Palmitic and Stearic Acids Inhibit Chaperone-Mediated Autophagy (CMA) in POMC-like Neurons In Vitro
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Culture
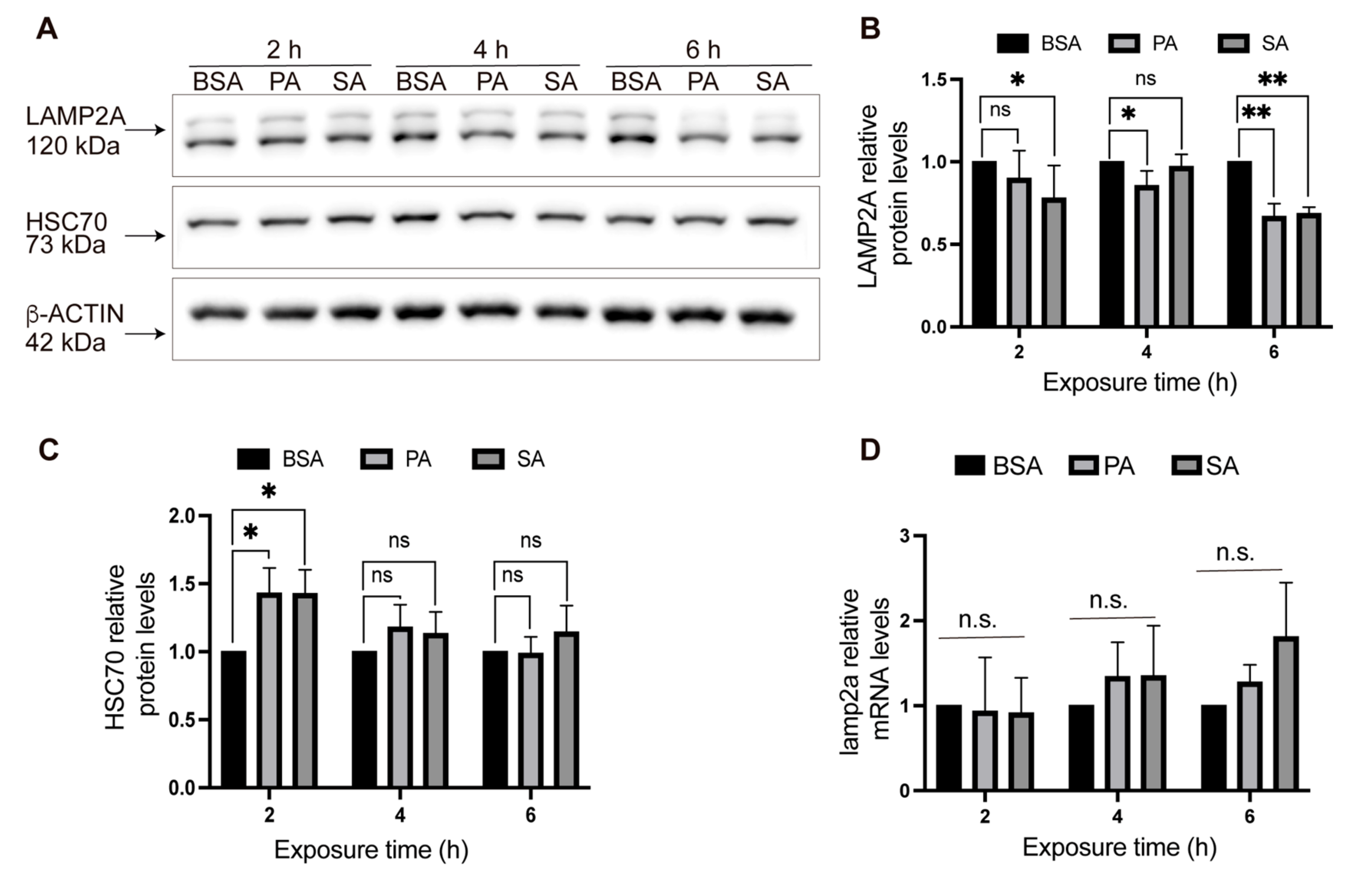
2.2. LAMP2A and HSC70 Protein Levels after PA and SA Treatment
2.3. RT-qPCR
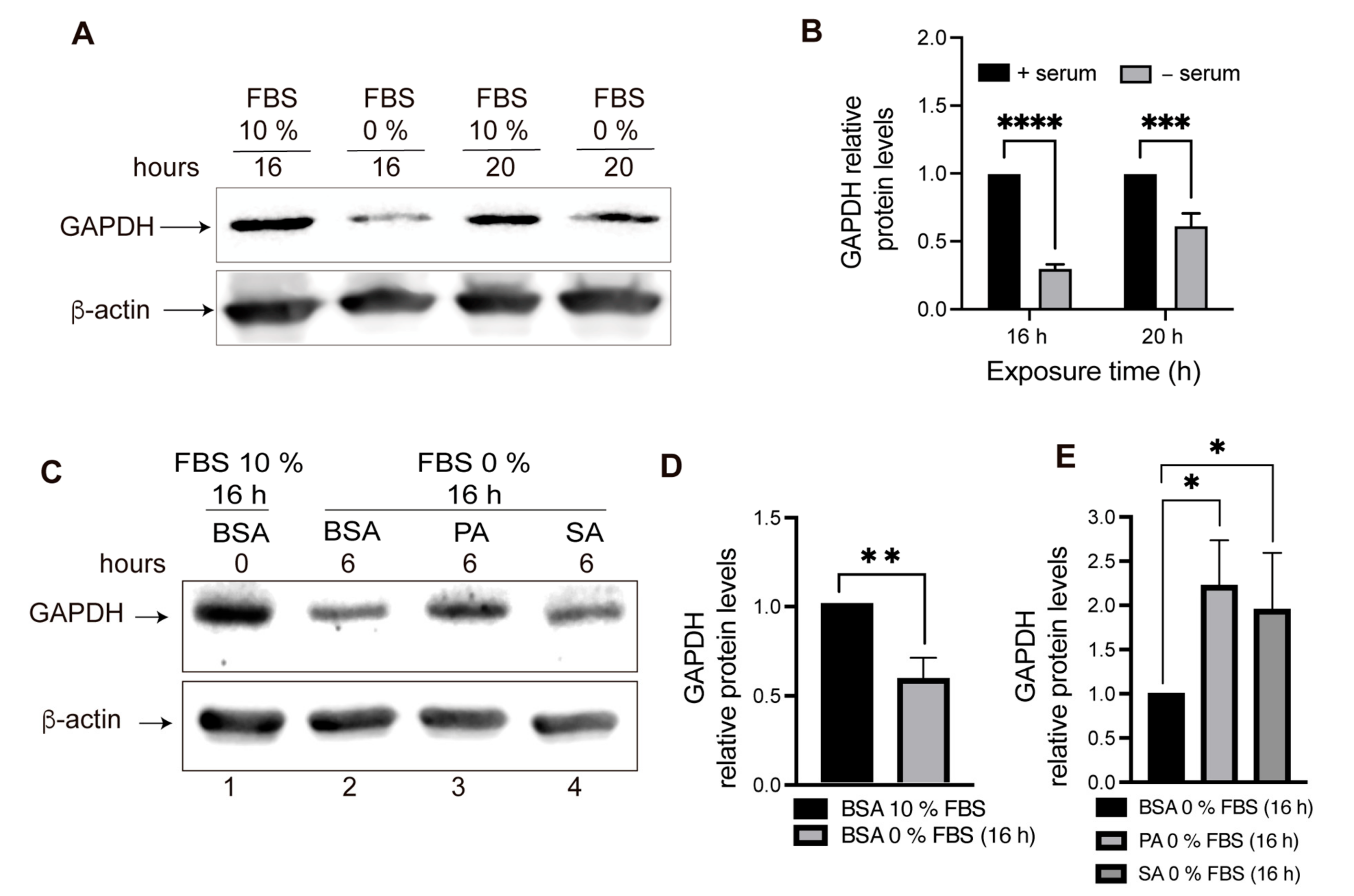
2.4. GAPDH Protein Level Evaluation after PA and SA Treatment
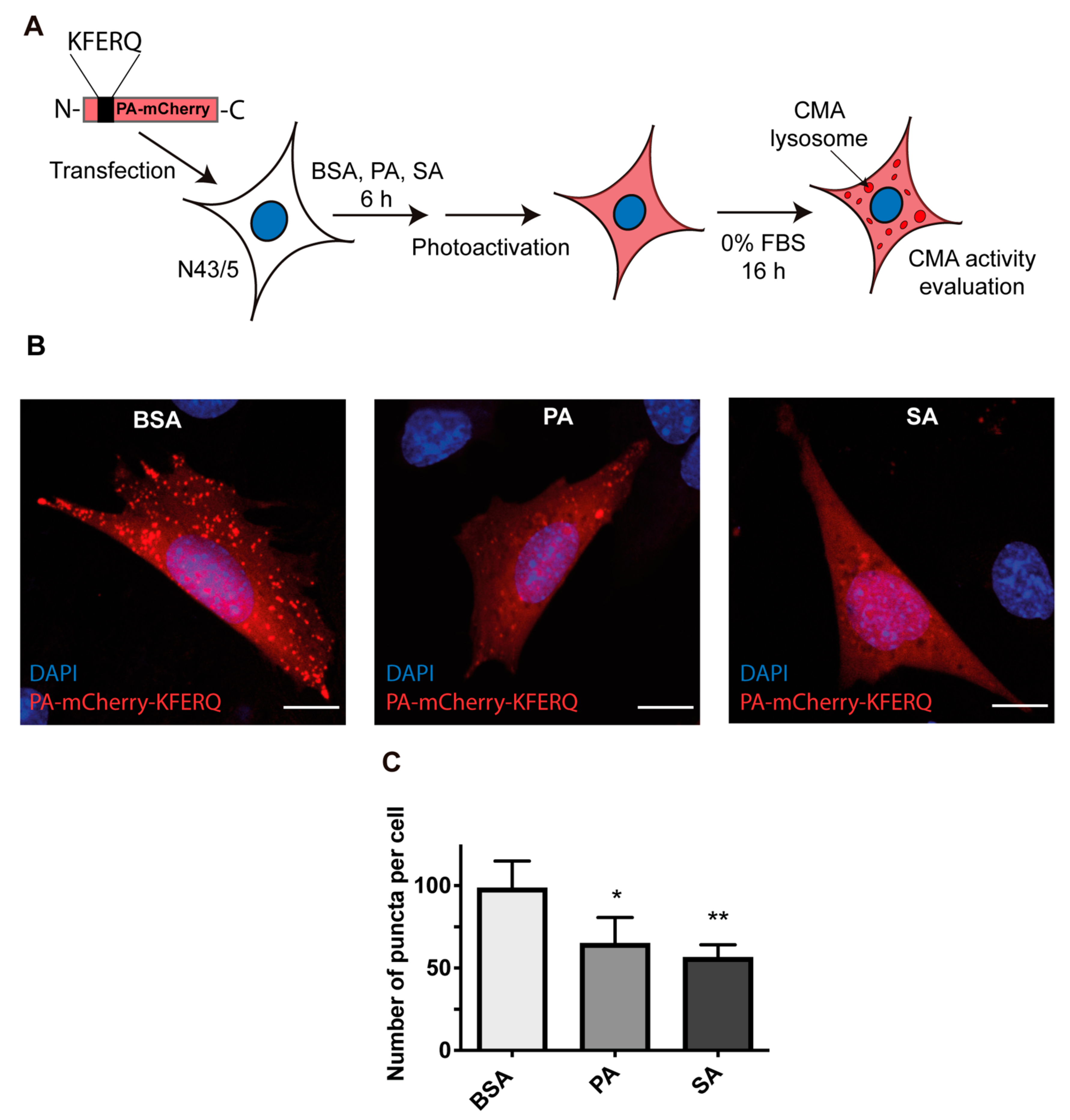
2.5. PA-mCherry-KFERQ Immunofluorescence Assay
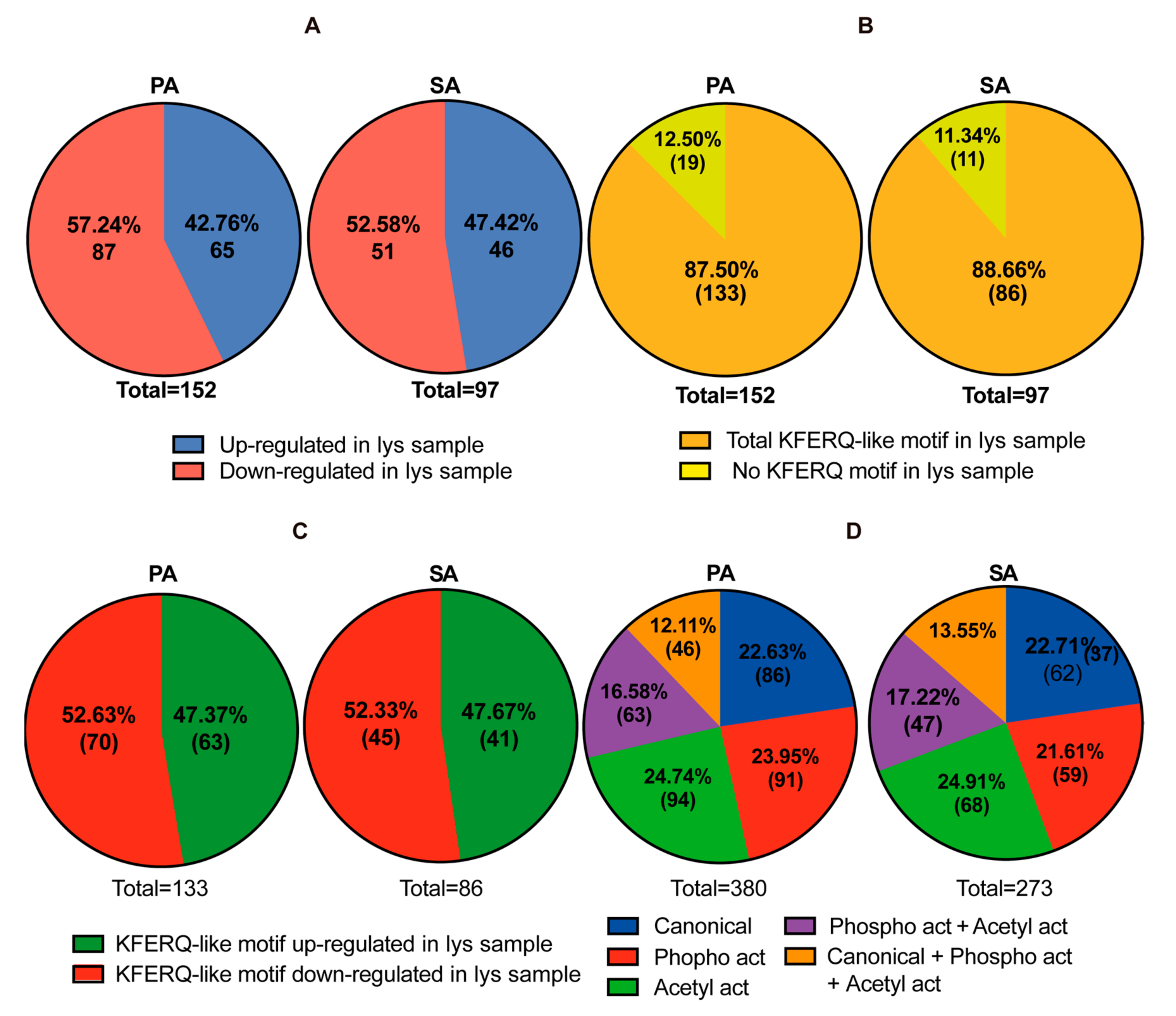
2.6. Lysosomal and Mitochondrial Fractionation and SILAC Analysis
2.7. Mass Spectrometry and Data Analysis
2.8. KFERQ-like Motifs Containing Proteins and STRING Analysis
2.9. Preparation of the N45/3 Cell Line with Reduced Levels of LAMP2A Protein
2.10. Insulin Treatment
2.11. Statistics
3. Results
3.1. Total and Lysosomal LAMP2A Protein Levels Decrease in POMC-like Neurons Treated with Palmitic and Stearic Acids
3.2. Chaperone-Mediated Autophagy (CMA) Activity Is Inhibited in POMC Neurons Exposed to Palmitic and Stearic Acids
3.3. Putative CMA Protein Substrates in Lysosomes Isolated from POMC-like Cells Treated with PA and SA
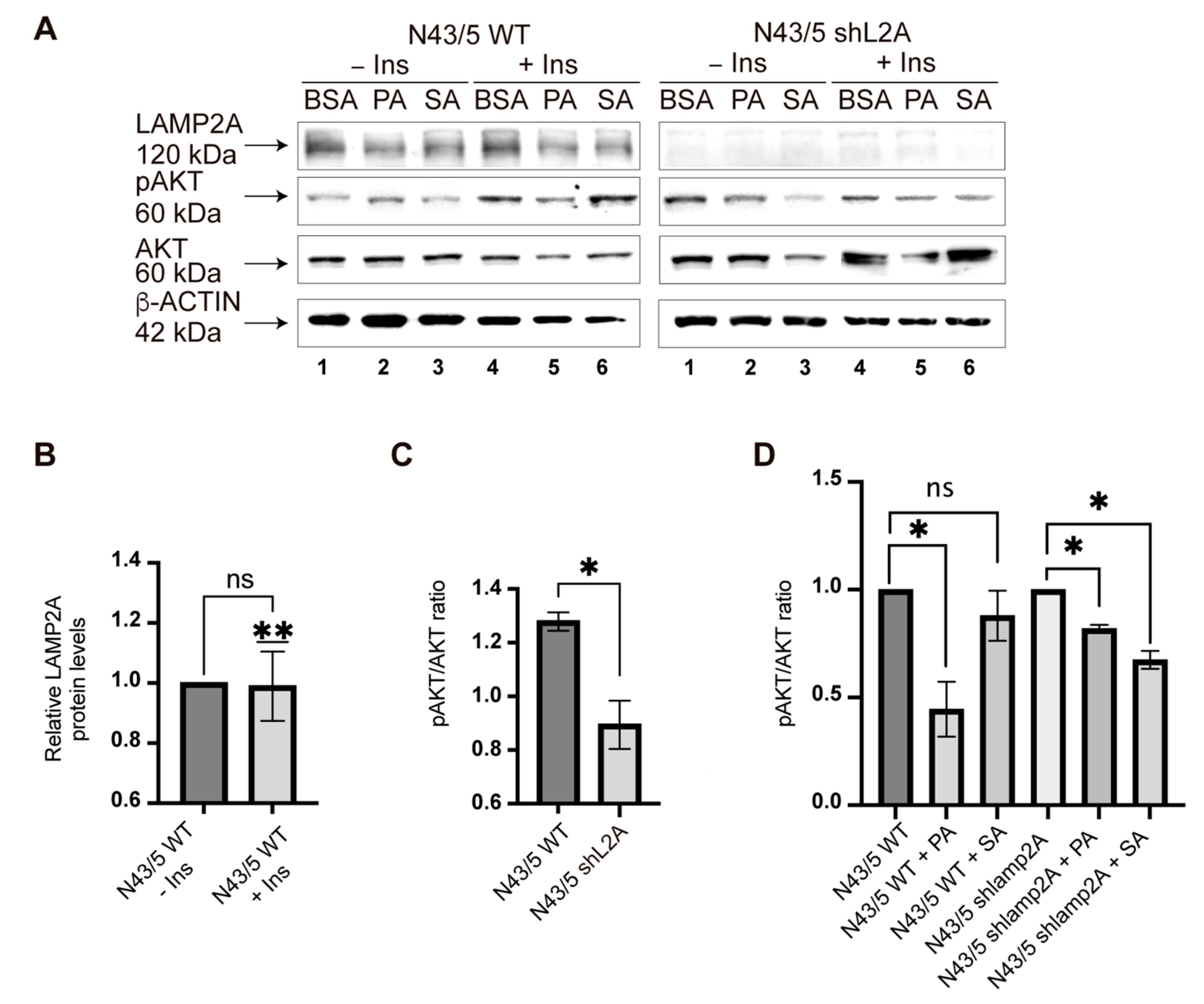
3.4. PA and SA Can Affect the Insulin Response in POMC-like Neurons by Inhibiting CMA Activity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Opie, L.H.; Walfish, P.G. Plasma Free Fatty Acid Concentrations in Obesity. N. Engl. J. Med. 1963, 268, 757–760. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, P.G. Obesity as a medical problem. Nature 2000, 404, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Sohn, J.-W. Network of hypothalamic neurons that control appetite. BMB Rep. 2015, 48, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Navas, C.; Morselli, E.; Clegg, D.J. Sexually dimorphic brain fatty acid composition in low and high fat diet-fed mice. Mol. Metab. 2016, 5, 680–689. [Google Scholar] [CrossRef]
- Vagena, E.; Ryu, J.K.; Baeza-Raja, B.; Walsh, N.M.; Syme, C.; Day, J.P.; Houslay, M.D.; Baillie, G.S. A high-fat diet promotes depression-like behavior in mice by suppressing hypothalamic PKA signaling. Transl. Psychiatry 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Valdearcos, M.; Robblee, M.M.; Benjamin, D.I.; Nomura, D.K.; Xu, A.W.; Koliwad, S.K. Microglia dictate the impact of saturated fat consumption on hypothalamic inflammation and neuronal function. Cell Rep. 2014, 9, 2124–2138. [Google Scholar] [CrossRef] [Green Version]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef]
- Onorati, A.V.; Dyczynski, M.; Ojha, R.; Amaravadi, R.K. Targeting autophagy in cancer. Cancer 2018, 124, 3307–3318. [Google Scholar] [CrossRef] [Green Version]
- Menzies, F.M.; Fleming, A.; Caricasole, A.; Bento, C.F.; Andrews, S.P.; Ashkenazi, A.; Füllgrabe, J.; Jackson, A.; Sanchez, M.J.; Karabiyik, C.; et al. Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 2017, 93, 1015–1034. [Google Scholar] [CrossRef] [Green Version]
- Bravo-San Pedro, J.M.; Kroemer, G.; Galluzzi, L. Autophagy and Mitophagy in Cardiovascular Disease. Circ. Res. 2017, 120, 1812–1824. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, M.-S. Autophagy—a key player in cellular and body metabolism. Nat. Rev. Endocrinol. 2014, 10, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Dice, J.F. Chaperone-mediated autophagy. Autophagy 2007, 3, 295–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandyopadhyay, U.; Cuervo, A.M. Entering the lysosome through a transient gate by chaperone-mediated autophagy. Autophagy 2008, 4, 1101–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarraberes, F.; Dice, J.F. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 2001, 114, 2491–2499. [Google Scholar] [CrossRef]
- Cuervo, A.M.; Knecht, E.; Terlecky, S.R.; Dice, J.F. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. Physiol. 1995, 269, C1200–C1208. [Google Scholar] [CrossRef]
- Kirchner, P.; Bourdenx, M.; Madrigal-Matute, J.; Tiano, S.; Diaz, A.; Bartholdy, B.A.; Will, B.; Cuervo, A.M. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 2019, 17, e3000301. [Google Scholar] [CrossRef]
- Arias, E.; Cuervo, A.M. Chaperone-mediated autophagy in protein quality control. Curr. Opin. Cell Biol. 2011, 23, 184–189. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Takabatake, Y.; Takahashi, A.; Kimura, T.; Namba, T.; Matsuda, J.; Minami, S.; Kaimori, J.-Y.; Matsui, I.; Matsusaka, T.; et al. High-Fat Diet–Induced Lysosomal Dysfunction and Impaired Autophagic Flux Contribute to Lipotoxicity in the Kidney. J. Am. Soc. Nephrol. 2016, 28, 1534–1551. [Google Scholar] [CrossRef]
- Che, Y.; Wang, Z.; Yuan, Y.; Zhang, N.; Jin, Y.; Wan, C.; Tang, Q. Role of autophagy in a model of obesity: A long-term high fat diet induces cardiac dysfunction. Mol. Med. Rep. 2018, 18, 3251–3261. [Google Scholar] [CrossRef]
- Portovedo, M.; Ignacio-Souza, L.M.; Bombassaro, B.; Coope, A.; Reginato, A.; Razolli, D.; Torsoni, M.; Torsoni, A.; Leal, R.; Velloso, L.A.; et al. Saturated Fatty Acids Modulate Autophagy’s Proteins in the Hypothalamus. PLoS ONE 2015, 10, e0119850. [Google Scholar] [CrossRef]
- Morselli, E.; Criollo, A.; Rodriguez-Navas, C.; Clegg, D.J. Chronic High Fat Diet Consumption Impairs Metabolic Health of Male Mice. Inflamm. Cell Signal. 2015, 1, e561. [Google Scholar] [CrossRef]
- Hernández-Cáceres, M.P.; Cereceda, K.; Hernández, S.; Li, Y.; Narro, C.; Rivera, P.; Silva, P.; Ávalos, Y.; Jara, C.; Burgos, P.; et al. Palmitic acid reduces the autophagic flux in hypothalamic neurons by impairing autophagosome-lysosome fusion and endolysosomal dynamics. Mol. Cell. Oncol. 2020, 7, 1789418. [Google Scholar] [CrossRef]
- Hernández-Cáceres, M.P.; Toledo-Valenzuela, L.; Díaz-Castro, F.; Ávalos, Y.; Burgos, P.; Narro, C.; Peña-Oyarzun, D.; Espinoza-Caicedo, J.; Cifuentes-Araneda, F.; Navarro-Aguad, F.; et al. Palmitic Acid Reduces the Autophagic Flux and Insulin Sensitivity Through the Activation of the Free Fatty Acid Receptor 1 (FFAR1) in the Hypothalamic Neuronal Cell Line N43/5. Front. Endocrinol. 2019, 10, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Li, P.; Fu, S.; Calay, E.; Hotamisligil, G.S. Defective Hepatic Autophagy in Obesity Promotes ER Stress and Causes Insulin Resistance. Cell Metab. 2010, 11, 467–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.S.; Chung, K.W.; Kim, J.W.; Kim, J.; Komatsu, M.; Tanaka, K.; Nguyen, Y.H.; Kang, T.M.; Yoon, K.H.; Kim, J.W.; et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008, 8, 318–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Navarro, J.A.; Kaushik, S.; Koga, H.; Dall’Armi, C.; Shui, G.; Wenk, M.R.; Di Paolo, G.; Cuervo, A.M. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA 2012, 109, E705–E714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portovedo, M.; Reginato, A.; Miyamoto, J.; Simino, L.; Hakim, M.; Campana, M.; Leal, R.; Ignácio-Souza, L.; Torsoni, M.; Magnan, C.; et al. Lipid excess affects chaperone-mediated autophagy in hypothalamus. Biochimie 2020, 176, 110–116. [Google Scholar] [CrossRef]
- Ormeño, F.; Hormazabal, J.; Moreno, J.; Riquelme, F.; Rios, J.; Criollo, A.; Albornoz, A.; Alfaro, I.E.; Budini, M. Chaperone Mediated Autophagy Degrades TDP-43 Protein and Is Affected by TDP-43 Aggregation. Front. Mol. Neurosci. 2020, 13, 19. [Google Scholar] [CrossRef] [Green Version]
- Massey, A.C.; Kaushik, S.; Sovak, G.; Kiffin, R.; Cuervo, A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA 2006, 103, 5805–5810. [Google Scholar] [CrossRef] [Green Version]
- Morselli, E.; Fuente-Martin, E.; Finan, B.; Kim, M.; Frank, A.; Garcia-Caceres, C.; Navas, C.R.; Gordillo, R.; Neinast, M.; Kalainayakan, S.P.; et al. Hypothalamic PGC-1alpha protects against high-fat diet exposure by regulating ERalpha. Cell Rep. 2014, 9, 633–645. [Google Scholar] [CrossRef] [Green Version]
- Arias, E. Methods to Study Chaperone-Mediated Autophagy. Methods Enzymol. 2017, 588, 283–305. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 2009, 452, 297–324. [Google Scholar]
- Koga, H.; Martinez-Vicente, M.; Macian, F.; Verkhusha, V.V.; Cuervo, A.M. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat. Commun. 2011, 2, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Ke, B.; Zhao, Z.; Ye, X.; Gao, Z.; Ye, J. Regulation of Insulin Degrading Enzyme Activity by Obesity-Associated Factors and Pioglitazone in Liver of Diet-Induced Obese Mice. PLoS ONE 2014, 9, e95399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sofer, Y.; Nash, Y.; Osher, E.; Fursht, O.; Goldsmith, G.; Nahary, L.; Shaklai, S.; Tordjman, K.M.; Serebro, M.; Touati, E.B.; et al. Insulin-degrading enzyme higher in subjects with metabolic syndrome. Endocrine 2021, 71, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Desbuquois, B.; Carré, N.; Burnol, A.F. Regulation of insulin and type 1 insulin-like growth factor signaling and action by the Grb10/14 and SH2B1/B2 adaptor proteins. FEBS J. 2013, 280, 794–816. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yoon, S.-O.; Poulogiannis, G.; Yang, Q.; Ma, X.M.; Villén, J.; Kubica, N.; Hoffman, G.R.; Cantley, L.C.; Gygi, S.P.; et al. Phosphoproteomic Analysis Identifies Grb10 as an mTORC1 Substrate That Negatively Regulates Insulin Signaling. Science 2011, 332, 1322–1326. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Bhattacharya, S.; Swamy, O.R.; Tandon, R.; Wang, Y.; Janda, R.; Riedel, H. Growth Factor Receptor-binding Protein 10 (Grb10) as a Partner of Phosphatidylinositol 3-Kinase in Metabolic Insulin Action. J. Biol. Chem. 2003, 278, 39311–39322. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.B.; Smith, M.S. Obesity Statistics. Prim. Care Clin. Off. Pract. 2016, 43, 121–135. [Google Scholar] [CrossRef]
- Silva Figueiredo, P.; Carla Inada, A.; Marcelino, G.; Maiara Lopes Cardozo, C.; de Cássia Freitas, K.; De Cássia Avellaneda Guimarães, R.; Pereira de Castro, A.; Aragão do Nascimento, V.; Aiko Hiane, P. Fatty Acids Consumption: The Role Metabolic Aspects Involved in Obesity and Its Associated Disorders. Nutrients 2017, 9, 1158. [Google Scholar] [CrossRef] [Green Version]
- Cuervo, A.M.; Mann, L.; Bonten, E.J.; D’Azzo, A.; Dice, J.F. Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 2003, 22, 47–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourdenx, M.; Martín-Segura, A.; Scrivo, A.; Rodriguez-Navarro, J.A.; Kaushik, S.; Tasset, I.; Diaz, A.; Storm, N.J.; Xin, Q.; Juste, Y.R.; et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell 2021, 184, 2696–2714.e25. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Kacal, M.; Ouchida, A.T.; Zhang, B.; Norberg, E.; Vakifahmetoglu-Norberg, H. Targetome analysis of chaperone-mediated autophagy in cancer cells. Autophagy 2019, 15, 1558–1571. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Deng, Y.; Yuan, F.; Xia, T.; Liu, H.; Li, Z.; Liu, Z.; Ying, H.; Liu, Y.; Zhai, Q.; et al. ATF4/ATG5 Signaling in Hypothalamic Proopiomelanocortin Neurons Regulates Fat Mass via Affecting Energy Expenditure. Diabetes 2017, 66, 1146–1158. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espinosa, R.; Gutiérrez, K.; Rios, J.; Ormeño, F.; Yantén, L.; Galaz-Davison, P.; Ramírez-Sarmiento, C.A.; Parra, V.; Albornoz, A.; Alfaro, I.E.; et al. Palmitic and Stearic Acids Inhibit Chaperone-Mediated Autophagy (CMA) in POMC-like Neurons In Vitro. Cells 2022, 11, 920. https://doi.org/10.3390/cells11060920
Espinosa R, Gutiérrez K, Rios J, Ormeño F, Yantén L, Galaz-Davison P, Ramírez-Sarmiento CA, Parra V, Albornoz A, Alfaro IE, et al. Palmitic and Stearic Acids Inhibit Chaperone-Mediated Autophagy (CMA) in POMC-like Neurons In Vitro. Cells. 2022; 11(6):920. https://doi.org/10.3390/cells11060920
Chicago/Turabian StyleEspinosa, Rodrigo, Karla Gutiérrez, Javiera Rios, Fernando Ormeño, Liliana Yantén, Pablo Galaz-Davison, César A. Ramírez-Sarmiento, Valentina Parra, Amelina Albornoz, Iván E. Alfaro, and et al. 2022. "Palmitic and Stearic Acids Inhibit Chaperone-Mediated Autophagy (CMA) in POMC-like Neurons In Vitro" Cells 11, no. 6: 920. https://doi.org/10.3390/cells11060920
APA StyleEspinosa, R., Gutiérrez, K., Rios, J., Ormeño, F., Yantén, L., Galaz-Davison, P., Ramírez-Sarmiento, C. A., Parra, V., Albornoz, A., Alfaro, I. E., Burgos, P. V., Morselli, E., Criollo, A., & Budini, M. (2022). Palmitic and Stearic Acids Inhibit Chaperone-Mediated Autophagy (CMA) in POMC-like Neurons In Vitro. Cells, 11(6), 920. https://doi.org/10.3390/cells11060920







