Mechanisms of RNA and Protein Quality Control and Their Roles in Cellular Senescence and Age-Related Diseases
Abstract
1. Cellular Senescence: Types and Characteristics
2. QC of RNA
2.1. QC of RNA in the Nucleus
2.2. QC of RNA in the Cytoplasm
3. QC of Proteins: Proteostasis
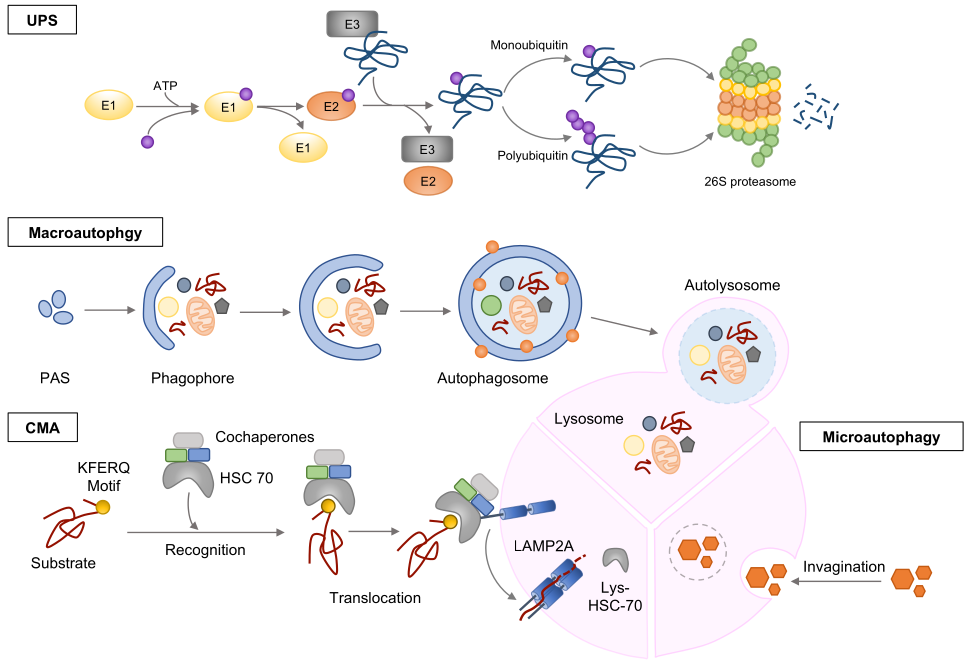
3.1. Ubiquitin-Proteasome System (UPS)
3.2. Autophagy
3.3. QC of RNAs and Proteins in Cellular Senescence and Aging
4. QC of RNAs and Proteins in Age-Related Diseases
4.1. Cancer
4.2. Neurodegenerative Diseases
4.2.1. Alzheimer’s Disease (AD)
4.2.2. Parkinson’s Disease (PD)
4.2.3. Huntington’s Disease (HD)
4.3. Muscle Diseases
4.4. Cardiovascular Diseases
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, W.; Hickson, L.J.; Eirin, A.; Kirkland, J.L.; Lerman, L.O. Cellular senescence: The good, the bad and the unknown. Nat. Rev. Nephrol. 2022, 18, 611–627. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef] [PubMed]
- Hemann, M.T.; Strong, M.A.; Hao, L.Y.; Greider, C.W. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 2001, 107, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- Di Micco, R.; Fumagalli, M.; Cicalese, A.; Piccinin, S.; Gasparini, P.; Luise, C.; Schurra, C.; Garre, M.; Nuciforo, P.G.; Bensimon, A.; et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444, 638–642. [Google Scholar] [CrossRef]
- Campaner, S.; Doni, M.; Hydbring, P.; Verrecchia, A.; Bianchi, L.; Sardella, D.; Schleker, T.; Perna, D.; Tronnersjö, S.; Murga, M.; et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat. Cell Biol. 2010, 12, 54–59. [Google Scholar] [CrossRef]
- Campisi, J. Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell 2005, 120, 513–522. [Google Scholar] [CrossRef]
- Camell, C.D.; Yousefzadeh, M.J.; Zhu, Y.; Prata, L.G.P.L.; Huggins, M.A.; Pierson, M.; Zhang, L.; O’Kelly, R.D.; Pirtskhalava, T.; Xun, P.; et al. Senolytics reduce coronavirus-related mortality in old mice. Science 2021, 373, eabe4832. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Schmitt, C.A.; Wang, B.; Demaria, M. Senescence and cancer-role and therapeutic opportunities. Nat. Rev. Clin. Oncol. 2022, 19, 619–636. [Google Scholar] [CrossRef]
- Coppé, J.P.; Desprez, P.Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lankhorst, L.; Bernards, R. Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer 2022, 22, 340–355. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Meljem, J.M.; Apps, J.R.; Fraser, H.C.; Martinez-Barbera, J.P. Paracrine roles of cellular senescence in promoting tumourigenesis. Br. J. Cancer 2018, 118, 1283–1288. [Google Scholar] [CrossRef] [PubMed]
- Lecot, P.; Alimirah, F.; Desprez, P.Y.; Campisi, J.; Wiley, C. Context-dependent effects of cellular senescence in cancer development. Br. J. Cancer 2016, 114, 1180–1184. [Google Scholar] [CrossRef]
- Doma, M.K.; Parker, R. RNA quality control in eukaryotes. Cell 2007, 131, 660–668. [Google Scholar] [CrossRef]
- Licatalosi, D.D.; Darnell, R.B. RNA processing and its regulation: Global insights into biological networks. Nat. Rev. Genet. 2010, 11, 75–87. [Google Scholar] [CrossRef]
- Wegener, M.; Müller-McNicoll, M. Nuclear retention of mRNAs-quality control, gene regulation and human disease. Semin. Cell Dev. Biol. 2018, 79, 131–142. [Google Scholar] [CrossRef]
- Boutz, P.L.; Bhutkar, A.; Sharp, P.A. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015, 29, 63–80. [Google Scholar] [CrossRef]
- Tutucci, E.; Stutz, F. Keeping mRNPs in check during assembly and nuclear export. Nat. Rev. Mol. Cell Biol. 2011, 12, 377–384. [Google Scholar] [CrossRef]
- Chlebowski, A.; Lubas, M.; Jensen, T.H.; Dziembowski, A. RNA decay machines: The exosome. Biochim. Biophys. Acta 2013, 1829, 552–560. [Google Scholar] [CrossRef]
- Kilchert, C.; Wittmann, S.; Vasiljeva, L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016, 17, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Zinder, J.C.; Lima, C.D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017, 31, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Kervestin, S.; Jacobson, A. NMD: A multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012, 13, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019, 20, 406–420. [Google Scholar] [CrossRef]
- Wang, Z.; Ballut, L.; Barbosa, I.; Le Hir, H. Exon Junction Complexes can have distinct functional flavours to regulate specific splicing events. Sci. Rep. 2018, 8, 9509. [Google Scholar] [CrossRef]
- Tan, K.; Stupack, D.G.; Wilkinson, M.F. Nonsense-mediated RNA decay: An emerging modulator of malignancy. Nat. Rev. Cancer 2022, 22, 437–451. [Google Scholar] [CrossRef]
- Van Hoof, A.; Frischmeyer, P.A.; Dietz, H.C.; Parker, R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 2002, 295, 2262–2264. [Google Scholar] [CrossRef]
- Frischmeyer, P.A.; van Hoof, A.; O’Donnell, K.; Guerrerio, A.L.; Parker, R.; Dietz, H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 2002, 295, 2258–2261. [Google Scholar] [CrossRef] [PubMed]
- Graille, M.; Séraphin, B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat. Rev. Mol. Cell Biol. 2012, 13, 727–735. [Google Scholar] [CrossRef]
- Harigaya, Y.; Parker, R. No-go decay: A quality control mechanism for RNA in translation. Wiley Interdiscip. Rev. RNA 2010, 1, 132–141. [Google Scholar] [CrossRef]
- Kong, J.; Liebhaber, S.A. A cell type-restricted mRNA surveillance pathway triggered by ribosome extension into the 3’ untranslated region. Nat. Struct. Mol. Biol. 2007, 14, 670–676. [Google Scholar] [CrossRef]
- Megel, C.; Morelle, G.; Lalande, S.; Duchêne, A.M.; Small, I.; Maréchal-Drouard, L. Surveillance and cleavage of eukaryotic tRNAs. Int. J. Mol. Sci. 2015, 16, 1873–1893. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M.; Cohen, R.E. Proteasomes and their kin: Proteases in the machine age. Nat. Rev. Mol. Cell Biol. 2004, 5, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.L. Functions of the proteasome: From protein degradation and immune surveillance to cancer therapy. Biochem. Soc. Trans. 2007, 35, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Bochtler, M.; Brandstetter, H.; Clausen, T.; Huber, R. Molecular machines for protein degradation. Chembiochem 2005, 6, 222–256. [Google Scholar] [CrossRef]
- Laney, J.D.; Hochstrasser, M. Substrate targeting in the ubiquitin system. Cell 1999, 97, 427–430. [Google Scholar] [CrossRef]
- Saez, I.; Vilchez, D. The Mechanistic Links Between Proteasome Activity, Aging and Age-related Diseases. Curr. Genom. 2014, 15, 38–51. [Google Scholar] [CrossRef]
- Ravalin, M.; Basu, K.; Gestwicki, J.E.; Craik, C.S. End-Binding E3 Ubiquitin Ligases Enable Protease Signaling. ACS Chem. Biol. 2021, 16, 2047–2056. [Google Scholar] [CrossRef]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Yu, H.; Mim, C.; Matouschek, A. Regulated protein turnover: Snapshots of the proteasome in action. Nat. Rev. Mol. Cell Biol. 2014, 15, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Bajorek, M.; Köhler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Debnath, J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015, 16, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the integrated stress response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef]
- Aman, Y.; Schmauck-Medina, T.; Hansen, M.; Morimoto, R.I.; Simon, A.K.; Bjedov, I.; Palikaras, K.; Simonsen, A.; Johansen, T.; Tavernarakis, N.; et al. Autophagy in healthy aging and disease. Nat. Aging 2021, 1, 634–650. [Google Scholar] [CrossRef]
- Hansen, M.; Rubinsztein, D.C.; Walker, D.W. Autophagy as a promoter of longevity: Insights from model organisms. Nat. Rev. Mol. Cell Biol. 2018, 19, 579–593. [Google Scholar] [CrossRef]
- Farré, J.C.; Subramani, S. Mechanistic insights into selective autophagy pathways: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2016, 17, 537–552. [Google Scholar] [CrossRef]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018, 20, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Klionsky, D.J.; Shen, H.M. The emerging mechanisms and functions of microautophagy. Nat. Rev. Mol. Cell Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Son, H.G.; Seo, M.; Ham, S.; Hwang, W.; Lee, D.; An, S.W.; Artan, M.; Seo, K.; Kaletsky, R.; Arey, R.N.; et al. RNA surveillance via nonsense-mediated mRNA decay is crucial for longevity in daf-2/insulin/IGF-1 mutant C. elegans. Nat. Commun. 2017, 8, 14749. [Google Scholar] [CrossRef] [PubMed]
- Mullani, N.; Porozhan, Y.; Mangelinck, A.; Rachez, C.; Costallat, M.; Batsché, E.; Goodhardt, M.; Cenci, G.; Mann, C.; Muchardt, C. Reduced RNA turnover as a driver of cellular senescence. Life Sci. Alliance 2021, 4, e202000809. [Google Scholar] [CrossRef]
- Bulteau, A.L.; Petropoulos, I.; Friguet, B. Age-related alterations of proteasome structure and function in aging epidermis. Exp. Gerontol. 2000, 35, 767–777. [Google Scholar] [CrossRef]
- Carrard, G.; Dieu, M.; Raes, M.; Toussaint, O.; Friguet, B. Impact of ageing on proteasome structure and function in human lymphocytes. Int. J. Biochem. Cell Biol. 2003, 35, 728–739. [Google Scholar] [CrossRef]
- Chondrogianni, N.; Stratford, F.L.; Trougakos, I.P.; Friguet, B.; Rivett, A.J.; Gonos, E.S. Central role of the proteasome in senescence and survival of human fibroblasts: Induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J. Biol. Chem. 2003, 278, 28026–28037. [Google Scholar] [CrossRef]
- Bulteau, A.L.; Szweda, L.I.; Friguet, B. Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 2002, 397, 298–304. [Google Scholar] [CrossRef]
- Husom, A.D.; Peters, E.A.; Kolling, E.A.; Fugere, N.A.; Thompson, L.V.; Ferrington, D.A. Altered proteasome function and subunit composition in aged muscle. Arch. Biochem. Biophys. 2004, 421, 67–76. [Google Scholar] [CrossRef]
- Dasuri, K.; Zhang, L.; Ebenezer, P.; Liu, Y.; Fernandez-Kim, S.O.; Keller, J.N. Aging and dietary restriction alter proteasome biogenesis and composition in the brain and liver. Mech. Ageing Dev. 2009, 130, 777–783. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Labbadia, J.; Morimoto, R.I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 2015, 84, 435–464. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Park, S.H.; Hartl, F.U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014, 24, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Ruano, D. Proteostasis Dysfunction in Aged Mammalian Cells. The Stressful Role of Inflammation. Front. Mol. Biosci. 2021, 8, 658742. [Google Scholar] [CrossRef]
- Tomaru, U.; Takahashi, S.; Ishizu, A.; Miyatake, Y.; Gohda, A.; Suzuki, S.; Ono, A.; Ohara, J.; Baba, T.; Murata, S.; et al. Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am. J. Pathol. 2012, 180, 963–972. [Google Scholar] [CrossRef]
- Dahlmann, B. Role of proteasomes in disease. BMC Biochem. 2007, 8, S3. [Google Scholar] [CrossRef]
- Tonoki, A.; Kuranaga, E.; Tomioka, T.; Hamazaki, J.; Murata, S.; Tanaka, K.; Miura, M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell Biol. 2009, 29, 1095–1106. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Husom, A.D.; Thompson, L.V. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005, 19, 644–646. [Google Scholar] [CrossRef]
- Hwang, J.S.; Hwang, J.S.; Chang, I.; Kim, S. Age-associated decrease in proteasome content and activities in human dermal fibroblasts: Restoration of normal level of proteasome subunits reduces aging markers in fibroblasts from elderly persons. J. Gerontol. A Biol. Sci. Med. Sci. 2007, 62, 490–499. [Google Scholar] [CrossRef]
- Huber, N.; Sakai, N.; Eismann, T.; Shin, T.; Kuboki, S.; Blanchard, J.; Schuster, R.; Edwards, M.J.; Wong, H.R.; Lentsch, A.B. Age-related decrease in proteasome expression contributes to defective nuclear factor-kappaB activation during hepatic ischemia/reperfusion. Hepatology 2009, 49, 1718–1728. [Google Scholar] [CrossRef]
- Grune, T. Oxidative stress, aging and the proteasomal system. Biogerontology 2000, 1, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Sakurai, T.; Usami, H.; Uchida, K. Oxidative modification of proteasome: Identification of an oxidation-sensitive subunit in 26 S proteasome. Biochemistry 2005, 44, 13893–13901. [Google Scholar] [CrossRef] [PubMed]
- Zmijewski, J.W.; Banerjee, S.; Abraham, E. S-glutathionylation of the Rpn2 regulatory subunit inhibits 26 S proteasomal function. J. Biol. Chem. 2009, 284, 22213–22221. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.M.; Netto, L.E.; Simões, V.; Santos, L.F.; Gozzo, F.C.; Demasi, M.A.; Oliveira, C.L.; Bicev, R.N.; Klitzke, C.F.; Sogayar, M.C.; et al. Redox control of 20S proteasome gating. Antioxid. Redox Signal. 2012, 16, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Kors, S.; Geijtenbeek, K.; Reits, E.; Schipper-Krom, S. Regulation of Proteasome Activity by (Post-)transcriptional Mechanisms. Front. Mol. Biosci. 2019, 6, 48. [Google Scholar] [CrossRef]
- Lipinski, M.M.; Zheng, B.; Lu, T.; Yan, Z.; Py, B.F.; Ng, A.; Xavier, R.J.; Li, C.; Yankner, B.A.; Scherzer, C.R.; et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2010, 107, 14164–14169. [Google Scholar] [CrossRef]
- Lapierre, L.R.; Kumsta, C.; Sandri, M.; Ballabio, A.; Hansen, M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy 2015, 11, 867–880. [Google Scholar] [CrossRef]
- Shi, Y.; Shen, H.M.; Gopalakrishnan, V.; Gordon, N. Epigenetic Regulation of Autophagy Beyond the Cytoplasm: A Review. Front. Cell Dev. Biol. 2021, 9, 675599. [Google Scholar] [CrossRef]
- Shi, L.; Yan, H.; An, S.; Shen, M.; Jia, W.; Zhang, R.; Zhao, L.; Huang, G.; Liu, J. SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Mol. Oncol. 2019, 13, 358–375. [Google Scholar] [CrossRef]
- Lindeboom, R.G.; Supek, F.; Lehner, B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 2016, 48, 1112–1118. [Google Scholar] [CrossRef]
- Pestova, T.V.; Lomakin, I.B.; Lee, J.H.; Choi, S.K.; Dever, T.E.; Hellen, C.U. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 2000, 403, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Leslie, N.R.; Downes, C.P. PTEN function: How normal cells control it and tumour cells lose it. Biochem. J. 2004, 382, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Floquet, C.; Deforges, J.; Rousset, J.P.; Bidou, L. Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 2011, 39, 3350–3362. [Google Scholar] [CrossRef] [PubMed]
- Gudikote, J.P.; Cascone, T.; Poteete, A.; Sitthideatphaiboon, P.; Wu, Q.; Morikawa, N.; Zhang, F.; Peng, S.; Tong, P.; Li, L.; et al. Inhibition of nonsense-mediated decay rescues p53β/γ isoform expression and activates the p53 pathway in MDM2-overexpressing and select p53-mutant cancers. J. Biol. Chem. 2021, 297, 101163. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Kang, D.; Han, N.; Lee, Y.; Hwang, H.J.; Lee, S.B.; You, J.S.; Min, B.S.; Park, H.J.; Ko, Y.G.; et al. A novel long noncoding RNA Linc-ASEN represses cellular senescence through multileveled reduction of p21 expression. Cell Death Differ. 2020, 27, 1844–1861. [Google Scholar] [CrossRef]
- Torres-Fernández, L.A.; Jux, B.; Bille, M.; Port, Y.; Schneider, K.; Geyer, M.; Mayer, G.; Kolanus, W. The mRNA repressor TRIM71 cooperates with Nonsense-Mediated Decay factors to destabilize the mRNA of CDKN1A/p21. Nucleic Acids Res. 2019, 47, 11861–11879. [Google Scholar] [CrossRef]
- Kim, B.C.; Lee, H.C.; Lee, J.J.; Choi, C.M.; Kim, D.K.; Lee, J.C.; Ko, Y.G.; Lee, J.S. Wig1 prevents cellular senescence by regulating p21 mRNA decay through control of RISC recruitment. EMBO J. 2012, 31, 4289–4303. [Google Scholar] [CrossRef]
- Kishor, A.; Ge, Z.; Hogg, J.R. hnRNP L-dependent protection of normal mRNAs from NMD subverts quality control in B cell lymphoma. EMBO J. 2019, 38, e99128. [Google Scholar] [CrossRef]
- Ryu, S.; Jung, M.; Kim, C.; Kang, H.; Han, S.; Cha, S.; Jeong, S.M.; Lee, E.K. Loss of RNA binding protein HuD facilitates the production of the senescence-associated secretory phenotype. Cell Death Dis. 2022, 13, 329. [Google Scholar] [CrossRef]
- Senft, D.; Qi, J.; Ronai, Z.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 2018, 18, 69–88. [Google Scholar] [CrossRef]
- Liu, Y.; Tao, S.; Liao, L.; Li, Y.; Li, H.; Li, Z.; Lin, L.; Wan, X.; Yang, X.; Chen, L. TRIM25 promotes the cell survival and growth of hepatocellular carcinoma through targeting Keap1-Nrf2 pathway. Nat. Commun. 2020, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Wu, X.; Nan, Y.; Chang, W.; Zhao, P.; Zhang, Y.; Su, D.; Liu, Z. TRIM32/USP11 Balances ARID1A Stability and the Oncogenic/Tumor-Suppressive Status of Squamous Cell Carcinoma. Cell Rep. 2020, 30, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Gu, X.; Cui, H. Emerging Roles of SKP2 in Cancer Drug Resistance. Cells 2021, 10, 1147. [Google Scholar] [CrossRef]
- Bornstein, G.; Bloom, J.; Sitry-Shevah, D.; Nakayama, K.; Pagano, M.; Hershko, A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 2003, 278, 25752–25757. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, L.M.; Yeh, K.H.; Lee, S.J.; Sun, H.; Zhang, H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 1999, 9, 661–664. [Google Scholar] [PubMed]
- Kossatz, U.; Dietrich, N.; Zender, L.; Buer, J.; Manns, M.P.; Malek, N.P. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev. 2004, 18, 2602–2607. [Google Scholar] [CrossRef]
- Wei, Z.; Jiang, X.; Qiao, H.; Zhai, B.; Zhang, L.; Zhang, Q.; Wu, Y.; Jiang, H.; Sun, X. STAT3 interacts with Skp2/p27/p21 pathway to regulate the motility and invasion of gastric cancer cells. Cell Signal. 2013, 25, 931–938. [Google Scholar] [CrossRef]
- Zhang, Y.; Zvi, Y.S.; Batko, B.; Zaphiros, N.; O’Donnell, E.F.; Wang, J.; Sato, K.; Yang, R.; Geller, D.S.; Koirala, P.; et al. Down-regulation of Skp2 expression inhibits invasion and lung metastasis in osteosarcoma. Sci. Rep. 2018, 8, 294. [Google Scholar] [CrossRef]
- Hume, S.; Grou, C.P.; Lascaux, P.; D’Angiolella, V.; Legrand, A.J.; Ramadan, K.; Dianov, G.L. The NUCKS1-SKP2-p21/p27 axis controls S phase entry. Nat. Commun. 2021, 12, 6959. [Google Scholar] [CrossRef]
- Huang, H.; Regan, K.M.; Wang, F.; Wang, D.; Smith, D.I.; van Deursen, J.M.; Tindall, D.J. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA 2005, 102, 1649–1654. [Google Scholar] [CrossRef]
- Li, C.; Du, L.; Ren, Y.; Liu, X.; Jiao, Q.; Cui, D.; Wen, M.; Wang, C.; Wei, G.; Wang, Y.; et al. SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination. J. Exp. Clin. Cancer Res. 2019, 38, 76. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Sun, D.; Zhang, X. The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int. 2019, 19, 216. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, J.; Cao, W.; Yang, L.; Chen, Q.; He, J.; Yi, Q.; Huang, H.; Zhang, E.; Cai, Z. The many substrates and functions of NEDD4-1. Cell Death Dis. 2019, 10, 904. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Bellon, M.; Ju, M.; Zhao, L.; Wei, M.; Fu, L.; Nicot, C. Clinical significance of FBXW7 loss of function in human cancers. Mol. Cancer 2022, 21, 87. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, M.; Mimori, K.; Ishii, H.; Yokobori, T.; Takatsuno, Y.; Sato, T.; Toh, H.; Onoyama, I.; Nakayama, K.I.; Baba, H.; et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: Clinical significance. Int. J. Cancer 2010, 126, 1828–1837. [Google Scholar] [CrossRef]
- Ibusuki, M.; Yamamoto, Y.; Shinriki, S.; Ando, Y.; Iwase, H. Reduced expression of ubiquitin ligase FBXW7 mRNA is associated with poor prognosis in breast cancer patients. Cancer Sci. 2011, 102, 439–445. [Google Scholar] [CrossRef]
- Yang, H.; Lu, X.; Liu, Z.; Chen, L.; Xu, Y.; Wang, Y.; Wei, G.; Chen, Y. FBXW7 suppresses epithelial-mesenchymal transition, stemness and metastatic potential of cholangiocarcinoma cells. Oncotarget 2015, 6, 6310–6325. [Google Scholar] [CrossRef]
- Yeh, C.H.; Bellon, M.; Nicot, C. FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef]
- Tan, P.; He, L.; Zhou, Y. TRIM59 deficiency curtails breast cancer metastasis through SQSTM1-selective autophagic degradation of PDCD10. Autophagy 2019, 15, 747–749. [Google Scholar] [CrossRef]
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chételat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer disease. Nat. Rev. Dis. Primers 2021, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Sivanandam, T.M.; Thakur, M.K. Amyloid precursor protein (APP) mRNA level is higher in the old mouse cerebral cortex and is regulated by sex steroids. J. Mol. Neurosci. 2011, 43, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.L.; Vassar, R. The Alzheimer’s disease beta-secretase enzyme, BACE1. Mol. Neurodegener. 2007, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.J.; Abdelmohsen, K.; Hutchison, E.R.; Mitchell, S.J.; Grammatikakis, I.; Guo, R.; Noh, J.H.; Martindale, J.L.; Yang, X.; Lee, E.K.; et al. HuD regulates coding and noncoding RNA to induce APP→Aβ processing. Cell Rep. 2014, 7, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Ni, H.; Yu, Y.; Zhang, M.; Wu, M.; Wang, Q.; Wang, L.; Xu, S.; Xu, Z.; Xu, C.; et al. BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs. J. Chem. Neuroanat. 2019, 98, 87–96. [Google Scholar] [CrossRef]
- Lee, E.K.; Kim, H.H.; Kuwano, Y.; Abdelmohsen, K.; Srikantan, S.; Subaran, S.S.; Gleichmann, M.; Mughal, M.R.; Martindale, J.L.; Yang, X.; et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 2010, 17, 732–739. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Ku, L.; Wilkinson, K.D.; Warren, S.T.; Feng, Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001, 29, 2276–2283. [Google Scholar] [CrossRef]
- Ghosh, A.; Mizuno, K.; Tiwari, S.S.; Proitsi, P.; Gomez Perez-Nievas, B.; Glennon, E.; Martinez-Nunez, R.T.; Giese, K.P. Alzheimer’s disease-related dysregulation of mRNA translation causes key pathological features with ageing. Transl. Psychiatry 2020, 10, 192. [Google Scholar] [CrossRef]
- Lee, J.H.; Ahn, N.H.; Choi, S.B.; Kwon, Y.; Yang, S.H. Natural Products Targeting Amyloid Beta in Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 2341. [Google Scholar] [CrossRef]
- Liu, Q.Y.; Lei, J.X.; Sikorska, M.; Liu, R. A novel brain-enriched E3 ubiquitin ligase RNF182 is up regulated in the brains of Alzheimer’s patients and targets ATP6V0C for degradation. Mol. Neurodegener. 2008, 3, 4. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, X.; Chaput, D.; Shin, M.-K.; Koh, Y.; Gan, L.; Pieper, A.A.; Woo, J.A.; Kang, D.E. X-linked ubiquitin-specific peptidase 11 increases tauopathy vulnerability in women. Cell 2022, 185, 3913–3930. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, T.; Tsutsui, A.; Tomomori-Sato, C.; Sato, S.; Saraf, A.; Washburn, M.P.; Florens, L.; Terada, T.; Shimizu, K.; Conaway, R.C.; et al. NRBP1-Containing CRL2/CRL4A Regulates Amyloid β Production by Targeting BRI2 and BRI3 for Degradation. Cell Rep. 2020, 30, 3478–3491. [Google Scholar] [CrossRef] [PubMed]
- Blair, L.J.; Nordhues, B.A.; Hill, S.E.; Scaglione, K.M.; O’Leary, J.C., 3rd; Fontaine, S.N.; Breydo, L.; Zhang, B.; Li, P.; Wang, L.; et al. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Investig. 2013, 123, 4158–4169. [Google Scholar] [CrossRef]
- Singh, A.K.; Pati, U. CHIP stabilizes amyloid precursor protein via proteasomal degradation and p53-mediated trans-repression of β-secretase. Aging Cell 2015, 14, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Sahara, N.; Murayama, M.; Mizoroki, T.; Urushitani, M.; Imai, Y.; Takahashi, R.; Murata, S.; Tanaka, K.; Takashima, A. In vivo evidence of CHIP up-regulation attenuating tau aggregation. J. Neurochem. 2005, 94, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Ambasta, R.K.; Veereshwarayya, V.; Rosen, K.M.; Kosik, K.S.; Band, H.; Mestril, R.; Patterson, C.; Querfurth, H.W. CHIP and HSPs interact with beta-APP in a proteasome-dependent manner and influence Abeta metabolism. Hum. Mol. Genet. 2007, 16, 848–864. [Google Scholar] [CrossRef] [PubMed]
- Joshi, V.; Amanullah, A.; Upadhyay, A.; Mishra, R.; Kumar, A.; Mishra, A. A Decade of Boon or Burden: What Has the CHIP Ever Done for Cellular Protein Quality Control Mechanism Implicated in Neurodegeneration and Aging? Front. Mol. Neurosci. 2016, 9, 93. [Google Scholar] [CrossRef]
- Du, F.; Yu, Q.; Yan, S.; Hu, G.; Lue, L.F.; Walker, D.G.; Wu, L.; Yan, S.F.; Tieu, K.; Yan, S.S. PINK1 signalling rescues amyloid pathology and mitochondrial dysfunction in Alzheimer’s disease. Brain 2017, 140, 3233–3251. [Google Scholar] [CrossRef]
- De Strooper, B. Loss-of-function presenilin mutations in Alzheimer disease. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007, 8, 141–146. [Google Scholar] [CrossRef]
- Pickford, F.; Masliah, E.; Britschgi, M.; Lucin, K.; Narasimhan, R.; Jaeger, P.A.; Small, S.; Spencer, B.; Rockenstein, E.; Levine, B.; et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J. Clin. Investig. 2008, 118, 2190–2199. [Google Scholar]
- Pavel, M.; Imarisio, S.; Menzies, F.M.; Jimenez-Sanchez, M.; Siddiqi, F.H.; Wu, X.; Renna, M.; O’Kane, C.J.; Crowther, D.C.; Rubinsztein, D.C. CCT complex restricts neuropathogenic protein aggregation via autophagy. Nat. Commun. 2016, 7, 13821. [Google Scholar] [CrossRef] [PubMed]
- Brehme, M.; Voisine, C.; Rolland, T.; Wachi, S.; Soper, J.H.; Zhu, Y.; Orton, K.; Villella, A.; Garza, D.; Vidal, M.; et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 2014, 9, 1135–1150. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Q.; Fang, F.; Florio, J.B.; Rockenstein, E.; Masliah, E.; Mobley, W.C.; Rissman, R.A.; Wu, C. T-complex protein 1-ring complex enhances retrograde axonal transport by modulating tau phosphorylation. Traffic 2018, 19, 840–853. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Luo, Y.; Zhang, X.N.; Yang, X.F.; Hong, X.Y.; Sun, D.S.; Li, X.C.; Hu, Y.; Li, X.G.; Zhang, J.F.; et al. MAPT/Tau accumulation represses autophagy flux by disrupting IST1-regulated ESCRT-III complex formation: A vicious cycle in Alzheimer neurodegeneration. Autophagy 2020, 16, 641–658. [Google Scholar] [CrossRef]
- Uddin, M.S.; Mamun, A.A.; Labu, Z.K.; Hidalgo-Lanussa, O.; Barreto, G.E.; Ashraf, G.M. Autophagic dysfunction in Alzheimer’s disease: Cellular and molecular mechanistic approaches to halt Alzheimer’s pathogenesis. J. Cell Physiol. 2019, 234, 8094–8112. [Google Scholar] [CrossRef]
- Poewe, W. Parkinson disease Primer—A true team effort. Nat. Rev. Dis. Primers 2020, 6, 31. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, Z.; Long, F.; Tan, J.; Gong, N.; Li, X.; Lin, C. The Roles of ATP13A2 Gene Mutations Leading to Abnormal Aggregation of α-Synuclein in Parkinson’s Disease. Front. Cell Neurosci. 2022, 16, 927682. [Google Scholar] [CrossRef]
- Hallacli, E.; Kayatekin, C.; Nazeen, S.; Wang, X.H.; Sheinkopf, Z.; Sathyakumar, S.; Sarkar, S.; Jiang, X.; Dong, X.; Di Maio, R.; et al. The Parkinson’s disease protein alpha-synuclein is a modulator of processing bodies and mRNA stability. Cell 2022, 185, 2035–2056. [Google Scholar] [CrossRef]
- Leroy, E.; Boyer, R.; Auburger, G.; Leube, B.; Ulm, G.; Mezey, E.; Harta, G.; Brownstein, M.J.; Jonnalagada, S.; Chernova, T.; et al. The ubiquitin pathway in Parkinson’s disease. Nature 1998, 395, 451–452. [Google Scholar] [CrossRef]
- Kabuta, T.; Furuta, A.; Aoki, S.; Furuta, K.; Wada, K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J. Biol. Chem. 2008, 283, 23731–23738. [Google Scholar] [CrossRef]
- Yasuda, T.; Miyachi, S.; Kitagawa, R.; Wada, K.; Nihira, T.; Ren, Y.R.; Hirai, Y.; Ageyama, N.; Terao, K.; Shimada, T.; et al. Neuronal specificity of alpha-synuclein toxicity and effect of Parkin co-expression in primates. Neuroscience 2007, 144, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Kabuta, T.; Setsuie, R.; Mitsui, T.; Kinugawa, A.; Sakurai, M.; Aoki, S.; Uchida, K.; Wada, K. Aberrant molecular properties shared by familial Parkinson’s disease-associated mutant UCH-L1 and carbonyl-modified UCH-L1. Hum. Mol. Genet. 2008, 17, 1482–1496. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Wu, R.M. LRRK 2 gene mutations in the pathophysiology of the ROCO domain and therapeutic targets for Parkinson’s disease: A review. J. Biomed. Sci. 2018, 25, 52. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.M.J.; Moreira, P.I.; Ambrósio, A.F.; Alves, C.H. PINK1/PARKIN signalling in neurodegeneration and neuroinflammation. Acta Neuropathol. Commun. 2020, 8, 189. [Google Scholar] [CrossRef]
- Bauer, P.O.; Goswami, A.; Wong, H.K.; Okuno, M.; Kurosawa, M.; Yamada, M.; Miyazaki, H.; Matsumoto, G.; Kino, Y.; Nagai, Y.; et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat. Biotechnol. 2010, 28, 256–263. [Google Scholar] [CrossRef]
- Koga, H.; Martinez-Vicente, M.; Arias, E.; Kaushik, S.; Sulzer, D.; Cuervo, A.M. Constitutive upregulation of chaperone-mediated autophagy in Huntington’s disease. J. Neurosci. 2011, 31, 18492–18505. [Google Scholar] [CrossRef] [PubMed]
- Anvar, S.Y.; Raz, Y.; Verway, N.; van der Sluijs, B.; Venema, A.; Goeman, J.J.; Vissing, J.; van der Maarel, S.M.; AC’t Hoen, P.; van Engelen, B.G.; et al. A decline in PABPN1 induces progressive muscle weakness in oculopharyngeal muscle dystrophy and in muscle aging. Aging (Albany N. Y.) 2013, 5, 412–426. [Google Scholar] [CrossRef]
- Riaz, M.; Raz, Y.; van Putten, M.; Paniagua-Soriano, G.; Krom, Y.D.; Florea, B.I.; Raz, V. PABPN1-Dependent mRNA Processing Induces Muscle Wasting. PLoS Genet. 2016, 12, e1006031. [Google Scholar] [CrossRef]
- Huo, F.; Liu, Q.; Liu, H. Contribution of muscle satellite cells to sarcopenia. Front. Physiol. 2022, 13, 892749. [Google Scholar] [CrossRef]
- Chen, W.; Datzkiw, D.; Rudnicki, M.A. Satellite cells in ageing: Use it or lose it. Open Biol. 2020, 10, 200048. [Google Scholar] [CrossRef]
- Hausburg, M.A.; Doles, J.D.; Clement, S.L.; Cadwallader, A.B.; Hall, M.N.; Blackshear, P.J.; Lykke-Andersen, J.; Olwin, B.B. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. eLife 2015, 4, e03390. [Google Scholar] [CrossRef] [PubMed]
- Sandler, H.; Kreth, J.; Timmers, H.T.; Stoecklin, G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011, 39, 4373–4386. [Google Scholar] [CrossRef]
- Abbadi, D.; Andrews, J.J.; Katsara, O.; Schneider, R.J. AUF1 gene transfer increases exercise performance and improves skeletal muscle deficit in adult mice. Mol. Ther. Methods Clin. Dev. 2021, 22, 222–236. [Google Scholar] [CrossRef]
- Bodine, S.C.; Baehr, L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E469–E484. [Google Scholar] [CrossRef] [PubMed]
- Foletta, V.C.; White, L.J.; Larsen, A.E.; Léger, B.; Russell, A.P. The role and regulation of MAFbx/atrogin-1 and MuRF1 in skeletal muscle atrophy. Pflug. Arch. 2011, 461, 325–335. [Google Scholar] [CrossRef]
- Clavel, S.; Coldefy, A.S.; Kurkdjian, E.; Salles, J.; Margaritis, I.; Derijard, B. Atrophy-related ubiquitin ligases, atrogin-1 and MuRF1 are up-regulated in aged rat Tibialis Anterior muscle. Mech. Ageing Dev. 2006, 127, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.K.; Gupta, S.K.; Bhatnagar, S.; Panguluri, S.K.; Darnay, B.G.; Choi, Y.; Kumar, A. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J. Cell Biol. 2010, 191, 1395–1411. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.K.; Bhatnagar, S.; Mishra, V.; Srivastava, S.; Darnay, B.G.; Choi, Y.; Kumar, A. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol. Cell Biol. 2012, 32, 1248–1259. [Google Scholar] [CrossRef] [PubMed]
- Franco-Romero, A.; Sandri, M. Role of autophagy in muscle disease. Mol. Asp. Med. 2021, 82, 101041. [Google Scholar] [CrossRef]
- Drummond, M.J.; Addison, O.; Brunker, L.; Hopkins, P.N.; McClain, D.A.; LaStayo, P.C.; Marcus, R.L. Downregulation of E3 ubiquitin ligases and mitophagy-related genes in skeletal muscle of physically inactive, frail older women: A cross-sectional comparison. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Carnio, S.; LoVerso, F.; Baraibar, M.A.; Longa, E.; Khan, M.M.; Maffei, M.; Reischl, M.; Canepari, M.; Loefler, S.; Kern, H.; et al. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep. 2014, 8, 1509–1521. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Thavarajah, T.; Gu, W.; Cai, J.; Xu, Q. Impact of miRNA in Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2018, 38, e159–e170. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, K.C.; Wu, W.; Subramaniam, S.; Shyy, J.Y.; Chiu, J.J.; Li, J.Y.; Chien, S. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc. Natl. Acad. Sci. USA 2011, 108, 10355–10360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Taylor, W.R.; Joseph, G.; Caracciolo, V.; Gonzales, D.M.; Sidell, N.; Seli, E.; Blackshear, P.J.; Kallen, C.B. mRNA-binding protein ZFP36 is expressed in atherosclerotic lesions and reduces inflammation in aortic endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1212–1220. [Google Scholar] [CrossRef]
- Li, M.; Sun, G.; Wang, P.; Wang, W.; Cao, K.; Song, C.; Sun, Y.; Zhang, Y.; Zhang, N. Research progress of Nedd4L in cardiovascular diseases. Cell Death Discov. 2022, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, V.; Daidié, D.; Li, H.; Pao, A.C.; LaGrange, L.P.; Wang, J.; Vandewalle, A.; Stockand, J.D.; Staub, O.; Pearce, D. Serum- and glucocorticoid-regulated kinase 1 regulates ubiquitin ligase neural precursor cell-expressed, developmentally down-regulated protein 4-2 by inducing interaction with 14-3-3. Mol. Endocrinol. 2005, 19, 3073–3084. [Google Scholar] [CrossRef]
- Rothman, A.M.; Arnold, N.D.; Pickworth, J.A.; Iremonger, J.; Ciuclan, L.; Allen, R.M.; Guth-Gundel, S.; Southwood, M.; Morrell, N.W.; Thomas, M.; et al. MicroRNA-140-5p and SMURF1 regulate pulmonary arterial hypertension. J. Clin. Investig. 2016, 126, 2495–2508. [Google Scholar] [CrossRef]
- Miyamoto, S. Autophagy and cardiac aging. Cell Death Differ. 2019, 26, 653–664. [Google Scholar] [CrossRef]
- Bravo-San Pedro, J.M.; Kroemer, G.; Galluzzi, L. Autophagy and Mitophagy in Cardiovascular Disease. Circ. Res. 2017, 120, 1812–1824. [Google Scholar] [CrossRef]
- Nakai, A.; Yamaguchi, O.; Takeda, T.; Higuchi, Y.; Hikoso, S.; Taniike, M.; Omiya, S.; Mizote, I.; Matsumura, Y.; Asahi, M.; et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007, 13, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Billia, F.; Hauck, L.; Konecny, F.; Rao, V.; Shen, J.; Mak, T.W. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc. Natl. Acad. Sci. USA 2011, 108, 9572–9577. [Google Scholar] [CrossRef]
- Chen, Y.; Dorn, G.W., 2nd. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 2013, 340, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Mita, Y.; Okawa, Y.; Ariyoshi, M.; Iwai-Kanai, E.; Ueyama, T.; Ikeda, K.; Ogata, T.; Matoba, S. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat. Commun. 2013, 4, 2308. [Google Scholar] [CrossRef] [PubMed]
- Pyo, J.O.; Yoo, S.M.; Ahn, H.H.; Nah, J.; Hong, S.H.; Kam, T.I.; Jung, S.; Jung, Y.K. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 2013, 4, 2300. [Google Scholar] [PubMed]
- Gu, J.; Hu, W.; Song, Z.P.; Chen, Y.G.; Zhang, D.D.; Wang, C.Q. Rapamycin Inhibits Cardiac Hypertrophy by Promoting Autophagy via the MEK/ERK/Beclin-1 Pathway. Front. Physiol. 2016, 7, 104. [Google Scholar] [CrossRef] [PubMed]

| RNA or Proteins | Mechanisms of Regulation | References |
|---|---|---|
| Cancer | ||
| Linc-ASEN | Degradation of p21 mRNA by cooperating with NMD factors, UPF1 and DCP1A. | [85] |
| TRIM71 | NMD-mediated p21 mRNA decay with NMD factors, UPF1 and SMG1 | [86] |
| Wig1 | Destabilize p21 mRNA by recruiting Ago2, a major component of RISC | [87] |
| HuD/ELAVL4 | Negatively regulation of CCL2 mRNA by directly binding to 3’UTR of the CCL2 mRNA | [89] |
| TRIM25 | Reduction of Keap1, a Nrf2 inhibitor, by its ubiquitination and degradation | [91] |
| TRIM32 | Decrease ARID1A protein level via ubiquitin-mediated degradation | [92] |
| Skp2 | Degradation of p21, p27, FOXO1 and PDCD4 through ubiquitination | [93,94,95,96,97,98,99,100,101] |
| MDM2 | Ubiquitination and degradation of p53 | [102] |
| NEDD4-1 | Degradation of PTEN, CNrasGEF, N-Myc, Her3 and Ras via K48-linked ubiquitination. Stabilization of MDM2 via K63-linked ubiquitination | [103] |
| FBXW7 | Ubiquitin-mediated degradation of cyclin E, Aurora A, Notch1, mTOR, c-Myc, Mcl-1 and Jun | [104,105,106,107,108] |
| TRIM59 | Increase the stability of PDCD10 by inhibiting RNFT1-mediated K63 ubiquitination and SQSTM1-mediated autophagic degradation | [110] |
| Neurodegenerative diseases-Alzheimer’s diseases (AD) | ||
| HuD/ELAVL4 | Increase the stability of APP mRNA, BACE1 mRNA, and BACE1-AS lncRNA | [114] |
| FMRP | Recruiting the APP mRNA into P-bodies to inhibits its translation. | [116,117,118] |
| hnRNPC | Binding to APP mRNA and enhancing APP translation. | [116] |
| RNF182 | Ubiquitination and degradation of ATP6V0C | [120] |
| USP11 | Increase of tau stability and aggregation. Involve in the process of tau acetylation (K281 and K274) | [121] |
| NBRP1 | Degradation of BRI2 and BRI3 | [122] |
| FKBP51 | Inhibition of tau clearance and increase of tau aggregation by cooperating with Hsp90. | [123] |
| CHIP | Ubiquitin-mediated degradation of Aβ42 peptide, phosphorylated tau and β-secretase 1 | [124,125,126,127] |
| PINK1 | Degradation of dysfunctional mitochondria via mitophagy | [128] |
| Presenilin-1 | Cleaves of βCTF to produce Aβ. | [129] |
| Beclin-1 | Beclin-1 mRNA and protein levels are also reduced in human aged brain, independent of AD pathology. Decreased Beclin-1 expression impairs autophagosome formation and leads to the accumulation of Aβ | [130] |
| CCT/TRiC | CCT expression is reduced in aging and neurodegenerative diseases. Decreased CCT promotes LC3 degradation, improper autophagosome formation, and tau folding | [131,132,133] |
| Neurodegenerative diseases-Parkinson’s disease (PD) | ||
| ATP13A2/PARK9 | Preventing the accumulation and aggregation of α-synuclein in the dopaminergic cells | [137] |
| UCHL1/PARK5 | Role as deubiquitinase or hydrolase enzymes in the UPS. | [139,140,141,142] |
| LRRK2 | Mutation of LRRK2 cause the abnormal mitochondrial dynamics and the aberrant au-tophagic-lysosomal pathway, leading to cell death and α-synuclein accumulation | [143] |
| PINK1, Parkin | S-nitrosylation of PINK1 and Parkin disrupt mitophagy, accumulating damaged mitochondria. | [144] |
| Neurodegenerative diseases-Huntington’s disease (HD) | ||
| HTT | Modulator of selective autophagy. Required for autophagy recognition and activating machinery (p62 and ULK1). | [145,146] |
| Muscle disease | ||
| PABPN1 | Regulation of distal polyadenylation site and proteasomal activity | [147,148] |
| TTP | Destabilize MyoD mRNA by recruiting the deadenylase components, Not1 and Caf1 | [151,152] |
| AUF1 | Maintenance and differentiation of MuSCs via the regulation of mRNA stability | [153] |
| MuRF1 | Regulation of MHC-related proteins to induce overall protein degradation | [154,155,156] |
| Atrogin-1 | Degradation of eIF3-f, myogenin, and MyoD to affect protein synthesis and myogenic differentiation | [154,155,156] |
| TRAF6 | Prevention of Beclin1, p62, and LC3B through ubiquitination | [157,158] |
| ATG7 | Atg7-deficient mice induces muscle atrophy, inflammation, and muscle weakness by mitochondrial dysfunction and oxidative stress | [160,161] |
| Cardiovascular disease | ||
| miR-21 | Repression of PPARa mRNA by directly binding to 3’UTR | [162,163] |
| miR-126 | Repression of proinflammatory TNF-α mRNA | [162] |
| ZFP36 | Regulation of MCP-1 and IL-6 mRNA stability | [164] |
| NEDD4L | Degradation of epithelial sodium channel (ENaC) through ubiquitination | [165,166] |
| SMURF1 | Degradation of BMPR2 through ubiquitination | [167] |
| ATG5 | Atg5-deficient mice exhibit cardiovascular diseases, including cardiac hypertrophy, contractile dysfunction, and left ventricular dilatation. Overexpression of Atg5 activates autophagy, delays cardiac aging, and extends the lifespan in mice | [170,174] |
| PINK1 | Knockout of Pink1 induces cardiac hypertrophy and left ventricular dysfunction through impaired mitochondrial biogenesis in mice | [171] |
| Parkin | Cardiomyocyte-specific overexpression of Parkin improves cardiac function and reduces the positivity of senescence-associated β-galactosidase (SA-β-Gal) through increasing mitophagy in aged mice | [173] |
| Mnf2 | Protein required for mitochondrial dynamics and associated with mitophagy | [172] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, D.; Baek, Y.; Lee, J.-S. Mechanisms of RNA and Protein Quality Control and Their Roles in Cellular Senescence and Age-Related Diseases. Cells 2022, 11, 4062. https://doi.org/10.3390/cells11244062
Kang D, Baek Y, Lee J-S. Mechanisms of RNA and Protein Quality Control and Their Roles in Cellular Senescence and Age-Related Diseases. Cells. 2022; 11(24):4062. https://doi.org/10.3390/cells11244062
Chicago/Turabian StyleKang, Donghee, Yurim Baek, and Jae-Seon Lee. 2022. "Mechanisms of RNA and Protein Quality Control and Their Roles in Cellular Senescence and Age-Related Diseases" Cells 11, no. 24: 4062. https://doi.org/10.3390/cells11244062
APA StyleKang, D., Baek, Y., & Lee, J.-S. (2022). Mechanisms of RNA and Protein Quality Control and Their Roles in Cellular Senescence and Age-Related Diseases. Cells, 11(24), 4062. https://doi.org/10.3390/cells11244062






