Soom, A. Biochemical changes in the follicular fluid of the dominant follicle of high producing dairy cows exposed to heat stress early post-partum. Anim. Reprod. Sci. 2010, 117, 189–200. [Google Scholar] [CrossRef]
- Baumgard, L.H.; Wheelock, J.B.; Sanders, S.R.; Moore, C.E.; Green, H.B.; Waldron, M.R.; Rhoads, R.P. Postabsorptive carbohydrate adaptations to heat stress and monensin supplementation in lactating Holstein cows1. J. Dairy Sci. 2011, 94, 5620–5633. [Google Scholar] [CrossRef]
- Shin, E.K.; Jeong, J.K.; Choi, I.S.; Kang, H.G.; Hur, T.Y.; Jung, Y.H.; Kim, I.H. Relationships among ketosis, serum metabolites, body condition, and reproductive outcomes in dairy cows. Theriogenology 2015, 84, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Leroy, J.L.M.R.; Vanholder, T.; Mateusen, B.; Christophe, A.; Opsomer, G.; de Kruif, A.; Genicot, G.; Van Soom, A. Non-esterified fatty acids in follicular fluid of dairy cows and their effect on developmental capacity of bovine oocytes in vitro. Reproduction 2005, 130, 485–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bromfield, J.J.; Sheldon, I.M. Lipopolysaccharide initiates inflammation in bovine granulosa cells via the TLR4 pathway and perturbs oocyte meiotic progression in vitro. Endocrinology 2011, 152, 5029–5040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saadeldin, I.M.; Swelum, A.A.A.; Elsafadi, M.; Mahmood, A.; Osama, A.; Shikshaky, H.; Alfayez, M.; Alowaimer, A.N.; Magdeldin, S. Thermotolerance and plasticity of camel somatic cells exposed to acute and chronic heat stress. J. Adv. Res. 2020, 22, 105–118. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Swelum, A.A.A.; Elsafadi, M.; Mahmood, A.; Alfayez, M.; Alowaimer, A.N. Differences between the tolerance of camel oocytes and cumulus cells to acute and chronic hyperthermia. J. Therm. Biol. 2018, 74, 47–54. [Google Scholar] [CrossRef]
- Fu, Y.; He, C.-J.; Ji, P.-Y.; Zhuo, Z.-Y.; Tian, X.-Z.; Wang, F.; Tan, D.-X.; Liu, G.-S. Effects of Melatonin on the Proliferation and Apoptosis of Sheep Granulosa Cells under Thermal Stress. Int. J. Mol. Sci. 2014, 15, 21090–21104. [Google Scholar] [CrossRef] [Green Version]
- Sammad, A.; Hu, L.; Luo, H.; Abbas, Z.; Umer, S.; Zhao, S.; Xu, Q.; Khan, A.; Wang, Y.; Zhu, H.; et al. Investigation of Metabolome Underlying the Biological Mechanisms of Acute Heat Stressed Granulosa Cells. Int. J. Mol. Sci. 2022, 23, 2146. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI Reagent). Cold Spring Harb. Protoc. 2010, 5, pdb-prot5439. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, J.; Schenkel, F.; Hu, L.; Khan, A.; Khan, M.Z.; Yu, Y.; Wang, Y.; Wang, Y. Genome-wide identification and functional prediction of long non-coding RNAs in Sprague-Dawley rats during heat stress. BMC Genom. 2021, 22, 122. [Google Scholar] [CrossRef] [PubMed]
- Dobson, H.; Smith, R.; Royal, M.; Knight, C.; Sheldon, I. The high-producing dairy cow and its reproductive performance. Reprod. Domest. Anim. 2007, 42 (Suppl. 2), 17–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vergara, C.F.; Döpfer, D.; Cook, N.B.; Nordlund, K.V.; McArt, J.A.A.; Nydam, D.V.; Oetzel, G.R. Risk factors for postpartum problems in dairy cows: Explanatory and predictive modeling. J. Dairy Sci. 2014, 97, 4127–4140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miqueo, E.; Chiarle, A.; Giuliodori, M.J.; Relling, A.E. Association between prepartum metabolic status and resumption of postpartum ovulation in dairy cows. Domest. Anim. Endocrinol. 2019, 69, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Al-Katanani, Y.M.; Webb, D.W.; Hansen, P.J. Factors affecting seasonal variation in 90-day nonreturn rate to first service in lactating Holstein cows in a hot climate. J. Dairy Sci. 1999, 82, 2611–2616. [Google Scholar] [CrossRef]
- Siatka, K.; Sawa, A.; Piwczyński, D.; Bogucki, M.; Krężel-Czopek, S. Factors affecting first insemination success in Polish Holstein-Fresian cows. Anim. Sci. Pap. Rep. 2018, 36, 275–285. [Google Scholar]
- Abdelnour, S.A.; Swelum, A.A.; Abd El-Hack, M.E.; Khafaga, A.F.; Taha, A.E.; Abdo, M. Cellular and functional adaptation to thermal stress in ovarian granulosa cells in mammals. J. Therm. Biol. 2020, 92, 102688. [Google Scholar] [CrossRef]
- Gendelman, M.; Aroyo, A.; Yavin, S.; Roth, Z. Seasonal effects on gene expression, cleavage timing, and developmental competence of bovine preimplantation embryos. Reproduction 2010, 140, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Abbas, Z.; Sammad, A.; Hu, L.; Fang, H.; Xu, Q.; Wang, Y. Glucose metabolism and dynamics of facilitative glucose transporters (Gluts) under the influence of heat stress in dairy cattle. Metabolites 2020, 10, 312. [Google Scholar] [CrossRef]
- Butler, W.R. Energy balance relationships with follicular development ovulation and fertility in postpartum dairy cows. Livest. Prod. Sci. 2003, 83, 211–218. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, W.Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. 2015, 35, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Yu-Wei, D.; Li, Z.; Xiong, S.; Huang, G.; Luo, Y.; Huo, T.; Zhou, M.; Zheng, Y. Paclitaxel induces apoptosis through the TAK1–JNK activation pathway. FEBS Open Bio 2020, 10, 1655–1667. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Vermeulen, K.; Berneman, Z.N.; Van Bockstaele, D.R. Cell cycle and apoptosis. Cell Prolif. 2003, 36, 165–175. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, C.; Elsheikh, N.A.H.; Li, C.; Yang, F.; Wang, G.; Li, L. HO-1 reduces heat stress-induced apoptosis in bovine granulosa cells by suppressing oxidative stress. Aging 2019, 11, 5535–5547. [Google Scholar] [CrossRef]
- Soto, P.; Smith, L.C. BH4 peptide derived from Bcl-xL and Bax-inhibitor peptide suppresses apoptotic mitochondrial changes in heat stressed bovine oocytes. Mol. Reprod. Dev. 2009, 76, 637–646. [Google Scholar] [CrossRef]
- Abdelnour, S.A.; Abd El-Hack, M.E.; Khafaga, A.F.; Arif, M.; Taha, A.E.; Noreldin, A.E. Stress biomarkers and proteomics alteration to thermal stress in ruminants: A review. J. Therm. Biol. 2019, 79, 120–134. [Google Scholar] [CrossRef]
- Azad, M.A.K.; Kikusato, M.; Sudo, S.; Amo, T.; Toyomizu, M. Time course of ROS production in skeletal muscle mitochondria from chronic heat-exposed broiler chicken. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2010, 157, 266–271. [Google Scholar] [CrossRef]
- Gu, Z.T.; Li, L.; Wu, F.; Zhao, P.; Yang, H.; Liu, Y.S.; Geng, Y.; Zhao, M.; Su, L. Heat stress induced apoptosis is triggered by transcription-independent p53, Ca2+ dyshomeostasis and the subsequent Bax mitochondrial translocation. Sci. Rep. 2015, 5, 11497. [Google Scholar] [CrossRef] [Green Version]
- Hou, C.H.; Lin, F.L.; Hou, S.M.; Liu, J.F. Hyperthermia induces apoptosis through endoplasmic reticulum and reactive oxygen species in human Osteosarcoma cells. Int. J. Mol. Sci. 2014, 15, 17380–17395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guérin, P.; El Mouatassim, S.; Ménézo, Y. Oxidative stress and protection against reactive oxygen species in the pre-implantation embryo and its surroundings. Hum. Reprod. Update 2001, 7, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; Xiong, Y.-M.; Tan, Y.-J.; Wang, L.; Li, R.; Zhang, Y.; Liu, X.-M.; Lin, X.-H.; Jin, L.; Hu, Y.-T.; et al. Melatonin rescues impaired penetration ability of human spermatozoa induced by mitochondrial dysfunction. Reproduction 2019, 158, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Bu, D.P.; Wang, J.Q.; Huo, X.K.; Guo, T.J.; Wei, H.Y.; Zhou, L.Y.; Rastani, R.R.; Baumgard, L.H.; Li, F.D. Effect of saturated fatty acid supplementation on production and metabolism indices in heat-stressed mid-lactation dairy cows. J. Dairy Sci. 2010, 93, 4121–4127. [Google Scholar] [CrossRef] [Green Version]
- Pedruzzi, L.M.; Stockler-Pinto, M.B.; Leite, M.; Mafra, D. Nrf2–keap1 system versus NF-κB: The good and the evil in chronic kidney disease? Biochimie 2012, 94, 2461–2466. [Google Scholar] [CrossRef]
- Wardyn, J.D.; Ponsford, A.H.; Sanderson, C.M. Dissecting molecular cross-talk between Nrf2 and NF-κB response pathways. Biochem. Soc. Trans. 2015, 43, 621–626. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, S.; Summers, C.M.; Pearce, S.C.; Gabler, N.K.; Valentine, R.J.; Baumgard, L.H.; Rhoads, R.P.; Selsby, J.T. Short-term heat stress altered metabolism and insulin signaling in skeletal muscle. J. Anim. Sci. 2018, 96, 154–167. [Google Scholar] [CrossRef] [Green Version]
- Hambright, H.G.; Meng, P.; Kumar, A.P.; Ghosh, R. Inhibition of PI3K/AKT/mTOR axis disrupts oxidative stress-mediated survival of melanoma cells. Oncotarget 2015, 6, 1949–2553. [Google Scholar] [CrossRef] [Green Version]
- Reddy, N.M.; Potteti, H.R.; Vegiraju, S.; Chen, H.J.; Tamatam, C.M.; Reddy, S.P. PI3K-AKT Signaling via Nrf2 Protects against Hyperoxia-Induced Acute Lung Injury, but Promotes Inflammation Post-Injury Independent of Nrf2 in Mice. PLoS ONE 2015, 10, e0129676. [Google Scholar] [CrossRef] [Green Version]
- Cantó, C.; Auwerx, J. PGC-1α, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 2009, 20, 98–105. [Google Scholar] [CrossRef] [Green Version]
- Villeneuve, N.F.; Sun, Z.; Chen, W.; Zhang, D.D. Nrf2 and p21 regulate the fine balance between life and death by controlling ROS levels. Cell Cycle 2009, 8, 3255–3256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.Y.; Chen, Z.L.; Zhang, T.; Wei, C.; Shi, W.Y. TGF-β and NF-κB signaling pathway crosstalk potentiates corneal epithelial senescence through an RNA stress response. Aging 2016, 8, 2337–2354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tominaga, K.; Suzuki, H.I. TGF-β signaling in cellular senescence and aging-related pathology. Int. J. Mol. Sci. 2019, 20, 5002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.E. Non-Smad pathways in TGF-β signaling. Cell Res. 2009, 19, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Castellanos, J.; Wang, C.; Sinha-Hikim, I.; Lue, Y.; Swerdloff, R.S.; Sinha-Hikim, A.P. Mitogen-activated protein kinase signaling in male germ cell apoptosis in the rat. Biol. Reprod. 2009, 80, 771–780. [Google Scholar] [CrossRef] [Green Version]
- Mihaylova, M.M.; Shaw, R.J. The AMP-activated protein kinase (AMPK) signaling pathway coordinates cell growth, autophagy, & metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar]
- Ehlén, Å.; Nodin, B.; Rexhepaj, E.; Brändstedt, J.; Uhlén, M.; Alvarado-Kristensson, M.; Pontén, F.; Brennan, D.J.; Jirström, K. RBM3-regulated genes promote DNA integrity and affect clinical outcome in epithelial ovarian cancer. Transl. Oncol. 2011, 4, 212. [Google Scholar] [CrossRef] [Green Version]
- Lauridsen, C. From oxidative stress to inflammation: Redox balance and immune system. Poult. Sci. 2019, 98, 4240–4246. [Google Scholar] [CrossRef]
- Hansson, E.; Glaser, J.; Jakobsson, K.; Weiss, I.; Wesseling, C.; Lucas, R.A.I.; Wei, J.L.K.; Ekström, U.; Wijkström, J.; Bodin, T.; et al. Pathophysiological mechanisms by which heat stress potentially induces kidney inflammation and chronic kidney disease in sugarcane workers. Nutrients 2020, 12, 1639. [Google Scholar] [CrossRef]
- Könner, A.C.; Brüning, J.C. Toll-like receptors: Linking inflammation to metabolism. Trends Endocrinol. Metab. 2011, 22, 16–23. [Google Scholar] [CrossRef]
- Chimenti, M.S.; Triggianese, P.; Conigliaro, P.; Candi, E.; Melino, G.; Perricone, R. The interplay between inflammation and metabolism in rheumatoid arthritis. Cell Death Dis. 2015, 6, e1887. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Samples | Mean Reads | Ratios (%) for Mapping | ||||
|---|---|---|---|---|---|---|
| Raw Reads | Valid Reads | Q20 (%) | Uniquely Mapped | Mapped to Multiple Loci | Mapped Reads | |
| Control | 43,955,930 | 43,547,344 | 97.99% | 91.92% | 1.27% | 93.19% |
| Control | 44,851,996 | 44,429,040 | 98.04% | 87.19% | 1.28% | 88.47% |
| Control | 44,353,984 | 43,930,404 | 97.86% | 93.37% | 1.27% | 94.64% |
| Treated | 51,330,838 | 50,826,320 | 97.87% | 93.58% | 1.29% | 94.87% |
| Treated | 41,539,088 | 41,174,768 | 97.95% | 93.25% | 1.28% | 94.53% |
| Treated | 50,037,758 | 49,578,770 | 98.03% | 92.40% | 1.26% | 93.66% |
| DEGs | LogFC | LogCPM | FDR | Description |
|---|---|---|---|---|
| RBM3 | 1.66 | 8.16 | 2 × 10−256 | RNA binding motif protein 3 |
| GREM1 | 0.72 | 6.91 | 5 × 10−38 | “gremlin 1, DAN family BMP antagonist” |
| ACTA2 | 0.80 | 7.10 | 5 × 10−37 | “actin alpha 2, smooth muscle” |
| IFI27 | 0.74 | 5.16 | 6 × 10−18 | putative ISG12(a) protein |
| LCP1 | 0.60 | 5.67 | 4 × 10−15 | lymphocyte cytosolic protein 1 |
| ARHGDIB | 0.86 | 3.50 | 5 × 10−11 | Rho GDP dissociation inhibitor beta |
| IQCK | 0.98 | 2.54 | 4 × 10−8 | IQ motif containing K |
| PCSK1 | 0.62 | 3.98 | 2 × 10−7 | proprotein convertase subtilisin/kexin type 1 |
| DTL | 0.61 | 3.74 | 8 × 10−7 | denticleless E3 ubiquitin protein ligase homolog |
| ADH6 | 0.77 | 3.04 | 1 × 10−6 | alcohol dehydrogenase 6 (class V) |
| PYGM | 1.91 | 0.08 | 3 × 10−6 | “glycogen phosphorylase, muscle associated” |
| C3AR1 | 0.75 | 2.90 | 4 × 10−6 | complement C3a receptor 1 |
| PREX2 | 0.84 | 3.25 | 5 × 10−6 | phosphatidylinositol-3,4,5-trisphosphate dependent Rac-exchange factor 2 |
| HERC5 | 0.73 | 3.17 | 6 × 10−6 | HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| SPARCL1 | 0.66 | 3.38 | 7 × 10−6 | SPARC like 1 |
| AOX1 | 0.64 | 3.64 | 1 × 10−5 | aldehyde oxidase 1 |
| ADAMTS20 | 1.81 | 0.24 | 2 × 10−5 | ADAM metallopeptidase with thrombospondin type 1 motif 20 |
| KASH5 | 1.84 | 0.33 | 2 × 10−5 | KASH Domain Containing 5 |
| LOXL4 | 0.80 | 2.89 | 2 × 10−5 | lysyl oxidase like 4 |
| CDKN2A | 0.67 | 3.42 | 3 × 10−5 | cyclin dependent kinase inhibitor 2A |
| LENG8 | −0.91 | 7.45 | 6 × 10−41 | leukocyte receptor cluster member 8 |
| DCHS1 | −0.97 | 5.88 | 5 × 10−36 | dachsous cadherin-related 1 |
| CCDC136 | −1.21 | 4.19 | 6 × 10−32 | coiled-coil domain containing 136 |
| PLXNA2 | −0.63 | 6.69 | 2 × 10−31 | plexin A2 |
| MPPE1 | −0.95 | 5.00 | 1 × 10−30 | metallophosphoesterase 1 |
| TBX18 | −0.65 | 6.37 | 5 × 10−27 | T-box transcription factor 18 |
| SMAD6 | −0.78 | 5.61 | 1 × 10−26 | SMAD family member 6 |
| ABCC5 | −0.74 | 5.41 | 4 × 10−25 | ATP binding cassette subfamily C member 5 |
| SCN9A | −1.00 | 4.03 | 2 × 10−22 | sodium voltage-gated channel alpha subunit 9 |
| RSRP1 | −1.07 | 4.17 | 2 × 10−18 | arginine and serine rich protein 1 |
| CROCC | −0.70 | 5.30 | 3 × 10−18 | “ciliary rootlet coiled-coil, rootletin” |
| EFNB2 | −0.81 | 4.55 | 5 × 10−17 | ephrin B2 |
| VEGFA | −0.71 | 8.52 | 3 × 10−16 | vascular endothelial growth factor A |
| DTX4 | −1.68 | 1.96 | 3 × 10−15 | deltex E3 ubiquitin ligase 4 |
| KLF7 | −0.74 | 4.37 | 2 × 10−13 | Kruppel like factor 7 |
| SEL1L3 | −0.73 | 4.70 | 2 × 10−13 | SEL1L family member 3 |
| PPP1R12B | −0.84 | 3.81 | 7 × 10−13 | protein phosphatase 1 regulatory subunit 12B |
| RGS9 | −1.09 | 3.03 | 9 × 10−13 | regulator of G protein signaling 9 |
| KIFC2 | −0.69 | 4.63 | 3 × 10−12 | kinesin family member C2 |
| GABBR1 | −0.63 | 4.31 | 2 × 10−10 | gamma-aminobutyric acid type B receptor subunit 1 |
| Genes | E.R. | LogFC | LogCPM | p-Value | Description |
|---|---|---|---|---|---|
| ADCY2 | 7 | −0.73 | 2.83 | 2 × 10−5 | adenylate cyclase 2 |
| RYR2 | 7 | −0.88 | 1.33 | 3 × 10−5 | ryanodine receptor 2 |
| MAOB | 6 | −0.82 | 1.47 | 4 × 10−3 | monoamine oxidase B |
| ATP2A1 | 5 | −1.50 | 0.80 | 3 × 10−7 | ATPase sarcoplasmic Ca2+ transporting 1 |
| VEGFA | 4 | −0.71 | 8.52 | 3 × 10−16 | vascular endothelial growth factor A |
| CACNB1 | 4 | −0.81 | 3.07 | 8 × 10−7 | Ca2+ voltage-gated channel subunit beta 1 |
| GNB3 | 4 | −0.91 | 1.09 | 5 × 10−3 | G protein subunit beta 3 |
| NOS2 | 4 | 0.95 | 0.16 | 2 × 10−2 | nitric oxide synthase 2 |
| AOC2 | 3 | −0.91 | 1.68 | 2 × 10−4 | amine oxidase copper containing 2 |
| KDR | 3 | −1.05 | 0.02 | 1 × 10−2 | kinase insert domain receptor |
| Top 10 Genes in Biological Process (BP) of Gene Ontology Enrichment Terms | |||||
|---|---|---|---|---|---|
| Genes | TER | LogFC | LogCPM | FDR | Description |
| SFRP1 | 61 | −0.66 | 1.83 | 0.011 | secreted frizzled related protein 1 |
| TEK | 54 | −0.97 | 1.22 | 0.001 | TEK receptor tyrosine kinase |
| VEGFA | 49 | −0.71 | 8.52 | 3 × 10−16 | vascular endothelial growth factor A |
| DRC1 | 13 | 0.94 | 0.17 | 0.016 | dynein regulatory complex subunit 1 |
| LRRC6 | 15 | −0.70 | 1.39 | 0.016 | leucine rich repeat containing 6 |
| LFNG | 15 | −0.62 | 4.30 | 8 × 10−7 | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| INSL3 | 10 | −1.40 | 0.40 | 3 × 10−5 | Insulin Like 3 |
| CATSPERD | 10 | −0.83 | 3.17 | 7 × 10−9 | cation channel sperm associated auxiliary subunit delta |
| NFKBIZ | 10 | −0.75 | 4.09 | 1 × 10−9 | NFKB inhibitor zeta |
| ACTA2 | 9 | 0.80 | 7.10 | 5 × 10−37 | “actin alpha 2, smooth muscle” |
| Top 5 genes in cellular component (CC) of gene ontology enrichment terms | |||||
| TEK | 7 | −0.97 | 1.22 | 0.001 | TEK receptor tyrosine kinase |
| ACTA2 | 5 | 0.80 | 7.10 | 5 × 10−37 | “actin alpha 2, smooth muscle” |
| SLC2A4 | 3 | 0.66 | 2.31 | 0.004 | solute carrier family 2 member 4 |
| CAVIN4 | 3 | −0.67 | 1.21 | 0.023 | caveolae associated protein 4 |
| TLR2 | 2 | 0.70 | 2.06 | 0.005 | toll like receptor 2 |
| Top 5 genes in molecular function (MF) of gene ontology enrichment terms | |||||
| IGF2 | 7 | −0.98 | 0.20 | 0.032 | insulin like growth factor 2 |
| OXT | 6 | −1.51 | 0.13 | 0.0002 | oxytocin |
| APLN | 6 | −1.01 | 0.49 | 0.003 | apelin |
| INSL3 | 6 | −1.40 | 0.40 | 3 × 10−5 | Insulin Like 3 |
| VEGFA | 5 | −0.71 | 8.52 | 3 × 10−16 | vascular endothelial growth factor A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sammad, A.; Luo, H.; Hu, L.; Zhu, H.; Wang, Y. Transcriptome Reveals Granulosa Cells Coping through Redox, Inflammatory and Metabolic Mechanisms under Acute Heat Stress. Cells 2022, 11, 1443. https://doi.org/10.3390/cells11091443
Sammad A, Luo H, Hu L, Zhu H, Wang Y. Transcriptome Reveals Granulosa Cells Coping through Redox, Inflammatory and Metabolic Mechanisms under Acute Heat Stress. Cells. 2022; 11(9):1443. https://doi.org/10.3390/cells11091443
Chicago/Turabian StyleSammad, Abdul, Hanpeng Luo, Lirong Hu, Huabin Zhu, and Yachun Wang. 2022. "Transcriptome Reveals Granulosa Cells Coping through Redox, Inflammatory and Metabolic Mechanisms under Acute Heat Stress" Cells 11, no. 9: 1443. https://doi.org/10.3390/cells11091443
APA StyleSammad, A., Luo, H., Hu, L., Zhu, H., & Wang, Y. (2022). Transcriptome Reveals Granulosa Cells Coping through Redox, Inflammatory and Metabolic Mechanisms under Acute Heat Stress. Cells, 11(9), 1443. https://doi.org/10.3390/cells11091443








