Restoring Age-Related Cognitive Decline through Environmental Enrichment: A Transcriptomic Approach
Abstract
1. Introduction
2. Materials & Methods
2.1. Animals
2.2. Barnes Maze Test
2.3. Tissue and RNA Preparation
2.4. RNA-seq
2.5. Data Analysis
2.6. qPCR
3. Results
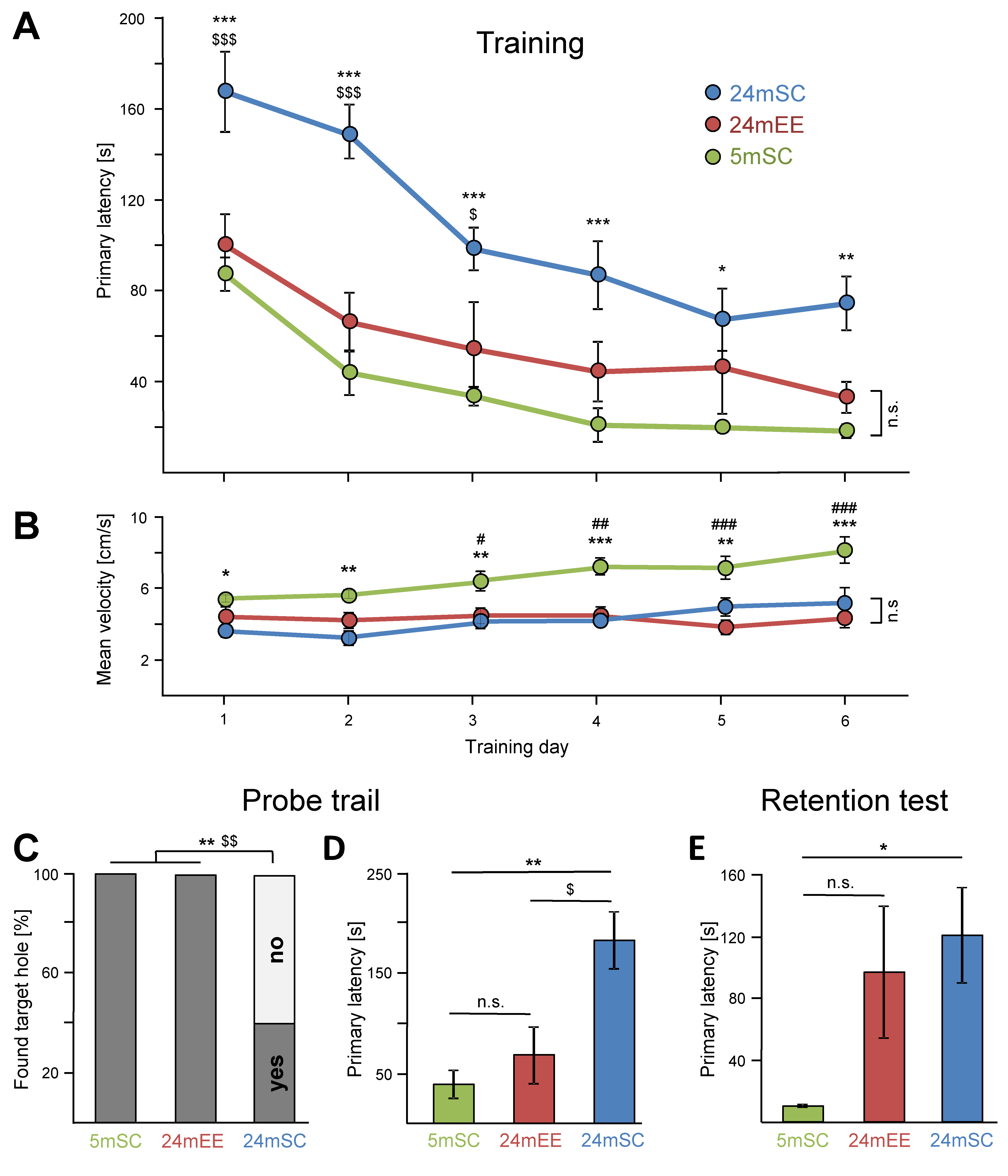
3.1. Performance in the Barnes Maze Test over the Training Days
3.2. Enriched Environment Improved Barnes Maze Performance in Aged Mice
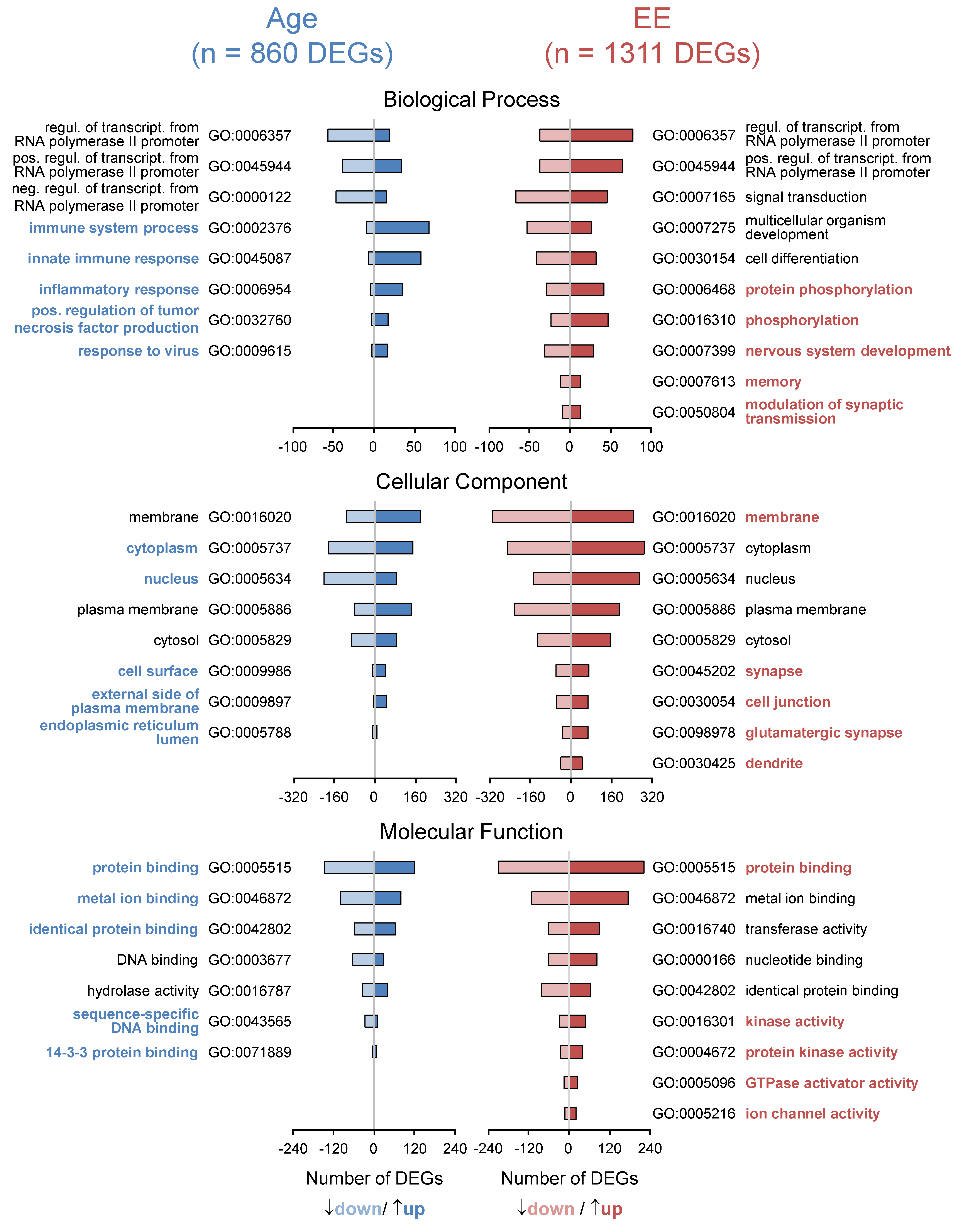
3.3. Aging of the Hippocampus Changes Gene Expression
3.4. Enriched Environment Changes Gene Expression in the Hippocampus
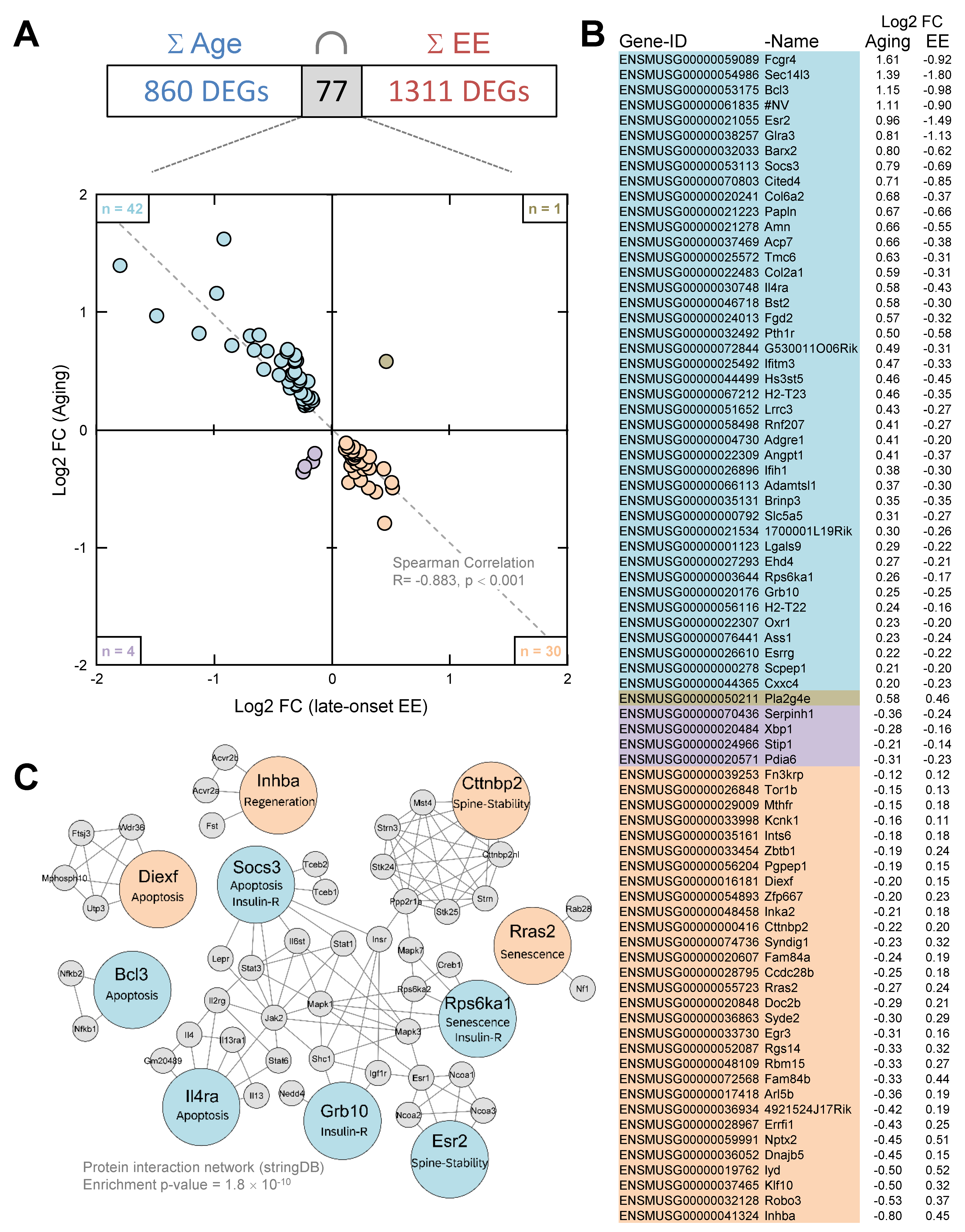
3.5. Enriched Environment Reverses Age-Related Gene Expression in the Hippocampus
4. Discussion
4.1. Enriched Environment Improves Cognitive Function in Aging
4.2. Age-Related Gene Expression Changes in the Hippocampus
4.3. Enriched Environment-Induced Gene Expression Changes in the Hippocampus
4.4. Reversal of Age-Related Gene Expression Changes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GBD2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.G.; Rahnama, N.P.; Silva, A.J. Investigation of age-related cognitive decline using mice as a model system: Behavioral correlates. Am. J. Geriatr. Psychiatry 2006, 14, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Oren, N.; Ash, E.L.; Shapira-Lichter, I.; Elkana, O.; Reichman-Eisikovits, O.; Chomsky, L.; Lerner, Y. Changes in Resting-State Functional Connectivity of the Hippocampus Following Cognitive Effort Predict Memory Decline at Older Age-A Longitudinal fMRI Study. Front. Aging Neurosci. 2019, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Small, S.A.; Schobel, S.A.; Buxton, R.B.; Witter, M.P.; Barnes, C.A. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat. Rev. Neurosci. 2011, 12, 585–601. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, I.; Hamilton, D.A.; Petropoulos, H.; Yeo, R.A.; Brooks, W.M.; Baumgartner, R.N.; Sutherland, R.J. The aging hippocampus: Cognitive, biochemical and structural findings. Cereb. Cortex 2003, 13, 1344–1351. [Google Scholar] [CrossRef]
- Cribbs, D.H.; Berchtold, N.C.; Perreau, V.; Coleman, P.D.; Rogers, J.; Tenner, A.J.; Cotman, C.W. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: A microarray study. J. Neuroinflamm. 2012, 9, 179. [Google Scholar] [CrossRef]
- Hu, W.T.; Howell, J.C.; Ozturk, T.; Gangishetti, U.; Kollhoff, A.L.; Hatcher-Martin, J.M.; Anderson, A.M.; Tyor, W.R. CSF Cytokines in Aging, Multiple Sclerosis, and Dementia. Front. Immunol. 2019, 10, 480. [Google Scholar] [CrossRef]
- Newcombe, E.A.; Camats-Perna, J.; Silva, M.L.; Valmas, N.; Huat, T.J.; Medeiros, R. Inflammation: The link between comorbidities, genetics, and Alzheimer’s disease. J. Neuroinflamm. 2018, 15, 276. [Google Scholar] [CrossRef]
- Sieber, M.W.; Claus, R.A.; Witte, O.W.; Frahm, C. Attenuated inflammatory response in aged mice brains following stroke. PLoS ONE 2011, 6, e26288. [Google Scholar] [CrossRef]
- Bishop, N.A.; Lu, T.; Yankner, B.A. Neural mechanisms of ageing and cognitive decline. Nature 2010, 464, 529–535. [Google Scholar] [CrossRef]
- van Praag, H.; Kempermann, G.; Gage, F.H. Neural consequences of environmental enrichment. Nat. Rev. Neurosci. 2000, 1, 191–198. [Google Scholar] [CrossRef]
- Lambert, K.; Eisch, A.J.; Galea, L.A.M.; Kempermann, G.; Merzenich, M. Optimizing brain performance: Identifying mechanisms of adaptive neurobiological plasticity. Neurosci. Biobehav. Rev. 2019, 105, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Petrosini, L.; De Bartolo, P.; Foti, F.; Gelfo, F.; Cutuli, D.; Leggio, M.G.; Mandolesi, L. On whether the environmental enrichment may provide cognitive and brain reserves. Brain Res. Rev. 2009, 61, 221–239. [Google Scholar] [CrossRef] [PubMed]
- van Uffelen, J.G.; Chin, A.P.M.J.; Hopman-Rock, M.; van Mechelen, W. The effects of exercise on cognition in older adults with and without cognitive decline: A systematic review. Clin. J. Sport Med. 2008, 18, 486–500. [Google Scholar] [CrossRef] [PubMed]
- Saez de Asteasu, M.L.; Martinez-Velilla, N.; Zambom-Ferraresi, F.; Casas-Herrero, A.; Izquierdo, M. Role of physical exercise on cognitive function in healthy older adults: A systematic review of randomized clinical trials. Ageing Res. Rev. 2017, 37, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Asl, N.A.; Sheikhzade, F.; Torchi, M.; Roshangar, L.; Khamnei, S. Long-term regular exercise promotes memory and learning in young but not in older rats. Pathophysiology 2008, 15, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Shan, G.; Gerstenberger, S. Fisher’s exact approach for post hoc analysis of a chi-squared test. PLoS ONE 2017, 12, e0188709. [Google Scholar] [CrossRef]
- Durlak, J.A. How to select, calculate, and interpret effect sizes. J. Pediatr Psychol. 2009, 34, 917–928. [Google Scholar] [CrossRef]
- Bentley, D.R.; Balasubramanian, S.; Swerdlow, H.P.; Smith, G.P.; Milton, J.; Brown, C.G.; Hall, K.P.; Evers, D.J.; Barnes, C.L.; Bignell, H.R.; et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008, 456, 53–59. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- HTSeq Software Package v0.5.4. Available online: https://htseq.readthedocs.io/en/release_0.11.1/history.html#version-0-5-4 (accessed on 20 February 2013).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yoav Benjamini, Y.H. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Sieber, M.W.; Guenther, M.; Kohl, M.; Witte, O.W.; Claus, R.A.; Frahm, C. Inter-age variability of bona fide unvaried transcripts Normalization of quantitative PCR data in ischemic stroke. Neurobiol. Aging 2010, 31, 654–664. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Takuma, K.; Ago, Y.; Matsuda, T. Preventive effects of an enriched environment on rodent psychiatric disorder models. J. Pharmacol. Sci 2011, 117, 71–76. [Google Scholar] [CrossRef]
- Garthe, A.; Roeder, I.; Kempermann, G. Mice in an enriched environment learn more flexibly because of adult hippocampal neurogenesis. Hippocampus 2016, 26, 261–271. [Google Scholar] [CrossRef]
- Nithianantharajah, J.; Hannan, A.J. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat. Rev. Neurosci. 2006, 7, 697–709. [Google Scholar] [CrossRef]
- Markham, J.A.; Greenough, W.T. Experience-driven brain plasticity: Beyond the synapse. Neuron Glia Biol. 2004, 1, 351–363. [Google Scholar] [CrossRef]
- Nithianantharajah, J.; Hannan, A.J. The neurobiology of brain and cognitive reserve: Mental and physical activity as modulators of brain disorders. Prog. Neurobiol. 2009, 89, 369–382. [Google Scholar] [CrossRef]
- Zhuang, Z.Q.; Zhang, Z.Z.; Zhang, Y.M.; Ge, H.H.; Sun, S.Y.; Zhang, P.; Chen, G.H. A Long-Term Enriched Environment Ameliorates the Accelerated Age-Related Memory Impairment Induced by Gestational Administration of Lipopolysaccharide: Role of Plastic Mitochondrial Quality Control. Front. Cell. Neurosci. 2020, 14, 559182. [Google Scholar] [CrossRef] [PubMed]
- Diniz, D.G.; de Oliveira, M.A.; de Lima, C.M.; Foro, C.A.; Sosthenes, M.C.; Bento-Torres, J.; da Costa Vasconcelos, P.F.; Anthony, D.C.; Diniz, C.W. Age, environment, object recognition and morphological diversity of GFAP-immunolabeled astrocytes. Behav. Brain Funct. 2016, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Ederer, M.L.; Gunther, M.; Best, L.; Lindner, J.; Kaleta, C.; Witte, O.W.; Simon, R.; Frahm, C. Voluntary Wheel Running in Old C57BL/6 Mice Reduces Age-Related Inflammation in the Colon but Not in the Brain. Cells 2022, 11, 566. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Su, S.; Cai, W.; Cao, J.; Miao, X.; Zang, W.; Gao, S.; Xu, Y.; Yang, J.; Tao, Y.X.; et al. Differentially Expressed Genes in the Brain of Aging Mice With Cognitive Alteration and Depression- and Anxiety-Like Behaviors. Front. Cell Dev. Biol. 2020, 8, 814. [Google Scholar] [CrossRef] [PubMed]
- Diniz, D.G.; Foro, C.A.; Rego, C.M.; Gloria, D.A.; de Oliveira, F.R.; Paes, J.M.; de Sousa, A.A.; Tokuhashi, T.P.; Trindade, L.S.; Turiel, M.C.; et al. Environmental impoverishment and aging alter object recognition, spatial learning, and dentate gyrus astrocytes. Eur. J. Neurosci. 2010, 32, 509–519. [Google Scholar] [CrossRef]
- Fuchs, F.; Cosquer, B.; Penazzi, L.; Mathis, C.; Kelche, C.; Majchrzak, M.; Barbelivien, A. Exposure to an enriched environment up to middle age allows preservation of spatial memory capabilities in old age. Behav. Brain Res. 2016, 299, 1–5. [Google Scholar] [CrossRef]
- Wang, X.; Meng, Z.X.; Chen, Y.Z.; Li, Y.P.; Zhou, H.Y.; Yang, M.; Zhao, T.T.; Gong, Y.L.; Wu, Y.; Liu, T. Enriched environment enhances histone acetylation of NMDA receptor in the hippocampus and improves cognitive dysfunction in aged mice. Neural Regen. Res. 2020, 15, 2327–2334. [Google Scholar] [CrossRef]
- Boehme, M.; Guenther, M.; Stahr, A.; Liebmann, M.; Jaenisch, N.; Witte, O.W.; Frahm, C. Impact of indomethacin on neuroinflammation and hippocampal neurogenesis in aged mice. Neurosci. Lett. 2014, 572, 7–12. [Google Scholar] [CrossRef]
- Leal, S.L.; Yassa, M.A. Neurocognitive Aging and the Hippocampus across Species. Trends Neurosci. 2015, 38, 800–812. [Google Scholar] [CrossRef]
- Barth, E.; Srivastava, A.; Stojiljkovic, M.; Frahm, C.; Axer, H.; Witte, O.W.; Marz, M. Conserved aging-related signatures of senescence and inflammation in different tissues and species. Aging 2019, 11, 8556–8572. [Google Scholar] [CrossRef]
- Frahm, C.; Srivastava, A.; Schmidt, S.; Mueller, J.; Groth, M.; Guenther, M.; Ji, Y.; Priebe, S.; Platzer, M.; Witte, O.W. Transcriptional profiling reveals protective mechanisms in brains of long-lived mice. Neurobiol. Aging 2017, 52, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Aramillo Irizar, P.; Schauble, S.; Esser, D.; Groth, M.; Frahm, C.; Priebe, S.; Baumgart, M.; Hartmann, N.; Marthandan, S.; Menzel, U.; et al. Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly. Nat. Commun. 2018, 9, 327. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Q.; Kong, Y.; Zhou, F.; Li, J.; Li, W.; Wang, K.; Wu, T.; Guan, Y.; Xie, J.; et al. Biosystems Study of the Molecular Networks Underlying Hippocampal Aging Progression and Anti-aging Treatment in Mice. Front. Aging Neurosci. 2017, 9, 393. [Google Scholar] [CrossRef]
- Rowe, W.B.; Blalock, E.M.; Chen, K.C.; Kadish, I.; Wang, D.; Barrett, J.E.; Thibault, O.; Porter, N.M.; Rose, G.M.; Landfield, P.W. Hippocampal expression analyses reveal selective association of immediate-early, neuroenergetic, and myelinogenic pathways with cognitive impairment in aged rats. J. Neurosci. 2007, 27, 3098–3110. [Google Scholar] [CrossRef] [PubMed]
- Norden, D.M.; Godbout, J.P. Review: Microglia of the aged brain: Primed to be activated and resistant to regulation. Neuropathol. Appl. Neurobiol. 2013, 39, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Ximerakis, M.; Lipnick, S.L.; Innes, B.T.; Simmons, S.K.; Adiconis, X.; Dionne, D.; Mayweather, B.A.; Nguyen, L.; Niziolek, Z.; Ozek, C.; et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 2019, 22, 1696–1708. [Google Scholar] [CrossRef]
- Blalock, E.M.; Chen, K.C.; Sharrow, K.; Herman, J.P.; Porter, N.M.; Foster, T.C.; Landfield, P.W. Gene microarrays in hippocampal aging: Statistical profiling identifies novel processes correlated with cognitive impairment. J. Neurosci. 2003, 23, 3807–3819. [Google Scholar] [CrossRef]
- Verbitsky, M.; Yonan, A.L.; Malleret, G.; Kandel, E.R.; Gilliam, T.C.; Pavlidis, P. Altered hippocampal transcript profile accompanies an age-related spatial memory deficit in mice. Learn. Mem. 2004, 11, 253–260. [Google Scholar] [CrossRef][Green Version]
- Rampon, C.; Jiang, C.H.; Dong, H.; Tang, Y.P.; Lockhart, D.J.; Schultz, P.G.; Tsien, J.Z.; Hu, Y. Effects of environmental enrichment on gene expression in the brain. Proc. Natl. Acad. Sci. USA 2000, 97, 12880–12884. [Google Scholar] [CrossRef]
- Gregoire, C.A.; Tobin, S.; Goldenstein, B.L.; Samarut, E.; Leclerc, A.; Aumont, A.; Drapeau, P.; Fulton, S.; Fernandes, K.J.L. RNA-Sequencing Reveals Unique Transcriptional Signatures of Running and Running-Independent Environmental Enrichment in the Adult Mouse Dentate Gyrus. Front. Mol. Neurosci. 2018, 11, 126. [Google Scholar] [CrossRef]
- Singhal, G.; Morgan, J.; Jawahar, M.C.; Corrigan, F.; Jaehne, E.J.; Toben, C.; Breen, J.; Pederson, S.M.; Hannan, A.J.; Baune, B.T. Short-term environmental enrichment, and not physical exercise, alleviate cognitive decline and anxiety from middle age onwards without affecting hippocampal gene expression. Cogn. Affect. Behav. Neurosci. 2019, 19, 1143–1169. [Google Scholar] [CrossRef] [PubMed]
- Singhal, G.; Morgan, J.; Jawahar, M.C.; Corrigan, F.; Jaehne, E.J.; Toben, C.; Breen, J.; Pederson, S.M.; Hannan, A.J.; Baune, B.T. The effects of short-term and long-term environmental enrichment on locomotion, mood-like behavior, cognition and hippocampal gene expression. Behav. Brain Res. 2019, 368, 111917. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, M.F.; Varayoud, J.; Moreno-Piovano, G.S.; Luque, E.H.; Ramos, J.G. Environmental enrichment attenuates the age-related decline in the mRNA expression of steroidogenic enzymes and reduces the methylation state of the steroid 5alpha-reductase type 1 gene in the rat hippocampus. Mol. Cell. Endocrinol. 2015, 412, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Neidl, R.; Schneider, A.; Bousiges, O.; Majchrzak, M.; Barbelivien, A.; de Vasconcelos, A.P.; Dorgans, K.; Doussau, F.; Loeffler, J.P.; Cassel, J.C.; et al. Late-Life Environmental Enrichment Induces Acetylation Events and Nuclear Factor kappaB-Dependent Regulations in the Hippocampus of Aged Rats Showing Improved Plasticity and Learning. J. Neurosci. 2016, 36, 4351–4361. [Google Scholar] [CrossRef] [PubMed]
- Singhal, G.; Morgan, J.; Jawahar, M.C.; Corrigan, F.; Jaehne, E.J.; Toben, C.; Breen, J.; Pederson, S.M.; Manavis, J.; Hannan, A.J.; et al. Effects of aging on the motor, cognitive and affective behaviors, neuroimmune responses and hippocampal gene expression. Behav. Brain Res. 2020, 383, 112501. [Google Scholar] [CrossRef] [PubMed]
- Park, C.S.; Valomon, A.; Welzl, H. Integrative Transcriptome Profiling of Cognitive Aging and Its Preservation through Ser/Thr Protein Phosphatase Regulation. PLoS ONE 2015, 10, e0130891. [Google Scholar] [CrossRef] [PubMed]
- Zocher, S.; Overall, R.W.; Lesche, M.; Dahl, A.; Kempermann, G. Environmental enrichment preserves a young DNA methylation landscape in the aged mouse hippocampus. Nat. Commun. 2021, 12, 3892. [Google Scholar] [CrossRef]
- Morse, S.J.; Butler, A.A.; Davis, R.L.; Soller, I.J.; Lubin, F.D. Environmental enrichment reverses histone methylation changes in the aged hippocampus and restores age-related memory deficits. Biology 2015, 4, 298–313. [Google Scholar] [CrossRef]
- Sharp, P.E.; Barnes, C.A.; McNaughton, B.L. Effects of aging on environmental modulation of hippocampal evoked responses. Behav. Neurosci. 1987, 101, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Rampon, C.; Tang, Y.P.; Shrom, D.; Jin, J.; Kyin, M.; Sopher, B.; Miller, M.W.; Ware, C.B.; Martin, G.M.; et al. Deficient neurogenesis in forebrain-specific presenilin-1 knockout mice is associated with reduced clearance of hippocampal memory traces. Neuron 2001, 32, 911–926. [Google Scholar] [CrossRef]
- Kumar, A.; Rani, A.; Tchigranova, O.; Lee, W.H.; Foster, T.C. Influence of late-life exposure to environmental enrichment or exercise on hippocampal function and CA1 senescent physiology. Neurobiol. Aging 2012, 33, 828.e1-17. [Google Scholar] [CrossRef] [PubMed]
- Stein, L.R.; O’Dell, K.A.; Funatsu, M.; Zorumski, C.F.; Izumi, Y. Short-term environmental enrichment enhances synaptic plasticity in hippocampal slices from aged rats. Neuroscience 2016, 329, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Pastalkova, E.; Serrano, P.; Pinkhasova, D.; Wallace, E.; Fenton, A.A.; Sacktor, T.C. Storage of spatial information by the maintenance mechanism of LTP. Science 2006, 313, 1141–1144. [Google Scholar] [CrossRef] [PubMed]
- Whitlock, J.R.; Heynen, A.J.; Shuler, M.G.; Bear, M.F. Learning induces long-term potentiation in the hippocampus. Science 2006, 313, 1093–1097. [Google Scholar] [CrossRef]
- Bilkey, D.K.; Cheyne, K.R.; Eckert, M.J.; Lu, X.; Chowdhury, S.; Worley, P.F.; Crandall, J.E.; Abraham, W.C. Exposure to complex environments results in more sparse representations of space in the hippocampus. Hippocampus 2017, 27, 1178–1191. [Google Scholar] [CrossRef]
- Ohline, S.M.; Abraham, W.C. Environmental enrichment effects on synaptic and cellular physiology of hippocampal neurons. Neuropharmacology 2019, 145, 3–12. [Google Scholar] [CrossRef]
- Schanzenbacher, C.T.; Sambandan, S.; Langer, J.D.; Schuman, E.M. Nascent Proteome Remodeling following Homeostatic Scaling at Hippocampal Synapses. Neuron 2016, 92, 358–371. [Google Scholar] [CrossRef]
- Desch, K.; Langer, J.D.; Schuman, E.M. Dynamic bi-directional phosphorylation events associated with the reciprocal regulation of synapses during homeostatic up- and down-scaling. Cell Rep. 2021, 36, 109583. [Google Scholar] [CrossRef]
- Kohman, R.A.; Rodriguez-Zas, S.L.; Southey, B.R.; Kelley, K.W.; Dantzer, R.; Rhodes, J.S. Voluntary wheel running reverses age-induced changes in hippocampal gene expression. PLoS ONE 2011, 6, e22654. [Google Scholar] [CrossRef]
- Foster, T.C. Role of estrogen receptor alpha and beta expression and signaling on cognitive function during aging. Hippocampus 2012, 22, 656–669. [Google Scholar] [CrossRef]
- Rettberg, J.R.; Yao, J.; Brinton, R.D. Estrogen: A master regulator of bioenergetic systems in the brain and body. Front. Neuroendocrinol. 2014, 35, 8–30. [Google Scholar] [CrossRef] [PubMed]
- Brann, D.W.; Lu, Y.; Wang, J.; Zhang, Q.; Thakkar, R.; Sareddy, G.R.; Pratap, U.P.; Tekmal, R.R.; Vadlamudi, R.K. Brain-derived estrogen and neural function. Neurosci. Biobehav. Rev. 2022, 132, 793–817. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, S.; Kalita, K.; Jaworski, J.; Mioduszewska, B.; Savonenko, A.; Markowska, A.; Merchenthaler, I.; Kaczmarek, L. Increased estrogen receptor beta expression correlates with decreased spine formation in the rat hippocampus. Hippocampus 2006, 16, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.K.; Hsueh, Y.P. Cortactin-binding protein 2 modulates the mobility of cortactin and regulates dendritic spine formation and maintenance. J. Neurosci. 2012, 32, 1043–1055. [Google Scholar] [CrossRef]
- Guo, J.F.; Zhang, L.; Li, K.; Mei, J.P.; Xue, J.; Chen, J.; Tang, X.; Shen, L.; Jiang, H.; Chen, C.; et al. Coding mutations in NUS1 contribute to Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2018, 115, 11567–11572. [Google Scholar] [CrossRef]
- Shih, P.Y.; Hsieh, B.Y.; Lin, M.H.; Huang, T.N.; Tsai, C.Y.; Pong, W.L.; Lee, S.P.; Hsueh, Y.P. CTTNBP2 Controls Synaptic Expression of Zinc-Related Autism-Associated Proteins and Regulates Synapse Formation and Autism-like Behaviors. Cell Rep. 2020, 31, 107700. [Google Scholar] [CrossRef]
- Li, J.Z.; Hao, X.H.; Wu, H.P.; Li, M.; Liu, X.M.; Wu, Z.B. An enriched environment delays the progression from mild cognitive impairment to Alzheimer’s disease in senescence-accelerated mouse prone 8 mice. Exp. Ther. Med. 2021, 22, 1320. [Google Scholar] [CrossRef]
- Sanz-Rodriguez, M.; Gruart, A.; Escudero-Ramirez, J.; de Castro, F.; Delgado-Garcia, J.M.; Wandosell, F.; Cubelos, B. R-Ras1 and R-Ras2 Are Essential for Oligodendrocyte Differentiation and Survival for Correct Myelination in the Central Nervous System. J. Neurosci. 2018, 38, 5096–5110. [Google Scholar] [CrossRef]
- Alcover-Sanchez, B.; Garcia-Martin, G.; Escudero-Ramirez, J.; Gonzalez-Riano, C.; Lorenzo, P.; Gimenez-Cassina, A.; Formentini, L.; de la Villa-Polo, P.; Pereira, M.P.; Wandosell, F.; et al. Absence of R-Ras1 and R-Ras2 causes mitochondrial alterations that trigger axonal degeneration in a hypomyelinating disease model. Glia 2021, 69, 619–637. [Google Scholar] [CrossRef]
- Chen, M.; Lyu, G.; Han, M.; Nie, H.; Shen, T.; Chen, W.; Niu, Y.; Song, Y.; Li, X.; Li, H.; et al. 3′ UTR lengthening as a novel mechanism in regulating cellular senescence. Genome Res. 2018, 28, 285–294. [Google Scholar] [CrossRef]
- Schmeer, C.; Kretz, A.; Wengerodt, D.; Stojiljkovic, M.; Witte, O.W. Dissecting Aging and Senescence-Current Concepts and Open Lessons. Cells 2019, 8, 1446. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, Y.; Du, G.; Qin, X.; Gao, L. Bioinformatic Analysis Reveals Key Genes and Pathways in Aging Brain of Senescence-accelerated Mouse P8 (SAMP8). CNS Neurol. Disord. Drug Targets 2018, 17, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, A.; Branca, C.; Talboom, J.S.; Shaw, D.M.; Turner, D.; Ma, L.; Messina, A.; Huang, Z.; Wu, J.; Oddo, S. Reducing Ribosomal Protein S6 Kinase 1 Expression Improves Spatial Memory and Synaptic Plasticity in a Mouse Model of Alzheimer’s Disease. J. Neurosci. 2015, 35, 14042–14056. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Selman, C.; Tullet, J.M.; Wieser, D.; Irvine, E.; Lingard, S.J.; Choudhury, A.I.; Claret, M.; Al-Qassab, H.; Carmignac, D.; Ramadani, F.; et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 2009, 326, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Um, S.H.; Frigerio, F.; Watanabe, M.; Picard, F.; Joaquin, M.; Sticker, M.; Fumagalli, S.; Allegrini, P.R.; Kozma, S.C.; Auwerx, J.; et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 2004, 431, 200–205. [Google Scholar] [CrossRef]
- Spinelli, M.; Fusco, S.; Grassi, C. Brain Insulin Resistance and Hippocampal Plasticity: Mechanisms and Biomarkers of Cognitive Decline. Front. Neurosci. 2019, 13, 788. [Google Scholar] [CrossRef]
- Kellar, D.; Craft, S. Brain insulin resistance in Alzheimer’s disease and related disorders: Mechanisms and therapeutic approaches. Lancet Neurol. 2020, 19, 758–766. [Google Scholar] [CrossRef]
- Arnold, S.E.; Arvanitakis, Z.; Macauley-Rambach, S.L.; Koenig, A.M.; Wang, H.Y.; Ahima, R.S.; Craft, S.; Gandy, S.; Buettner, C.; Stoeckel, L.E.; et al. Brain insulin resistance in type 2 diabetes and Alzheimer disease: Concepts and conundrums. Nat. Rev. Neurol. 2018, 14, 168–181. [Google Scholar] [CrossRef]
- Plasschaert, R.N.; Bartolomei, M.S. Tissue-specific regulation and function of Grb10 during growth and neuronal commitment. Proc. Natl. Acad. Sci. USA 2015, 112, 6841–6847. [Google Scholar] [CrossRef]
- Ma, L.; Wei, Q.; Deng, H.; Zhang, Q.; Li, G.; Tang, N.; Xie, J.; Chen, Y. Growth factor receptor-bound protein 10-mediated negative regulation of the insulin-like growth factor-1 receptor-activated signalling pathway results in cognitive disorder in diabetic rats. J. Neuroendocrinol. 2013, 25, 626–634. [Google Scholar] [CrossRef]
- Cao, L.; Wang, Z.; Wan, W. Suppressor of Cytokine Signaling 3: Emerging Role Linking Central Insulin Resistance and Alzheimer’s Disease. Front. Neurosci. 2018, 12, 417. [Google Scholar] [CrossRef]
- Smith, P.D.; Sun, F.; Park, K.K.; Cai, B.; Wang, C.; Kuwako, K.; Martinez-Carrasco, I.; Connolly, L.; He, Z. SOCS3 deletion promotes optic nerve regeneration in vivo. Neuron 2009, 64, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Williams, P.R.; He, Z. SOCS3: A common target for neuronal protection and axon regeneration after spinal cord injury. Exp. Neurol. 2015, 263, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Park, K.K.; Belin, S.; Wang, D.; Lu, T.; Chen, G.; Zhang, K.; Yeung, C.; Feng, G.; Yankner, B.A.; et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature 2011, 480, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Park, K.K.; Liu, K.; Hu, Y.; Smith, P.D.; Wang, C.; Cai, B.; Xu, B.; Connolly, L.; Kramvis, I.; Sahin, M.; et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 2008, 322, 963–966. [Google Scholar] [CrossRef] [PubMed]
- Higami, Y.; Shimokawa, I. Apoptosis in the aging process. Cell Tissue Res. 2000, 301, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Isaev, N.K.; Genrikhs, E.E.; Oborina, M.V.; Stelmashook, E.V. Accelerated aging and aging process in the brain. Rev. Neurosci. 2018, 29, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Young, D.; Lawlor, P.A.; Leone, P.; Dragunow, M.; During, M.J. Environmental enrichment inhibits spontaneous apoptosis, prevents seizures and is neuroprotective. Nat. Med. 1999, 5, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Birch, A.M.; Kelly, A.M. Lifelong environmental enrichment in the absence of exercise protects the brain from age-related cognitive decline. Neuropharmacology 2019, 145, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Mangan, D.F.; Robertson, B.; Wahl, S.M. IL-4 enhances programmed cell death (apoptosis) in stimulated human monocytes. J. Immunol. 1992, 148, 1812–1816. [Google Scholar]
- Bailey, D.P.; Kashyap, M.; Mirmonsef, P.; Bouton, L.A.; Domen, J.; Zhu, J.; Dessypris, E.N.; Ryan, J.J. Interleukin-4 elicits apoptosis of developing mast cells via a Stat6-dependent mitochondrial pathway. Exp. Hematol. 2004, 32, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Aoudjehane, L.; Podevin, P.; Scatton, O.; Jaffray, P.; Dusanter-Fourt, I.; Feldmann, G.; Massault, P.P.; Grira, L.; Bringuier, A.; Dousset, B.; et al. Interleukin-4 induces human hepatocyte apoptosis through a Fas-independent pathway. FASEB J. 2007, 21, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.W.; Kuhn, H.; Hennig, B.; Toborek, M. IL-4 induces apoptosis of endothelial cells through the caspase-3-dependent pathway. FEBS Lett. 2000, 485, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Orian, J.M.; Keating, P.; Downs, L.L.; Hale, M.W.; Jiang, X.; Pham, H.; LaFlamme, A.C. Deletion of IL-4Ralpha in the BALB/c mouse is associated with altered lesion topography and susceptibility to experimental autoimmune encephalomyelitis. Autoimmunity 2015, 48, 208–221. [Google Scholar] [CrossRef]
- Wightman, D.P.; Jansen, I.E.; Savage, J.E.; Shadrin, A.A.; Bahrami, S.; Holland, D.; Rongve, A.; Borte, S.; Winsvold, B.S.; Drange, O.K.; et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 2021, 53, 1276–1282. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Long, J.; Pan, G.; He, T.; Anichtchik, O.; Belshaw, R.; Albani, D.; Edison, P.; Green, E.K.; et al. Systematic Analysis and Biomarker Study for Alzheimer’s Disease. Sci. Rep. 2018, 8, 17394. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, H.; Xie, N.; Liu, D.; Jiang, Y.; Liu, Z.; Ye, D.; Liu, S.; Chen, X.; Li, C.; et al. Bcl-3 promotes TNF-induced hepatocyte apoptosis by regulating the deubiquitination of RIP1. Cell Death Differ. 2022, 29, 1176–1186. [Google Scholar] [CrossRef]
- Palmer, S.; Chen, Y.H. Bcl-3, a multifaceted modulator of NF-kappaB-mediated gene transcription. Immunol. Res. 2008, 42, 210–218. [Google Scholar] [CrossRef]
- Mattson, M.P.; Meffert, M.K. Roles for NF-kappaB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006, 13, 852–860. [Google Scholar] [CrossRef]
- Tao, T.; Shi, H.; Guan, Y.; Huang, D.; Chen, Y.; Lane, D.P.; Chen, J.; Peng, J. Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation. Cell Res. 2013, 23, 620–634. [Google Scholar] [CrossRef]
- Tao, T.; Sondalle, S.B.; Shi, H.; Zhu, S.; Perez-Atayde, A.R.; Peng, J.; Baserga, S.J.; Look, A.T. The pre-rRNA processing factor DEF is rate limiting for the pathogenesis of MYCN-driven neuroblastoma. Oncogene 2017, 36, 3852–3867. [Google Scholar] [CrossRef] [PubMed]
- Checler, F.; Alves da Costa, C. p53 in neurodegenerative diseases and brain cancers. Pharmacol. Ther. 2014, 142, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, C.K.; Zeng, A.; Alegre, D.; Hu, D.; Gotting, K.; Ortega Granillo, A.; Wang, Y.; Robb, S.; Schnittker, R.; et al. Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science 2020, 369, eaaz3090. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmidt, S.; Haase, M.; Best, L.; Groth, M.; Lindner, J.; Witte, O.W.; Kaleta, C.; Frahm, C. Restoring Age-Related Cognitive Decline through Environmental Enrichment: A Transcriptomic Approach. Cells 2022, 11, 3864. https://doi.org/10.3390/cells11233864
Schmidt S, Haase M, Best L, Groth M, Lindner J, Witte OW, Kaleta C, Frahm C. Restoring Age-Related Cognitive Decline through Environmental Enrichment: A Transcriptomic Approach. Cells. 2022; 11(23):3864. https://doi.org/10.3390/cells11233864
Chicago/Turabian StyleSchmidt, Silvio, Madlen Haase, Lena Best, Marco Groth, Julia Lindner, Otto W. Witte, Christoph Kaleta, and Christiane Frahm. 2022. "Restoring Age-Related Cognitive Decline through Environmental Enrichment: A Transcriptomic Approach" Cells 11, no. 23: 3864. https://doi.org/10.3390/cells11233864
APA StyleSchmidt, S., Haase, M., Best, L., Groth, M., Lindner, J., Witte, O. W., Kaleta, C., & Frahm, C. (2022). Restoring Age-Related Cognitive Decline through Environmental Enrichment: A Transcriptomic Approach. Cells, 11(23), 3864. https://doi.org/10.3390/cells11233864




