Legume Lectins with Different Specificities as Potential Glycan Probes for Pathogenic Enveloped Viruses
Abstract
:1. Introduction
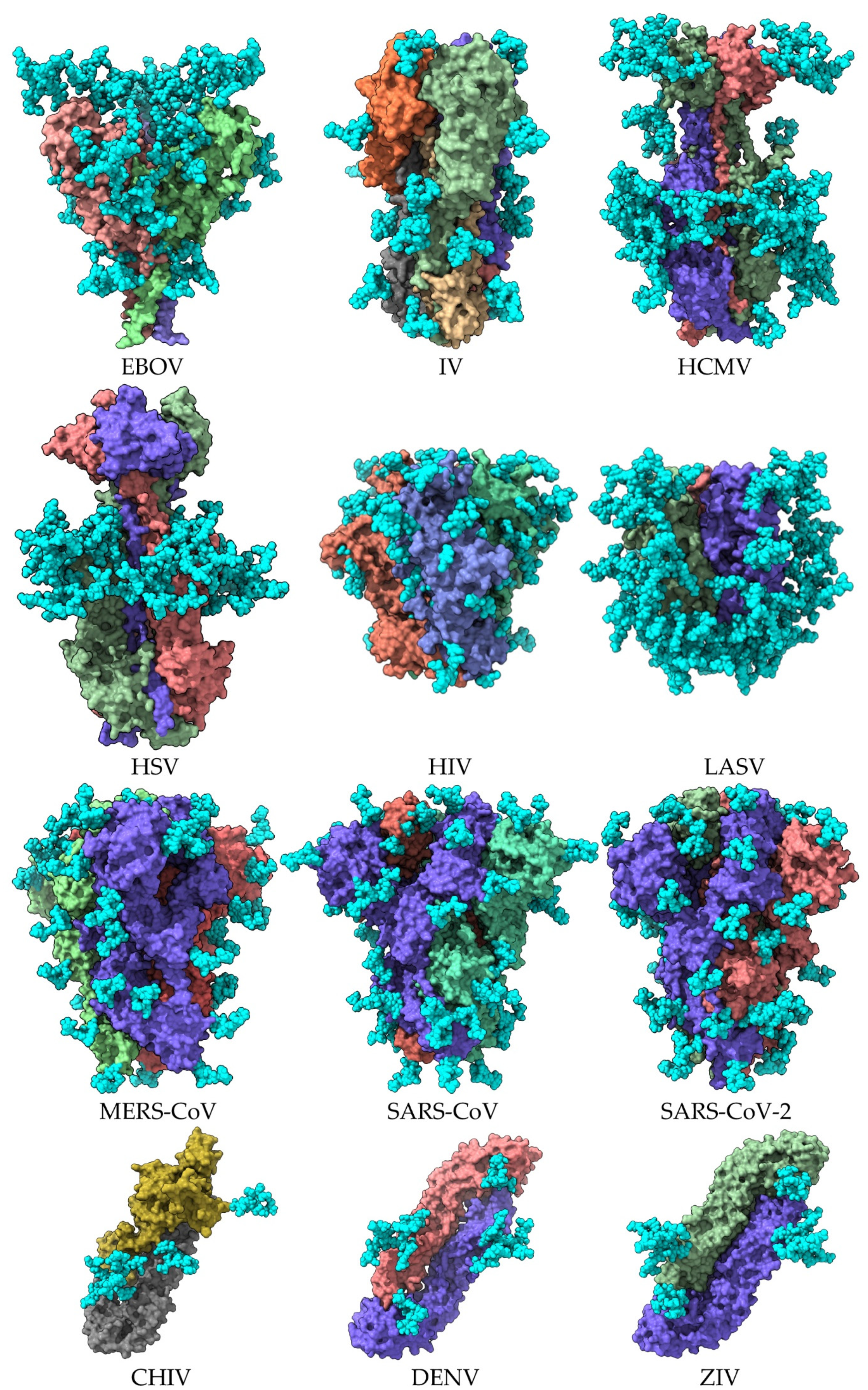
| Enveloped Virus | Homotrimer | Monomer | PDB Entry * | Reference |
|---|---|---|---|---|
| Ebola virus (EBOV) | Homotrimer | 7JPH | [7] | |
| Influenza virus (IV) | Hemagglutinin A Homotrimer | hemagglutinin A | 6Y5G | [8] |
| Human cytomegalovirus (HCMV) | Homotrimer | B glycoprotein | 5CXF | [9] |
| Herpes simplex virus (HSV) | Homotrimer | B glycoprotein | 2GUM | [3] |
| Human immunodeficiency virus (HIV) | Homotrimer | gp140 | 4TVP | [10] |
| Lassa virus (LASV) | Homotrimer | GPC glycoprotein | 5VK2 | [11] |
| Middle east respiratory syndrome coronavirus (MERS-CoV) | Spike | S protein | 5W9H | [12] |
| Severe acute respiratory syndrome coronavirus-1 (SAR-CoV) | Spike | S protein | 6ACD | [13] |
| Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) | Spike | S protein | 6VXX | [14] |
| Chikungunya virus (CHIV) | Homodimer | E protein | 3N40 | [4] |
| Dengue virus (DENV) | Homodimer | E protein | 1UZG | [5] |
| Zika virus (ZIV) | Homodimer | E protein | 57BUB | [6] |
2. The Glycan Shield of Pathogenic Enveloped Viruses
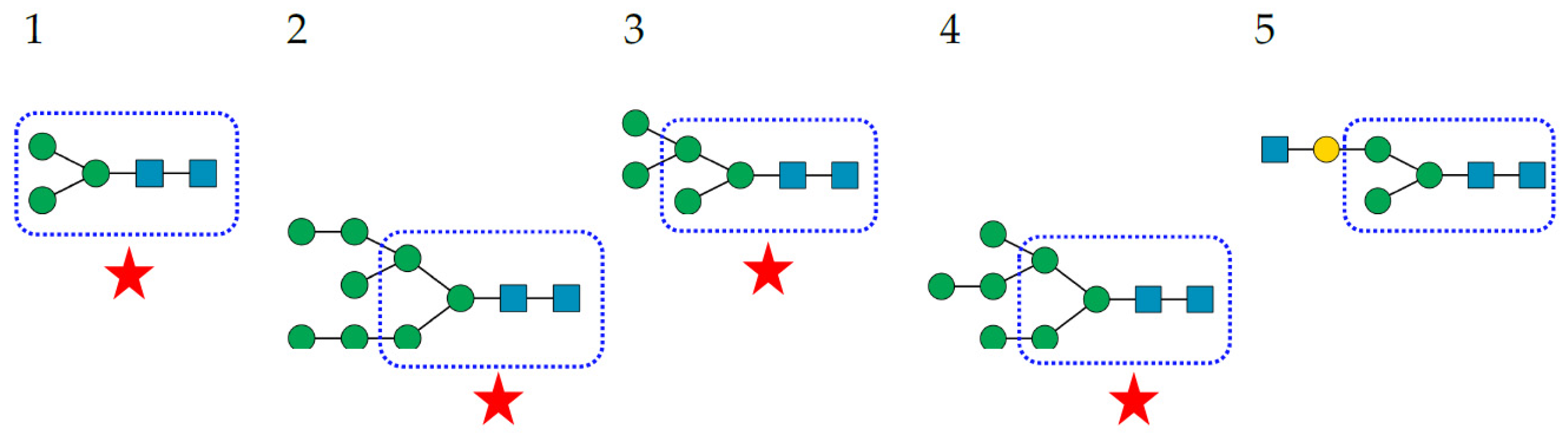
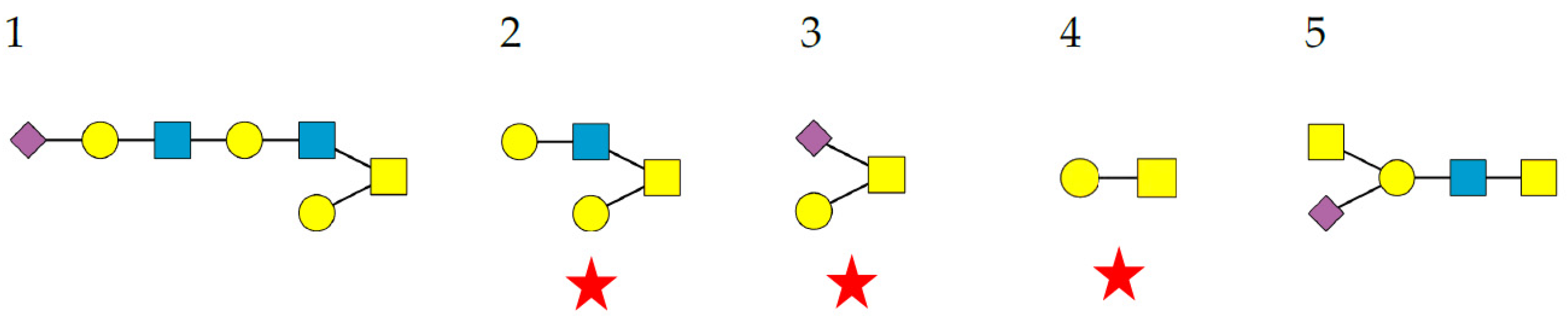
- Complex-type N-glycans are most abundant on all the envelope proteins except for the gp120 and hemagglutinin from HIV and IV, respectively, which predominantly contain high-mannose-type N-glycans. Complex glycans exhibit a high diversity in their glycan structure, including bi-, tri-, and tetra-antennary glycans which are often sialylated on their terminal Gal residues and fucosylated on sub-terminal GlcNAc residues. Most of these complex N-glycans possess an α1,6-fucosylated Man3GlcNAc2 core;
- High-mannose N-glycans are less abundant and offer less diversity than complex N-glycans because they consist exclusively of Man residues. High-mannose N-glycans from enveloped viruses include oligomannosides containing 4–9 (Man4–9) Man residues, and all of them possess a non-fucosylated Man3GlcNAc2 core;
- Hybrid N-glycans are least abundant on enveloped viruses.
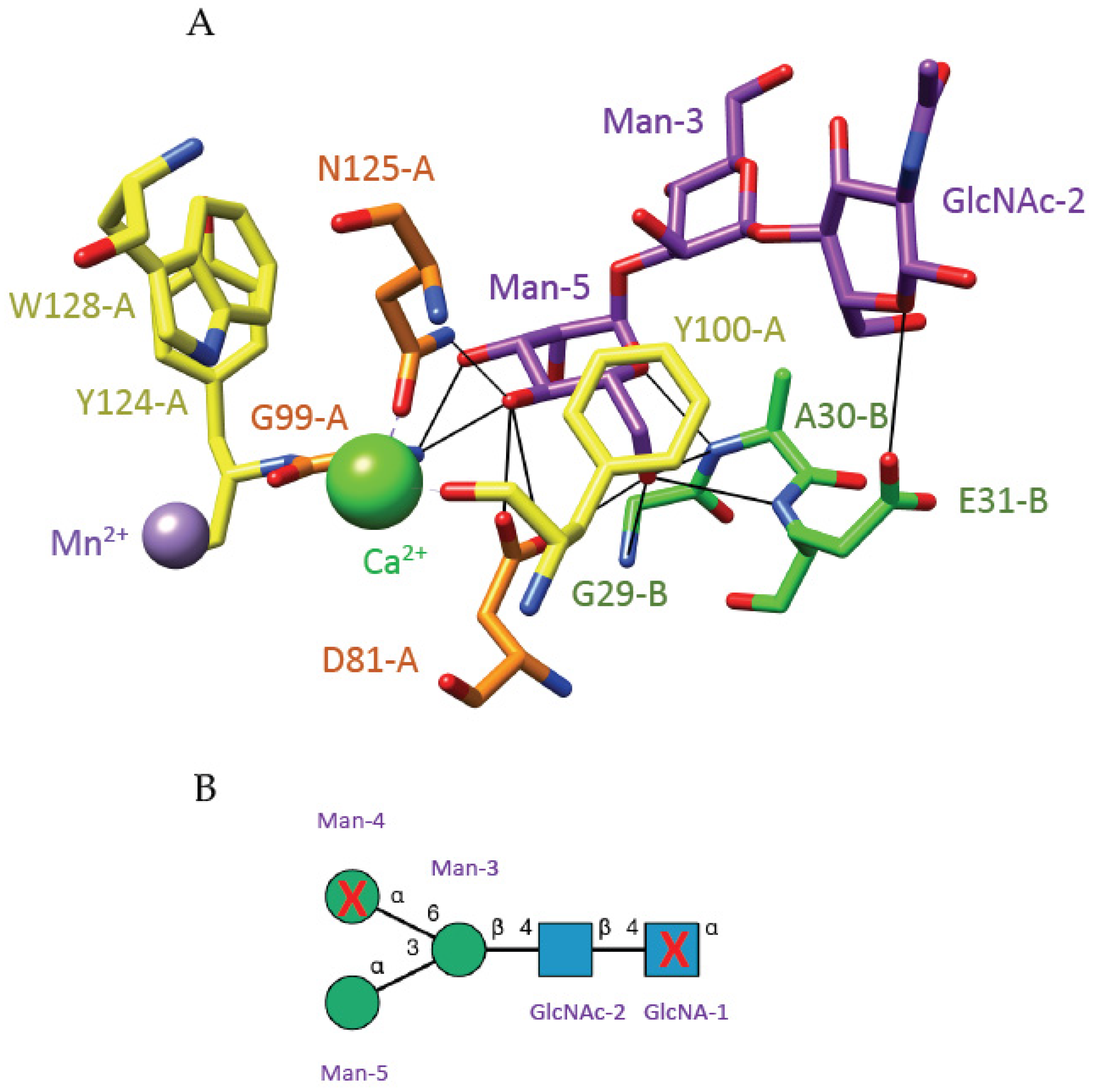
3. Plant Lectins with Different Specificities Are Potential CBAs for Pathogenic Enveloped Viruses
4. Man-Specific and Neu5Ac-Specific Lectins as Potential CBAs for Pathogenic Enveloped Viruses
| Lectin | Plant Species | Carbohydrate- Binding Specificity | Targeted Envelope Protein | Virus | Ref. |
|---|---|---|---|---|---|
| APA | Allium porum | Man | S-protein | SARS-CoV | [41] |
| AUA | Allium ursinum | Man | S-protein | SARS-CoV | [41] |
| BanLec | Musa acuminata | Man | gp120 | HIV | [42,43,44] |
| hemagglutinin | IV | [45] | |||
| E-glycoprotein | HCMV | [46] | |||
| E-glycoprotein | EBOV | [46,47] | |||
| E-glycoprotein | LASV | [46] | |||
| Con A | Canavalia ensiformis | Man | gp120 | HIV | [48] |
| S-protein | SARS-CoV-2 | [49] | |||
| Succinyl-Con A | Canavalia ensiformis | Man | S-protein | Mers-CoV | [50] |
| S-protein | SARS-CoV | [50] | |||
| S-protein | SARS-CoV-2 | [50] | |||
| ConBr | Canavalia brasiliensis | Man | S-protein | SARS-CoV-2 | [51] |
| ConM | Canavalia maritima | Man | S-protein | SARS-CoV-2 | [51] |
| CLA | Cladastris lutea | Man | S-protein | SARS-CoV | [41] |
| CHA | Cymbidium hybrid | Man | β-glycoprotein | HCMV | [52] |
| E-glycoprotein | HCV | [52] | |||
| gp120 | HIV | [52] | |||
| S-protein | SARS-CoV | [53] | |||
| DSL | Datura stramonium | Neu5Ac-Gal/GalNAc | S-protein | MERS-CoV | [50] |
| S-protein | SARS-CoV | [50] | |||
| S-protein | SARS-CoV-2 | [50] | |||
| DLasL | Dioclea lasiocarpa | Man | S-protein | SARS-CoV-2 | [51] |
| DSclerL | Dioclea sclerocarpa | Man | S-protein | SARS-CoV-2 | [51] |
| EHA | Epipactis helleborine | Man | β-glycoprotein | HCMV | [52] |
| gp120 | HIV | [52] | |||
| hemagglutinin | IV | [52] | |||
| GNA | Galanthus nivalis | Man | β-glycoprotein | HCMV | [53] |
| E-glycoprotein | HCV | [54,55] | |||
| gp120 | HIV | [53] | |||
| S-protein | SARS-CoV | [41] | |||
| S-protein | SARS-CoV-2 | [56] | |||
| hemagglutinin | IV | [57] | |||
| HHA | Hyppeastrum hybrid | Man | β-glycoprotein | HCMV | [53] |
| E-glycoprotein | HCV | [52] | |||
| gp120 | HIV | [52] | |||
| S-protein | SARS-CoV | [41] | |||
| hemagglutinin | IV | [57] | |||
| Horcolin | Hordeum vulgare | Man | gp120 | HIV | [58] |
| IRA | Iris hybrid | GalNAc/Gal | S-protein | SARS-CoV | [41] |
| FRIL | Lablab purpureus | Man | S-protein | SARS-CoV | [59] |
| hemagglutinin | IV | [59] | |||
| LcA | Lens culinaris | Man | S-protein | MERS-CoV | [50] |
| S-protein | SARS-CoV | [50] | |||
| S-protein | SARS-CoV-2 | [50] | |||
| LOA | Listera ovata | Man | β-glycoprotein | HCMV | [53] |
| gp120 | HIV | [53] | |||
| MAL | Maackia amurensis | Neu5Ac | S-protein | SARS-CoV-2 | [60] |
| Morniga-G | Morus nigra | Gal | S-protein | SARS-CoV | [41] |
| Morniga-M | Morus nigra | Man | S-protein | SARS-CoV | [41] |
| NPA | Narcissus pseudonarcissus | Man | β-glycoprotein | HCMV | [53] |
| gp120 | HIV | [53] | |||
| Nictaba | Nicotiana tabacum | (GlcNAc)n | S-protein | SARS-CoV | [41,61] |
| Orysata | Oryza sativa | Man | gp120 | HIV | [62] |
| S-protein | SARS-CoV | [62] | |||
| PHA | Phaseolus vulgaris | Complex glycans | S-protein | MERS-CoV | [50] |
| S-protein | SARS-CoV | [50] | |||
| S-protein | SARS-CoV-2 | [50] | |||
| PCA | Polygonatum cyrtonema | Man | gp120 | HIV | [63] |
| SSL | Sambucus sieboldiana | Neu5Ac-Gal/GalNAc | S-protein | SARS-CoV | [50] |
| S-protein | SARS-CoV-2 | [50] | |||
| TLC II | Tulipa hybrid | Man | S-protein | SARS-CoV | [41] |
| TDL | Typhonium divaricatum | Man | E-glycoprotein | HSV | [64] |
| UDA | Urtica dioica | (GlcNAc)n | β-glycoprotein | HCMV | [52] |
| E-glycoprotein | HCV | [51] | |||
| gp120 | HIV | [52] | |||
| S-protein | SARS-CoV | [41] | |||
| hemagglutinin | IV | [57] | |||
| ML II | Vicum album | Gal/GalNAc | S-protein | SARS-CoV | [41] |
| ML III | Viscum album | Gal/GalNAc | S-protein | SARS-CoV | [41] |
| WGA | Triticum aestivum | GlcNAc/Neu5Ac | S-protein | MERS-CoV | [50] |
| S-protein | SARS-CoV | [41,50] | |||
| S-protein | SARS-CoV-2 | [50,65] | |||
| GNAmaize | Zea mays | Man | S-protein | SARS-CoV | [66] |
| Lectin | Algal Species | Carbohydrate- Binding Specificity | Targeted Envelope Protein | Virus | Ref. |
| AML | Amantia multifida | Fetuin, mannan | hemagglutinin | IV | [52] |
| E-glycoprotein | HSV | [52] | |||
| gp120 | HIV | [52] | |||
| BSL | Bryothamnion seaforthii | Fetuin, mucin | gp120 | HIV | [52] |
| E-glycoprotein | HSV | [52] | |||
| hemagglutinin | IV | [52] | |||
| ESA-2 | Eucheuma serra | Man | hemagglutinin | IV | [70] |
| GCL | Grateloupia chiangii | Man | hemagglutinin | IV | [71] |
| E-glycoprotein | HSV | [71] | |||
| Griffithsin | Griffithsia sp. | Man | gp120 | HIV | [72] |
| E-glycoprotein | HCV | [73] | |||
| S-protein | MERS-CoV | [74] | |||
| S-protein | SARS-CoV | [68] | |||
| S-protein | SARS-CoV-2 | [75] | |||
| HML | Hypnea musciformis | Thyroglobulin, mucin | gp120 | HIV | [52] |
| hemagglutinin | IV | [52] | |||
| E-glycoprotein | HSV | [51] | |||
| HTL-40 | Halimeda renschii | Man | hemagglutinin | IV | [76] |
| KAA-2 | Kappaphycus alvarezii | Man | hemagglutinin | IV | [77] |
| gp120 | HIV | [78] | |||
| MEL | Meristiella echinocarpa | Mannan | hemagglutinin | IV | [52] |
| hemagglutinin | IV | [52] | |||
| SfL | Solieria filiformis | Mannan | E-glycoprotein | HSV | [52] |
| gp120 | HIV | [52] | |||
| hemagglutinin | IV | [52] | |||
| BCA | Boodlea coacta | Man | E-glycoprotein | HSV | [79] |
| hemagglutinin | IV | [79] | |||
| Lectin | Cyanobacterial Species | Carbohydrate- Binding Specificity | Targeted Envelope Protein | Virus | Ref. |
| MVN | Microcystis aeruginosa | Man | gp120 | HIV | [80,81] |
| E-glycoprotein | HSV | [81] | |||
| MVL | Microcystis viridis | Man | gp120 | HIV | [82] |
| E-glycoprotein | HCV | [83] | |||
| Cyanovirin-N | Nostoc ellipsosporum | Man | gp120 | HIV | [49,84] |
| (CV-N) | E-glycoprotein 1,2 | EBOV | [85,86] | ||
| hemagglutinin | IV | [86] | |||
| E-glycoprotein | HCV | [87] | |||
| E-glycoprotein | HSV | [88] | |||
| S-protein | SARS-CoV-2 | [89] | |||
| Oscillatoria agardhii | Man | gp120 | HIV | [90] | |
| OAA | Scytonema varium | Man | gp120 | HIV | [91] |
| SVN | Scytonema varium | E-glycoprotein | DENV | [92] | |
| E-glycoprotein | EBOV | [93] |
5. How Can the Infectivity of Pathogenic Enveloped Viruses Be Affected by Lectins?
6. Biomedical Perspectives for Antiviral Lectins
7. Bioinformatics
8. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rey, F.A.; Lok, S.-M. Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell 2018, 172, 1319–1334. [Google Scholar] [CrossRef] [PubMed]
- Thorley, J.A.; McKeating, J.A.; Rappoport, J.Z. Mechanisms of viral entry: Seaking in the front door. Protoplasma 2010, 244, 15–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heldwein, E.E.; Lou, H.; Bender, F.C.; Cohen, G.H.; Eisenberg, R.J.; Harrison, S.C. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 2006, 313, 217–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voss, J.E.; Vaney, M.-C.; Duquerroy, S.; Vonrhein, C.; Girard-Blanc, C.; Crublet, E.; Thompson, A.; Bricogne, G.; Rey, F.A. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 2010, 468, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 2005, 79, 1223–1231. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Loy, T.; Ng, T.S.; Lim, X.-N.; Chew, S.V.; Tan, T.Y.; Xu, M.; Kostyuchenko, V.A.; Tukijan, F.; Shi, J.; et al. A human antibody neutralizes different flaviviruses by using different mechanisms. Cell Rep. 2020, 31, 107584. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Chaudhary, A.; Lin, X.; Sou, C.; Alkutkar, T.; Kumar, S.; Ngo, T.; Kosviner, E.; Ozorowski, G.; Stanfield, R.L.; et al. Single-component multilayer self-assembling nanoparticles presenting rationally designed glycoprotein trimers as Ebola virus vaccines. Nat. Commun. 2021, 12, 2633. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.J.; Gamblin, S.; Rosenthal, P.B.; Skehel, J. Structural transitions in influenza haemagglutinin at membrane fusion pH. Nature 2020, 583, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Burke, H.G.; Heldwein, E.E. Crystal structure of the Human cytomegalovirus glycoprotein B. PLoS Pathog. 2015, 11, e1005227. [Google Scholar] [CrossRef] [Green Version]
- Pancera, M.; Zhou, T.; Druz, A.; Georgiev, I.S.; Soto, C.; Gorman, J.; Huang, J.; Acharya, P.; Chuang, G.-Y.; Ofek, G.; et al. Structure and immune recognition of trimeric prefusion HIV-1 Env. Nature 2014, 514, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Hastie, K.; Zandonatti, M.A.; Kleinfelter, L.M.; Heinrich, M.L.; Rowland, M.M.; Chandran, K.; Branco, L.M.; Robinson, J.E.; Garry, R.F.; Saphire, E.O. Structural basis for antibody-mediated neutralization of Lassa virus. Science 2017, 356, 923–928. [Google Scholar] [CrossRef] [Green Version]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Gui, M.; Wang, X.; Xiang, Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018, 14, e1007236. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Chon, B.G.; Gautam, S.; Peng, W.; Huang, Y.; Goli, M.; Mechref, Y. Direct comparison of N-glycans and their isomers derived from spike glycoprotein 1 of MERS-CoV, SARS-CoV, and SARS-CoV-2. J. Proteom. Res. 2021, 20, 4357–4365. [Google Scholar]
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Bagdonaite, I.; Thompson, A.J.; Wang, X.; Søgaard, M.; Fougeroux, C.; Frank, M.; Diedrich, J.K.; Yates III, J.R.; Salanti, A.; Vakhrushev, S.Y.; et al. Site-specific O-glycosylation analysis of SARS-CoV-2 spike protein produced in insect and Human cells. Viruses 2021, 13, 551. [Google Scholar] [CrossRef]
- Van Damme, E.J.M. 35 years in plant lectin research: A journey from basic science to applications in agriculture and medicine. Glycoconj. J. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Collar, A.L.; Clarke, E.C.; Anaya, E.; Merrill, D.; Yarborough, S.; Anthony, S.M.; Kuhn, J.H.; Merle, C.; Theisen, M.; Bradfute, S.B. Comparison of N- and O-linked glycosylation patterns of ebolavirus glycoproteins. Virology 2017, 502, 39–47. [Google Scholar] [CrossRef]
- Luo, S.; Hu, K.; He, S.; Wang, P.; Zhang, M.; Huang, X.; Du, T.; Zheng, C.; Liu, Y.; Hu, Q. Contribution of N-linked glycans on HSV-2 gG to cell-cell fusion and viral entry. Virology 2015, 483, 72–82. [Google Scholar] [CrossRef] [Green Version]
- Smargiasso, N.; Nader, J.; Rioux, S.; Mazzucchelli, G.; Boutry, M.; De Pauw, E.; Chaumont, F.; Navarre, C. Exploring the N-glycosylation profile of glycoprotein B from Human cytomegalovirus expressed in CHO and Nicotiana tabacum BY-2 cells. Int. J. Mol. Sci. 2019, 20, 3741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behrens, A.-J.; Vasiljevic, S.; Pritchard, L.K.; Harvey, D.J.; Andev, R.S.; Krumm, S.A.; Struwe, W.B.; Cupo, A.; Kumar, A.; Zitzmann, N.; et al. Composition and antigenic effects of individual glycan sites of a trimeric HIV-1 envelope glycoprotein. Cell Resp. 2016, 14, 2695–2706. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liu, S.; Gao, Y.; Tian, S.; Yang, Y.; Ma, N. Comparison of N-linked glycosylation on hemagglutinins derived from chicken embryos and MDCK cells: A case of the production of a trivalent seasonal influenza vaccine. Appl. Microbiol. Biotechnol. 2021, 105, 3559–3572. [Google Scholar] [CrossRef]
- Lancaster, C.; Pristatsky, P.; Van Hoang, M.; Casimiro, D.R.; Schwartz, R.M.; Rustandi, R.; Ha, S. Characterization of N-glycosylation profiles from mammalian and insect cell derived chikungunya VLP. J. Chromatogr. B Analyt. Technol. Life Sci. 2016, 1032, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Raghwani, J.; Allen, J.D.; Seabright, G.E.; Li, S.; Moser, F.; Huiskonen, J.T.; Strecker, T.; Bowden, T.A.; Crispin, M. Structure of the Lassa virus glycan shield provides a model for immunological resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 7320–7325. [Google Scholar] [CrossRef] [Green Version]
- Pralow, A.; Nikolay, A.; Leon, A.; Genzel, Y.; Rapp, E.; Reich, U. Site-specific N-glycosylation analysis of animal cell culture-derived Zika virus proteins. Sci. Rep. 2021, 11, 5147. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Hatmal, M.M.; Alshaer, W.; Al-Hatamleh, M.A.I.; Hatmal, M.; Smadi, O.; Taha, M.O.; Oweida, A.J.; Boer, J.C.; Mohamud, R.; Plebanski, M. Comprehensive structural and molecular comparison of spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and their interactions with ACE2. Cells 2020, 9, 2638. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Praissman, J.L.; Grant, O.C.; Xiao, T.; Rosenbalm, K.E.; Aoki, K.; Kellman, B.P.; Bridger, R.; Barouch, D.H.; Brindley, M.A.; et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and Human ACE2 receptor. Cell Host Microbe 2020, 28, 586–601. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, D.; Wang, Y.; Su, W.; Liu, G.; Dong, W. The importance of glycans of viral and host proteins in enveloped virus infection. Front. Immunol. 2021, 12, 638573. [Google Scholar] [CrossRef]
- Peumans, W.J.; Van Damme, E.J.M.; Barre, A.; Rougé, P. Classification of plant lectins in families of structurally and evolu- tionary related proteins. Adv. Exp. Med. Biol. 2001, 491, 27–54. [Google Scholar] [PubMed]
- Debray, H.; Decout, D.; Strecker, G.; Spik, G.; Montreuil, J. Specificity of twelve lectins towards oligosaccharides and glyco peptides related to N-glycosylproteins. Eur. J. Biochem. 1981, 117, 41–55. [Google Scholar] [CrossRef]
- Debray, H.; Rougé, P. The fine sugar specificity of the Lathyrus ochrus seed lectin and isolectins. FEBS Lett. 1984, 176, 120–124. [Google Scholar] [CrossRef] [Green Version]
- Einspahr, H.; Pareks, E.H.; Suguna, K.; Subramanian, E.; Suddath, F.L. The crystal structure of pea lectin at 3.0-Å resolution. J. Biol. Chem. 1986, 261, 16518–16527. [Google Scholar] [CrossRef]
- Foriers, A.; Van Driessche, E.; De Neve, R.; Kanarek, L.; Strosberg, A.D. The subunit structure and N-terminal sequences of theα- and β-subunits of the lentil lectin (Lens culinaris). FEBS Lett. 1977, 75, 237–240. [Google Scholar] [CrossRef] [Green Version]
- Bourne, Y.; Abergel, C.; Cambillau, C.; Frey, M.; Rougé, P.; Fontecilla-Camps, J.C. X-ray crystal structure determination and refinement at 1.9 Å resolution of isolectin I from the seeds of Lathyrus ochrus. J. Mol. Biol. 1990, 214, 571–584. [Google Scholar] [CrossRef]
- Reeke, G.N., Jr.; Becker, J.W. Three-dimensional structure of favin: Saccharide binding-cyclic permutation in leguminous lectins. Science 1986, 234, 1108–1111. [Google Scholar] [CrossRef] [PubMed]
- Hardman, K.D.; Ainsworth, C.F. Structure of concanavalin A at 2.4-Å resolution. Biochemistry 1972, 11, 4910–4919. [Google Scholar] [CrossRef]
- Bourne, Y.; Mazurier, J.; Legrand, D.; Rougé, P.; Montreuil, J.; Spik, G.; Cambillau, C. Structures of a legume lectin complexed with the human lactotransferrin N2 fragment, and with an isolated biantennary glycopeptide: Role of the fucose moiety. Structure 1994, 15, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Bourne, Y.; Rougé, P.; Cambillau, C. X-ray structure of (α-Man(1-3)β-Man(1-4)GlcNAc)-lectin complex at 2.1-Å resolution. The role of water in sugar-lectin interaction. J. Biol. Chem. 1990, 265, 18161–18165. [Google Scholar] [CrossRef]
- Rougé, P.; Peumans, W.J.; Van Damme, E.J.M.; Barre, A.; Singh, T.; Wu, J.H.; Wu, A.M. Glycotope structures and intramole-cular affinity factors of plant lectins for Tn/T antigens. Adv. Exp. Med. Biol. 2011, 705, 143–154. [Google Scholar] [PubMed] [Green Version]
- Keyaerts, E.; Vijgen, L.; Pannecouque, C.; Van Damme, E.; Peumans, W.; Egberink, H.; Balzarini, J.; Van Ranst, M. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antivir. Res. 2007, 75, 179–187. [Google Scholar] [CrossRef]
- Swanson, M.D.; Winter, H.C.; Goldstein, I.J.; Markovitz, D.M. A lectin isolated from banana is a potent inhibitor of HIV replication. J. Biol. Chem. 2010, 285, 8646–8655. [Google Scholar] [CrossRef] [Green Version]
- Swanson, M.D.; Boudreaux, D.M.; Salmon, L.; Clugh, J.; Winter, H.C.; Meagher, J.L.; André, S.; Murphy, P.V.; Oscarson, S.; Roy, R.; et al. Engineering a therapeutic lectin by uncoupling mitogenicity from antiviral activity. Cell 2015, 163, 746–758. [Google Scholar] [CrossRef] [Green Version]
- Mazalovska, M.; Kouokam, J.C. Lectins as promising therapeutics for the prevention and treatment of HIV and other potential coinfections. BioMed. Res. Int. 2018, 8, 3750646. [Google Scholar] [CrossRef]
- Covés-Datson, E.M.; King, S.R.; Legendre, M.; Gupta, A.; Chan, S.M.; Gitlin, E.; Kulkarni, V.V.; Pantaleón García, J.; Smee, D.F.; Lipka, E.; et al. A molecularly engineered antiviral banana lectin inhibits fusion and is efficacious against influenza virus infection in vivo. Proc. Natl. Acad. Sci. USA 2020, 117, 2122–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Covés-Datson, E.M.; King, S.R.; Legendre, M.; Swanson, M.D.; Gupta, A.; Claes, S.; Meagher, J.L.; Boonen, A.; Zhang, L.; Kalveram, B.; et al. Targeted disruption of pi-pi stacking in Malaysian banana lectin reduces mitogenicity while preserving antiviral activity. Sci. Rep. 2021, 11, 656. [Google Scholar] [CrossRef]
- Covés-Datson, E.M.; Dyall, J.; DeWald, L.E.; King, S.R.; Dube, D.; Legendre, M.; Nelson, E.; Drews, K.C.; Gross, R.; Gerhardt, D.M.; et al. Inhibition of Ebola virus by a molecularly engineered banana lectin. PLoS Negl. Trop. Dis. 2019, 13, e0007595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witvrouw, M.; Fikkert, V.; Hantson, A.; Pannecouque, C.; O’Keefe, B.R.; McMahon, J.; Stamatatos, L.; de Clercq, E.; Bolmstedt, A. Resistance to human immunodeficiency virus type 1 to the high-mannose binding agents cyanovirin N and concanavalin A. J. Virol. 2005, 79, 7777–7784. [Google Scholar] [CrossRef] [Green Version]
- Jang, H.; Lee, D.-H.; Kang, H.G.; Lee, S.J. Concanavalin A targeting N-linked glycans in spike proteins influence viral interactions. Dalton Trans. 2020, 49, 13538–13543. [Google Scholar] [CrossRef]
- Wang, W.; Li, Q.; Wu, J.; Hu, Y.; Wu, G.; Yu, C.; Xu, K.; Liu, X.; Wang, Q.; Huang, W.; et al. Lentil lectin from Lens culinaris exhibit broad antiviral activities against SARS-CoV-2 variants. Emerg. Microbes Infect. 2021, 10, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Gondim, A.C.S.; da Silva, S.R.; Mathys, L.; Noppen, S.; Liekens, S.; Sampaio, A.H.; Nagano, C.S.; Costa Rocha, C.R.; Nasci Mento, K.S.; Cavada, B.S.; et al. Potent antiviral activity of carbohydrate-specific algal and leguminous lectins from the Brazilian biodiversity. Med. Chem. Commun. 2019, 10, 390–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balzarini, J.; Neyts, J.; Schols, D.; Hosoya, M.; Van Damme, E.; Peumans, W.; De Clercq, E. The mannose-specific plant lectins from Cymbidium hybrid and Epipactis helleborine and the (N-acetylglucosamine)n-specific plant lectin from Urtica dioica are potent and selective inhibitors of human immunodeficiency virus and cytomegalovirus replication in vitro. Antivir. Res. 1992, 18, 191–207. [Google Scholar]
- Balzarini, J.; Schols, D.; Neyts, J.; Van Damme, E.; Peumans, W.; De Clercq, E. Alpha-(1-3)- and alpha-(1-6)-D-mannose-specific plant lectins are markedly inhibitory to human immunodeficiency virus and cytomegalovirus infections in vitro. Antimicrob. Agents Chemother. 1991, 35, 410–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertaux, C.; Daelemans, D.; Meertens, L.; Cormier, E.G.; Reinus, J.F.; Peumans, W.J.; Van Damme, E.J.M.; Igarashi, T.; Oki, T.; Schols, D.; et al. Entry of hepatitis C virus and human immunodeficiency virus is selectively inhibited by carbohydrate-binding agents but not by polyanions. Virology 2007, 366, 40–50. [Google Scholar] [CrossRef]
- Ashfaq, U.; Masoud, M.; Khaliq, S.; Nawaz, Z.; Riazuddin, S. Inhibition of hepatitis C virus 3a genotype entry through Galanthus nivalis agglutinin. Virol. J. 2011, 8, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Xu, X.; Liu, J.; Li, J.; Sun, Y.; Liu, Z.; Liu, J.; Van Damme, E.; Balzarini, J.; Bao, J. A novel mannose-binding tuber lectin from Typhonium divaricatum (L.) Decne (family Araceae) with antiviral activity against HSV-II and anti-proliferative effect on human cancer cell lines. J. Biochem. Mol. Biol. 2007, 40, 358–367. [Google Scholar] [CrossRef]
- Amundson, D.E.; Shah, U.; de Necochea-Campion, R.; Jacobs, M.; LaRosa, S.P.; Fisher, C.J., Jr. Removal of COVID-19 spike protein, whole virus, exosomes, and exosomal microRNAs by the Hemopurifier® lectin-affinity cartridge in critically III patients with COVID-19 infection. Front. Med. 2021, 8, 744141. [Google Scholar] [CrossRef] [PubMed]
- Jayaprakash, N.G.; Singh, A.; Vivek, R.; Yadav, S.; Pathak, S.; Trivedi, J.; Jayaraman, N.; Nandi, D.; Mitra, D.; Surolia, A. The barley lectin, horcolin, binds high-mannose glycans in a multivalent fashion, enabling high-affinity, specific inhibition of cellular HIV infection. J. Biol. Chem. 2020, 295, 12111–12129. [Google Scholar] [CrossRef]
- Liu, Y.-M.; Shahed-Al-Mahmud, M.D.; Chen, X.; Chen, T.-H.; Liao, K.-S.; Lo, J.M.; Wu, Y.-M.; Ho, M.-C.; Wu, C.-Y.; Wong, C.-H.; et al. A carbohydrate-binding protein from the edible lallab beans effectively blocks the infections of influenza viruses and SARS-CoV-2. Cell Rep. 2020, 32, 108016. [Google Scholar] [CrossRef]
- Sheehan, S.A.; Hamilton, K.L.; Retzbach, E.P.; Balachandran, P.; Krishnan, H.; Leone, P.; Lopez-Gonzalez, M.; Suryavanshi, S.; Kumar, P.; Russo, R.; et al. Evidence that Maackia amurensis seed lectin (MASL) exerts pleiotropic actions on oral squamous cells with potential to inhibit SARS-CoV-2 infection and COVID-19 disease progression. Exp. Cell Res. 2021, 403, 112594. [Google Scholar] [CrossRef] [PubMed]
- Gordts, S.C.; Renders, M.; Férir, G.; Huskens, D.; Van Damme, E.J.M.; Peumans, W.; Balzarini, J.; Schols, D. NICTABA and UDA, two GlcNAc-binding lectins with unique antiviral activity profiles. J. Antimicrob. Chemother. 2015, 70, 1674–1685. [Google Scholar] [CrossRef] [Green Version]
- Al Atalah, B.; Fouquaert, E.; Vanderschaeghe, D.; Proost, P.; Balzarini, J.; Smith, D.F.; Rougé, P.; Lasanajak, Y.; Callewaert, N.; Van Damme, E.J.M. Expression analysis of the nucleocytoplasmic lectin ‘Orysata’ from rice in Pichia pastoris. FEBS J. 2011, 278, 2064–2079. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Liu, J.-Z.; Wu, C.-F.; Li, J.; Dai, L.; Van Damme, E.; Balzarini, J.; De Clercq, E.; Chen, F.; Bao, J.-K. Anti-HIV I/II activity and molecular cloning of a novel mannose/sialic acid-binding lectin from rhizome of Polygonatum cyrtonema Hua. Acta Biochim. Biophys. Sin. 2006, 38, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Vanderlinden, E.; Van Winkel, N.; Naesens, L.; Van Damme, E.J.M.; Persoons, L.; Schols, D. In vitro characterization of the carbohydrate-binding agents HHA, GNA, and UDA as inhibitors of influenza A and B virus replication. Antimicrob. Agents Chemother. 2021, 65, e01732-20. [Google Scholar] [CrossRef] [PubMed]
- Auth, J.; Fröba, M.; Grobe, M.; Rauch, P.; Ruetalo, N.; Schindler, M.; Morokutti-Kurz, M.; Fraf, P.; Dolischka, A.; Prieschl- Grassauer, E.; et al. Lectin from Triticum vulgaris (WGA) inhibits infection with SARS-CoV-2 and its variants of concern alpha and beta. Int. J. Mol. Sci. 2021, 22, 10205. [Google Scholar] [CrossRef]
- Mori, T.; O’Keefe, B.R.; Sowder, R.C., II; Bringans, S.; Gardella, R.; Berg, S.; Cochran, P.; Turpin, J.A.; Buckheit, R.W., Jr.; McMahon, J.B.; et al. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 2005, 280, 9345–9353. [Google Scholar] [CrossRef] [Green Version]
- O’Keefe, B.R.; Giomarelli, B.; Barnard, D.L.; Shenoy, S.R.; Chan, P.K.S.; McMahon, J.B.; Palmer, K.E.; Barnett, B.W.; Mey Erholz, D.K.; Wohlford-Lenane, C.L.; et al. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J. Virol. 2010, 84, 2511–2521. [Google Scholar] [CrossRef] [Green Version]
- Moulaei, T.; Shenoy, S.R.; Giomarelli, B.; Thomas, C.; McMahon, J.B.; Dauter, Z.; O’Keefe, B.R.; Wlodawer, A. Monomerizationof viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity. Structure 2010, 18, 1104–1115. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Morimoto, K.; Kubo, T.; Sakaguchi, T.; Nishizono, A.; Hirayama, M.; Hori, K. Entry inhibition of influenza viruses with high mannose binding lectin ESA-2 from the red alga Eucheuma serra through the recognition of viral hemagglutinin. Mar. Drugs 2015, 13, 3454–3465. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.-J.; Han, J.-W.; Jeon, H.; Cho, K.; Kim, J.-H.; Lee, D.-S.; Han, J.W. Characterization of a novel mannose binding lectin with antiviral activities from red alga, Grateloupia chiangii. Biomolecules 2020, 10, 333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.; Hoorelbeke, B.; Kagiampakis, I.; Demeler, B.; Balzarini, J.; Liwang, P.J. The griffithsin dimer is required for high-potency inhibition of HIV-1: Evidence for manipulation of the structure of gp120 as part of the griffithsin dimer mechanism. Antimicrob. Agents Chemother. 2013, 57, 3976–3989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meuleman, P.; Albecka, A.; Belouzard, S.; Vercauteren, K.; Verhoye, L.; Wychowski, C.; Leroux-Roels, G.; Palmer, K.E.; Dubuisson, J. Griffithsin has antiviral activity against hepatitis C virus. Antimicrob. Agents Chemother. 2011, 55, 5159–5167. [Google Scholar] [CrossRef] [Green Version]
- Millet, J.K.; Séron, K.; Labitt, R.N.; Danneels, A.; Palmer, K.E.; Whittaker, G.R.; Dubuisson, J.; Belouzard, S. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antivir. Res. 2016, 133, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Alsaidi, S.; Cornejal, N.; Mahoney, O.; Melo, C.; Verma, N.; Bonnaire, T.; Chang, T.; O’Keefe, B.R.; Sailer, J.; Zydowsky, T.M.; et al. Griffithsin and carrageenan combination results in antiviral synergy against SARS- CoV-1 and 2 in a pseudoviral model. Mar. Drugs 2021, 19, 418. [Google Scholar] [CrossRef]
- Mu, J.; Hirayama, M.; Sato, Y.; Morimoto, K.; Hori, K. A novel high-mannose specific lectin from the green alga Halimeda renschii exhibits a potent anti-influenza virus activity through high-affinity binding to the viral hemagglutinin. Mar. Drugs 2017, 15, 255. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Morimoto, K.; Hirayama, M.; Hori, K. High-mannose-specific lectin (KAA-2) from the red alga Kappaphycus alvarezii potently inhibits influenza virus infection in a strain-independent manner. Biochem. Bipophys. Res. Commun. 2011, 405, 291–296. [Google Scholar] [CrossRef]
- Hirayama, M.; Shibata, H.; Imamura, K.; Sakaguchi, T.; Hori, K. High-mannose specific lectin and its recombinants from a Carrageenophyta Kappaphycus alvarezii represent a potent anti-HIV activity through high-affinity binding to the viral envelope glycoprotein gp120. Mar Biotechnol. 2016, 18, 144–160. [Google Scholar] [CrossRef]
- Sato, Y.; Hirayama, M.; Morimoto, K.; Yamamoto, N.; Okuyama, S.; Hori, K. High mannose-binding lectin with preference for the cluster of α1-2-mannose from the green alga Boodlea coacta is a potent entry inhibitor of HIV-1 and influenza viruses. J. Biol. Chem. 2011, 286, 19446–19458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huskens, D.; Férir, G.; Vermeire, K.; Kehr, J.-C.; Balzarini, J.; Dittmann, E.; Schols, D. Microvirin, a novel α(1,2)-mannose-specific lectin isolated from Microcystis aeruginosa, has anti-HIV-1 activity comparable with that of cyanovirin-N but a much higher safety profile. J. Biol. Chem. 2010, 285, 24845–24854. [Google Scholar] [CrossRef] [Green Version]
- Shahid, M.; Qadir, A.; Yang, J.; Ahmad, I.; Zahid, H.; Mirza, S.; Windisch, M.P.; Shahzad-Ul-Hussan, S. An engineered microvirin with identical structural domains potently inhibits human immunodeficiency virus and hepatitis C virus cellular entry. Viruses 2020, 12, 199. [Google Scholar] [CrossRef] [Green Version]
- Ziólkowska, N.E.; Wlodawer, A. Structural studies of algal lectins with anti-HIV activity. Acta Biochim. Pol. 2006, 53, 617–626. [Google Scholar] [CrossRef] [Green Version]
- Kachko, A.; Loesgen, S.; Shahzad-Ul-Hussan, S.; Tan, W.; Zubkova, I.; Takeda, K.; Wells, F.; Rubin, S.; Bewley, C.A.; Major, M.E. Inhibition of hepatitis C virus by the cyanobacterial protein Microcystis viridis lectin: Mechanistic differences between the high-mannose specific lectins MVL, CV-N, and GNA. Mol. Pharm. 2013, 10, 4590–4602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shenoy, S.R.; O’Keefe, B.R.; Bolmstedt, A.J.; Cartner, L.K.; Boyd, M.R. Selective interactions of the human immunodeficiency virus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J. Pharmacol. Exp. Ther. 2001, 297, 704–710. [Google Scholar] [PubMed]
- Barrientos, L.G.; O’Keefe, B.R.; Bray, M.; Sanchez, A.; Gronenborn, A.M.; Boyd, M.R. Cyanovirin-N binds to the viral surface glycoprotein, GP1,2 and inhibits infectivity of Ebola virus. Antivir. Res. 2003, 58, 47–56. [Google Scholar] [CrossRef]
- Maier, I.; Schiestl, R.H.; Kontaxis, G. Cyanovirin-N binds viral envelope proteins at the low-affinity carbohydrate binding site without direct virus neutralization ability. Molecules 2021, 26, 3621. [Google Scholar] [CrossRef]
- Helle, F.; Wychowski, C.; Vu-Dac, N.; Gustafson, K.R.; Voisset, C.; Dubuisson, J. Cyanovirin-N inhibits hepatitis C virus entry by binding to envelope protein glycans. J. Biol. Chem. 2006, 281, 15177–25183. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, V.; Shukla, S.; Shukla, D. A sugar binding protein cyanovirin-N blocks herpes simplex virus type-1 entry and cell fusion. Antivir. Res. 2009, 84, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Naidoo, D.; Kar, P.; Roy, A.; Mutanda, T.; Bwapwa, J.; Sen, A.; Anandraj, A. Structural insight into the binding of cyanovirin- N with the spike glycoprotein, Mpro and PLpro of SARS-CoV-2: Protein-protein interactions, dynamics simulations and free energy calculations. Molecules 2021, 26, 5114. [Google Scholar] [CrossRef]
- Sato, Y.; Okuyama, S.; Hori, K. Primary structure and carbohydrate binding specificity of a potent anti-HIV lectin isolated from the filamentous cyanobacterium Oscillatoria agardhii. J. Biol. Chem. 2007, 282, 11021–11029. [Google Scholar] [CrossRef] [Green Version]
- Bokesch, H.R.; O’Keefe, B.R.; McKee, T.C.; Pannell, L.K.; Patterson, G.M.L.; Gardella, R.S.; Sowder, R.C., II; Turpin, J.; Watson, K.; Buckheit, R.W., Jr.; et al. A potent novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Biochemistry 2003, 42, 2578–2584. [Google Scholar] [CrossRef]
- Siqueira, A.S.; Lima, A.R.J.; de Sousa, R.C.; Santos, A.S.; da Silva Gonçalves Vianez Júnior, J.L.; Costa Gonçalves, E. Anti-dengue virus activity of scytovirin and evaluation of point mutation effects by molecular dynamics and binding free energy calculations. Biochem. Biophys. Res. Commun. 2017, 490, 1033–1038. [Google Scholar] [CrossRef]
- Garrison, A.R.; Giomarelli, B.G.; Lear-Rooney, C.M.; Saucedo, C.J.; Yellayi, S.; Krumpe, L.R.H.; Rose, M.; Paragas, J.; Bray, M.; Olinger, G.G.; et al. The cyanobacterial lectin scytovirin displays potent in vitro and in vivo activity against Zaire Ebola virus. Antivir. Res. 2014, 112, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Koharudin, L.M.; Furey, W.; Gronenborn, A.M. Novel fold and carbohydrate specificity of the potent anti-HIV cyanobacterial lectin from Oscillatoria agardhii. J. Biol. Chem. 2011, 286, 1588–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazur-Marzec, H.; Ceglowska, M.; Konkel, R.; Pyrc, K. Antiviral cyanometabolites—A review. Biomolecules 2021, 11, 474. [Google Scholar] [CrossRef]
- François, K.O.; Balzarini, J. Potential of carbohydrate-binding agents as therapeutics against enveloped viruses. Med. Res. Rev. 2012, 32, 349–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, T.; Kobayashi, S.; Yoshida, O.; Ishii, S.; Abe, Y.; Yamamoto, N. Effects of succinylated concanavalin A on infectivity and syncytial formation of human immunodeficiency virus. Med. Microbiol. Immunol. 1990, 179, 225–235. [Google Scholar] [CrossRef]
- Banks, W.A.; Ibrahimi, F.; Farr, S.A.; Flood, S.A.; Morley, J.E. Effects of wheat germ agglutinin and aging on the regional brain uptake of HIV-1 gp120. Life Sci. 1999, 65, 81–89. [Google Scholar] [CrossRef]
- Balzarini, J.; Van Laethem, K.; Hatse, S.; Vermeire, K.; De Clercq, E.; Peumans, W.; Van Damme, E.; Vandamme, A.M.; Bolmstedt, A.; Schols, D. Profile of resistance of human immunodeficiency virus to mannose-specific plant lectins. J. Virol. 2004, 78, 10617–10627. [Google Scholar] [CrossRef] [Green Version]
- Balzarini, J. Targeting the glycans of gp120: A novel approach aimed at the Achilles heel of HIV. Lancet Infect. Dis. 2005, 5, 727–731. [Google Scholar] [CrossRef]
- Turville, S.G.; Vermeire, K.; Balzarini, J.; Schols, D. Sugar-binding proteins potently inhibit dendritic cell human immuno- deficiency virus type 1 (HIV-1) infection and dendritic-cell-directed HIV-1 transfer. J. Virol. 2005, 79, 13519–13527. [Google Scholar] [CrossRef] [Green Version]
- Balzarini, J.; Van Herrewege, Y.; Vermeire, K.; Vanham, G.; Schols, D. Carbohydrate-binding agents efficiently prevent dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN)-directed HIV-1 transmission to T lymphocytes. Mol. Pharmacol. 2007, 71, 3–11. [Google Scholar] [CrossRef]
- Auwerx, J.; François, K.O.; Vanstreels, E.; Van Laethem, K.; Daelemans, D.; Schols, D.; Balzarini, J. Capture and transmission of HIV-1 by the C-type lectin L-SIGN (DC-SIGNR) is inhibited by carbohydrate-binding agents and polyanions. Antivir. Res. 2009, 83, 61–70. [Google Scholar] [CrossRef]
- Hoorelbeke, B.; Van Damme, E.J.M.; Rougé, P.; Schols, D.; Van Laethem, K.; Fouquaert, E.; Balzarini, J. Differences in the mannose oligomer specificities of the closely related lectins from Galanthus nivalis and Zea mays strongly determine their eventual anti-HIV activity. Retrovirology 2011, 8, 10. [Google Scholar] [CrossRef] [Green Version]
- Kumaki, Y.; Wandersee, M.K.; Smith, A.J.; Zhou, Y.; Simmons, G.; Nelson, N.M.; Bailey, K.W.; Vest, Z.G.; Li, J.K.-K.; Chan, P.K.-S.; et al. Inhibition of severe acute respiratory syndrome coronavirus replication in a lethal SARS-CoV BALB/c mouse model by stinging nettle lectin, Urtica dioica agglutinin. Antivir. Res. 2011, 90, 22–32. [Google Scholar] [CrossRef]
- Barre, A.; Van Damme, E.J.M.; Simplicien, M.; Le Poder, S.; Konjklowski, B.; Benoist, H.; Peyrade, D.; Rougé, P. Man-specific lectins from plants, fungi, algae and cyanobacteria, as potential blockers for SARS-CoV, MERS-CoV and SARS-CoV-2 (COVID-19) coronaviruses: Biomedical perspectives. Cells 2021, 10, 1619. [Google Scholar] [CrossRef]
- Ahmed, N.; Jahan, R.; Nissapatorn, V.; Wilairatana, P.; Rahmatullah, M. Plant lectins as prospective antiviral biomolecules in the search for COVID-19 eradication strategies. Miomed. Pharmacother. 2022, 146, 112507. [Google Scholar] [CrossRef]
- Singh, R.S.; Walia, A.K.; Khattar, J.S.; Singh, D.P.; Kennedy, J.F. Cyanobacterial lectins characteristics and their role as antiviral agents. Int. J. Biol. Macromol. 2017, 102, 475–496. [Google Scholar] [CrossRef]
- Singh, R.S.; Walia, A.K. Lectins from red algae and their biomedical potential. J. Appl. Phycol. 2018, 30, 1833–1858. [Google Scholar] [CrossRef] [Green Version]
- Breitenbach Barroso Coelho, L.C.; Marcelino dos Santos Silva, P.; Felix de Oliveira, W.; De Moura, M.C.; Viana Pontual, E.; Soares Gomes, F.; Guedes Paiva, P.M.; Napoleão, T.H.; dos Santos Correia, M.T. Lectins as antimicrobial agents. J. Appl. Microbiol. 2018, 125, 1238–1252. [Google Scholar] [CrossRef] [Green Version]
- Carneiro, D.C.; Fernandez, L.G.; Monteiro-Cunha, J.P.; Benevides, R.G.; Cunha Lima, S.T. A patent review of the antimicrobial applications of lectins: Perspectives on therapy of infectious diseases. J. Appl. Microbiol. 2021; online ahead of print. [Google Scholar] [CrossRef]
- Martinez, D.; Amaral, D.; Markovitz, D.; Pinto, L. The use of lectins as tools to combat SARS-CoV-2. Curr. Pharm. Des. 2021, 27, 4212–4222. [Google Scholar] [CrossRef]
- Peumans, W.J.; Van Damme, E.J.M. Lectins as plant defense proteins. Plant Physiol. 1995, 109, 347–352. [Google Scholar] [CrossRef]
- Boisseau, C.; Moisand, A.; Père, D.; Rougé, P. Immunocytochemical localization of the Lathyrus ochrus (L.) DC. Seed lectin in seeds and seedlings. Plant Sci. 1985, 42, 25–34. [Google Scholar] [CrossRef]
- Rougé, P.; Sousa-Cavada, B. Isolation and partial characterization of two isolectins from Lathyrus ochrus (L.) DC. Seeds. Plant Sci. Lett. 1984, 37, 21–27. [Google Scholar]
- Fuqua, J.L.; Wanga, V.; Palmer, K.E. Improving the large-scale purification of the HIV microbicide, griffithsin. BMC Biotechnol. 2015, 15, 12. [Google Scholar] [CrossRef] [Green Version]
- Fuqua, J.L.; Hamorsky, K.; Khalsa, G.; Matoba, N.; Palmer, K.E. Bulk production of the antiviral lectin griffithsin. Plant Biotechnol. 2015, 13, 1160–1168. [Google Scholar] [CrossRef]
- Alam, A.; Jiang, L.; Kittleson, G.A.; Steadman, K.D.; Nandl, S.; Fuqua, J.L.; Palmer, K.E.; Tusé, D.; McDonald, K.A. Technoeconomic modeling of plant-based griffithsin manufacturing. Front. Bieng. Biotechnol. 2018, 6, 102. [Google Scholar] [CrossRef] [Green Version]
- Hoelscher, M.; Tiller, N.; The, A.Y.-H.; Wu, G.-Z.; Ma, J.K.-C.; Bock, R. High-level expression of the HIV entry inhibitor griffithsin from the plastid genome and retention of biological activity in dried tobacco leaves. Plant Mol. Biol. 2018, 97, 357–370. [Google Scholar] [CrossRef] [Green Version]
- Eapen, P.; Cates, J.; Mundell, R.; Palmer, K.E.; Fuqua, J.L. In preparation for outdoor pharming: Griffithsin can be expressed in Nicotiana excelsiana and retains activity after storage and silage. Front. Bioeng. Biotechnol. 2020, 8, 199. [Google Scholar] [CrossRef]
- Decker, J.S.; Menacho-Melgar, R.; Lynch, M.D. Low-cost, large-scale production of the anti-viral lectin griffithsin. Front. Bioeng. Biotechnol. 2020, 8, 1020. [Google Scholar] [CrossRef]
- Lueken, K.; Mazarguil, H.; Rougé, P. The identification of two peptide sequences of light subunits of the Lathyrus ochrus isolectins containing a sequential epitope. Immunol. Lett. 1988, 19, 309–312. [Google Scholar] [CrossRef]
- Lueken, K.; Liboz, T.; Mazarguil, H.; Rougé, P. Localization of amino acid sequence stretches containing a continuous epitope on the surface of the two Lathyrus ochrus isolectins. Immunol. Lett. 1989, 23, 223–226. [Google Scholar] [CrossRef]
- Kolberg, J.; Ayouba, A.; Rougé, P. Production and characterization of a mouse monoclonal antibody specific for lentil lectin. Biol. Chem. Hoppe-Seyler 1991, 372, 57–61. [Google Scholar]
- Lueken, K.; Kolberg, J.; Cambillau, C.; Bourne, Y.; Rougé, P. Monoclonal antibody 117, C-11 recognizes three exposed regions on the surface of the Lathyrus ochrus isolectin I. Immunol. Lett. 1991, 30, 47–52. [Google Scholar] [CrossRef]
- Koshte, V.L.; Aalbers, M.; Calkhoven, P.G.; Aalberse, R.C. The potent IgG4-inducing antigen in banana is a mannose-binding lectin, BanLec-I. Int. Arch. Allergy Immunol. 1992, 97, 17–24. [Google Scholar] [CrossRef]
- Tchernychev, B.; Rabinkov, A.; Mirelman, D.; Wilchek, M. Natural antibodies to dietary proteins—The existence of natural antibodies to alliinase (alliin lyase) and mannose-specific lectin from garlic (Allium sativum) in human serum. Immunol. Lett. 1995, 47, 53–57. [Google Scholar] [CrossRef]
- Bewley, C.A.; Gustafson, K.R.; Boyd, M.R.; Covell, D.G.; Bax, A.; Clore, G.M.; Gronenborn, A.M. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat. Struct. Biol. 1998, 5, 571–578. [Google Scholar] [CrossRef]
- Moulaei, T.; Botos, I.; Ziólkowska, N.E.; Bokesch, H.; Krumpe, L.R.; McKee, T.C.; O’Keefe, B.R.; Dauter, Z.; Wlodawer, A. Atomic-resolution crystal structure of the antiviral lectin scytovirin. Protein Sci. 2007, 16, 2756–2760. [Google Scholar] [CrossRef] [Green Version]
- Micevicz, E.D.; Cole, A.L.; Jung, C.L.; Phillips, M.L.; Pratikhya, P.; Sharma, S.; Waring, A.J.; Cole, A.M.; Ruchala, P. Grifonin-I: A small HIV-1 entry inhibitor derived from the algal lectin, griffithsin. PLoS ONE 2010, 5, e14360. [Google Scholar] [CrossRef] [Green Version]
- Cooper, H.L.; Rubin, A.D. Synthesis of nonribosomal RNA by lymphocytes: A response to phytohemagglutinin treatment. Science 1966, 152, 516–518. [Google Scholar] [CrossRef]
- Phillips, B.; Weisrose, E. The mitogenic response of human B lymphocytes to phytohaemagglutinin. Clin. Exp. Immunol. 1974, 16, 383–392. [Google Scholar]
- Lis, H.; Sharon, N. Biological properties of lectins. In The Lectins, Properties, Functions, and Applications in Biology and Medicine; Liener, I.E., Sharon, N., Goldstein, I.J., Eds.; Academic Press Inc.: Orlando, FL, USA; San Diego, CA, USA; New York, NY, USA; Austin TX, USA, 1986; pp. 265–291. [Google Scholar]
- Liu, T.; Wu, L.; Wang, H.; Chen, J.; Yang, C.; Bao, J.; Wu, C. Role of reactive oxygen species-mediated MAPK and NF-κB activation on Polygonatum cyrtonema lectin-induced apoptosis and autophagy in human lung adenocarcinoma A549 cells. J. Biochem. 2016, 160, 315–324. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, S.R.; Ray, U.; Chatterjee, B.; Roy, S.S. Targeted apoptosis in ovarian cancer cells through mitochondrial dysfunction in response to Sambucus nigra agglutinin. Cell Death Dis. 2017, 8, e2762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poiroux, G.; Barre, A.; Van Damme, E.J.M.; Benoist, H.; Rougé, P. Plant lectins targeting O-glycans at the cell surface as tools for cancer diagnosis, prognosis and therapy. Int. J. Mol. Sci. 2017, 18, 1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naik, S.; Rawat, R.S.; Khandai, S.; Kumar, M.; Jena, S.S.; Vijayalakshmi, M.A.; Kumar, S. Biochemical characterization of lectin from Indian hyacinth plant bulbs with potential inhibitory action against human cancer cells. Int. J. Biol. Macromol. 2017, 105, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Yayeh, T.; Leem, Y.-H.; Park, E.-M.; Ito, Y.; Oh, S. Concanavalin A induces cortical neuron apoptosis by causing ROS accumulation and tyrosine kinase activation. Neurochem. Res. 2017, 42, 3504–3514. [Google Scholar] [CrossRef]
- Islam, F.; Gopalan, V.; Lam, A.K.-Y.; Rashel Kabir, S. Pea lectin inhibits cell growth by inducing apoptosis in SW480 and SW48 cell lines. Int. J. Biol. Macromol. 2018, 117, 1050–1057. [Google Scholar] [CrossRef] [Green Version]
- Poiroux, G.; Barre, A.; Simplicien, M.; Pelofy, S.; Segui, B.; Van Damme, E.J.M.; Rougé, P.; Benost, H. Morniga-G, a T/Tn-specific lectin, induces leukemic cell death via caspase and DR5 receptor-dependent pathways. Int. J. Mol. Sci. 2019, 20, 230. [Google Scholar] [CrossRef] [Green Version]
- Bhutia, S.; Panda, P.K.; Sinha, N.; Praharaj, P.P.; Bhol, C.S.; Panigrahi, D.P.; Mahapatra, K.K.; Saha, S.; Patra, S.; Mishra, S.R.; et al. Plant lectins in cancer therapeutics: Targeting apoptosis and autophagy-dependent cell dath. Pharmacol. Res. 2019, 144, 8–18. [Google Scholar] [CrossRef]
- Lalli, R.C.; Kaur, K.; Chakraborti, A.; Srinivasan, R.; Ghosh, S. Maackia amurensis agglutinin induces apoptosis in cultured resistant human non-small lung cancer cells. Glycoconj. J. 2019, 36, 473–485. [Google Scholar] [CrossRef]
- Rashidbaghan, A.; Mostafaie, A.; Yazdani, Y.; Mansouri, K. Urtica dioica agglutinin (a plant lectin) has a caspase-dependent apoptosis induction effect on the acute lymphoblastic leukemia cell line. Cell Mol. Biol. 2020, 66, 121–126. [Google Scholar] [CrossRef]
- Balzarini, J.; Van Laethem, K.; Peumans, W.J.; Van Damme, E.J.; Bolmstedt, A.; Cago, F.; Schols, D. Mutational pathways, resistance profile, and side effects of cyanovirin relative to human immunodeficiency virus type 1 strains with N-glycan deletions in their gp120 envelopes. J. Virol. 2006, 80, 8411–8421. [Google Scholar] [CrossRef] [Green Version]
- Huskens, D.; Vermeire, K.; Vandermeulebroucke, E.; Balzarini, J. Safety concerns for the potential use of cyanovirin-N as a microbicidal anti-HIV agent. Int. J. Biochem. Cell Biol. 2008, 40, 2802–2814. [Google Scholar] [CrossRef]
- Balzarini, J.; François, K.O.; Van Laethem, K.; Hoorelbeke, B.; Renders, M.; Auwers, J.; Liekens, S.; Oki, T.; Igarashi, Y.; Schold, D. Pradimicin S, a highly-soluble non-peptidic small-size carbohydrate-binding antibiotic, is an anti-HIV drug lead for both microbicidal and systemic use. Antimicrob. Agents Chemother. 2010, 54, 1425–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Büttner, S.; Koch, B.; Dolnik, O.; Eickmann, M.; Freiwald, T.; Rudolf, S.; Engel, J.; Becker, S.; Ronco, C.; Geiger, H. Extracorporeal virus elimination for the treatment of severe Ebola virus disease—First experience with lectin affinity plasmapheresis. Blood Purif. 2014, 38, 286–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, B.; Schult-Dietrich, P.; Büttner, S.; Dilmaghani, B.; Lohmann, D.; Baer, P.C.; Dietrich, U.; Geiger, H. Lectin affinity plasmapheresis for middle east respiratory syndrome-coronavirus and Marburg virus glycoprotein elimination. Blood Purif. 2018, 46, 126–133. [Google Scholar] [CrossRef]
- Leblanc, J.-F.; Germain, M.; Delage, G.; O’Brien, S.; Drews, S.J.; Lewin, A. Risk of transmission of severe acute respiratory syndrome coronavirus 2 by transfusion: A literature review. Transfusion 2020, 60, 3046–3054. [Google Scholar] [CrossRef]
- Gaussen, A.; Hornby, L.; Rockl, G.; O’Brien, S.; Delage, G.; Sapir-Pichhadze, R.; Drews, S.J.; Weiss, M.J.; Lewin, A. Evidence of SARS-CoV-2 infection in cells, tissues and organs and the risk of transmission through transplantation. Transplantation 2021, 105, 1405–1422. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Böhm, M.; Bohne-Lang, A.; Frank, M.; Loss, A.; Rojas-Macias, M.A.; Lütteke, T. Glycosciences.DB: An annotated data collec- tion linking glycomics and proteomics data (2018 update). Nucleic Acids Res. 2019, 47, D1195–D1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, K.; Zhou, Y.; Neelamegham, S. DrawGlycan-SNFG: A robust tool to render glycans and glycopeptides with frag mentation information. Glycobiology 2017, 27, 200–205. [Google Scholar]
- Day, C.J.; Bailly, B.; Guillon, P.; Dirr, L.; Jen, F.E.-C.; Spillings, B.L.; Mak, J.; Itzstein, M.; Haselhorst, T.; Jennings, M.P. Mul- tidisciplinary approaches identify compounds that bind to human ACE2 or SARS-CoV-2 spike protein as candidates to block SARS-CoV-2-ACE2 receptor interactions. mBio 2021, 12, e03681-20. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, N.; Lin, C.-W.; Inoue, R.; Khoo, K.-H.; Kawasaki, N.; Ma, B.Y.; Oka, S.; Ishiguro, M.; Sawada, T.; Ishida, H.; et al. Highly fucosylated N-glycan ligand for mannan-binding protein expressed specifically on CD26(DPPIV) isolated from a human colorectal carcinoma cell line, SW1116. Glycobiology 2009, 19, 437–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, J.D.; Watanabe, Y.; Chawla, H.; Newby, M.L.; Crispin, M. Subtle influence of ACE2 glycan processing on SARS-CoV-2 recognition. J. Mol. Biol. 2021, 433, 166762. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barre, A.; Van Damme, E.J.M.; Klonjkowski, B.; Simplicien, M.; Sudor, J.; Benoist, H.; Rougé, P. Legume Lectins with Different Specificities as Potential Glycan Probes for Pathogenic Enveloped Viruses. Cells 2022, 11, 339. https://doi.org/10.3390/cells11030339
Barre A, Van Damme EJM, Klonjkowski B, Simplicien M, Sudor J, Benoist H, Rougé P. Legume Lectins with Different Specificities as Potential Glycan Probes for Pathogenic Enveloped Viruses. Cells. 2022; 11(3):339. https://doi.org/10.3390/cells11030339
Chicago/Turabian StyleBarre, Annick, Els J. M. Van Damme, Bernard Klonjkowski, Mathias Simplicien, Jan Sudor, Hervé Benoist, and Pierre Rougé. 2022. "Legume Lectins with Different Specificities as Potential Glycan Probes for Pathogenic Enveloped Viruses" Cells 11, no. 3: 339. https://doi.org/10.3390/cells11030339
APA StyleBarre, A., Van Damme, E. J. M., Klonjkowski, B., Simplicien, M., Sudor, J., Benoist, H., & Rougé, P. (2022). Legume Lectins with Different Specificities as Potential Glycan Probes for Pathogenic Enveloped Viruses. Cells, 11(3), 339. https://doi.org/10.3390/cells11030339









