1. Introduction
Anaerobic clostridial bacteria are causative for a number of severe human infectious diseases. A number of their proteinaceous toxins are crucial virulence factors of these pathogens. In many cases the cytoskeleton of host cells is the preferred target of these virulence factors. An important group are the toxins CDT from
Clostridioides difficile, C2I from
Clostridioides botulinum and Iota from
Clostridioides perfringens [
1,
2,
3]. These toxins are binary in structure and consist of an enzyme component with ADP-ribosyltransferase (ART) activity and a separate membrane binding component, which is responsible for cellular toxin binding and uptake [
4]. The membrane binding component attaches to a specific receptor, oligomerizes, and binds the ART containing component. Subsequently, this complex containing both toxin components is endocytosed. After acidification of the endosome the membrane binding component forms a pore allowing release of the ART containing component through the endosomal membrane into the cytosol (for a review see [
5]). There the ART containing toxins ADP-ribosylate actin at Arg177 leading to inhibition of its polymerizability and the partial disassembly of intracellular actin filaments ([
3]; reviewed in [
1,
2]).
Recently, hypervirulent and multidrug resistant strains of
C. difficile were recognized, which during antibiotic treatment can multiply and colonize the colon causing severe diarrhea and pseudomembranous colitis accompanied by high morbidity and mortality [
5,
6,
7]. These
C. difficile strains are a main cause of hospitalization associated infections [
7,
8]. In addition to the prototypical Rho/Ras-glycosylating toxins TcdA and TcdB they produce the binary
C. difficile toxin CDT composed of the ART containing CDTa and membrane binding component CDTb [
6,
8,
9,
10,
11].
However, the precise pathogenic mechanisms of clostridial ADP-ribosylating toxins are still not completely understood. Therefore, we searched for additional substrate proteins of the bacterial ADP-ribosyltransferases. To this aim we analyzed ART-activity containing toxins including the
C. difficile toxin CDT, C2I toxin from
C. botulinum, and Iota toxin from
C. perfringens for their ability to ADP-ribosylate the actin-related proteins of the Arp2/3 complex. The ubiquitous Arp2/3 complex consists of seven subunits, resides underneath the plasma membrane and is responsible for the stabilization of the cortical F-actin beneath the plasma membrane and its attachment to cell junctions. Furthermore, a number of stimulating signaling pathways converge on Arp2/3 complex, which subsequently regulates the formation of branched actin-filament networks within lamellipodia of migratory eukaryotic cells [
12,
13].
Our biochemical data showed that the actin-related protein 2 (Arp2) of the Arp2/3 complex is ADP-ribosylated by bacterial ADP-ribosyltransferases, in particular by the binary ART-activity containing CDT toxin of C. difficile toxin. In order to bridge the gap between protein modification by pathogens and possible clinical consequences, we subsequently infected human colon derived Caco2 cells and excised mouse colon pieces with this toxin to analyze its effects under conditions more closely resembling an in vivo situation. The results obtained from these experiments confirmed the ADP-ribosylation of Arp2 and at the same time that of actin at an apparently equal amount. Obviously, their modifications resulted in dramatic alterations in cell and tissue morphology suggesting that the ART-activity containing CDT toxin of C. difficile might suffice to provoke the pathological changes leading to the typical severe colitis.
2. Materials and Methods
2.1. Materials
Fetal calf serum (FCS) and media were obtained from Gibco (Deisenhofen/Germany). The monoclonal anti-actin antibody (clone AC74) was purchased from Sigma-Aldrich (Poole, Dorset, UK) and FITC-labelled anti-rabbit IgG from Amersham (Amersham Life Science/UK). TRITC-phalloidin was obtained from Molecular Probes (Eugene, OR, USA). The fluorescent etheno-NAD was obtained from Sigma-Aldrich (Munich, Germany)
2.2. Protein Expression and Analysis
Rabbit skeletal muscle actin was prepared from dried acetone powder obtained from fresh rabbit psoas muscle as described [
14]. G-actin was stored in G-buffer (5 mM HEPES-OH, pH 7.4, 0.1 mM CaCl
2, 0.5 mM NaN
3, and 0.2 mM ATP, pH 7.4). The Arp2/3 complex was purified from
Acanthamoeba castellani and its mammalian variant from pig brain as described [
15,
16]. The activating C-terminal VCA domain of WASp (containing Verprolin-like, Central and Acidic regions) from N-WASP (comprising residues 392 to 501) was expressed in
Escherichia coli and purified according to [
15]; both were kindly supplied by Prof. D.L. Barber and Dr. A. Schoenichen (University of California, San Francisco, CA, USA). The clostridial toxins (CDT, C2I and Iota-a toxin from
Clostridioides difficile, botulinum and perfrigens, respectively) with ADP-ribosyl-transferase (ART) activities were prepared recombinantly as described previously [
17,
18]. The components a and b of the CDT toxin from
Clostridioides difficile (CDTa containing the ART and CDTb the membrane translocator activity) were expressed recombinantly in
E. coli and due to their N-terminal His-tag purified by Ni-NTA affinity binding [
17]. The TccC3 toxin from
Photorhabdus luminescense was prepared as detailed [
19] and the FH2 segments of the formins mDia1 (comprising residues 826–1163) and mDia3 (comprising residues 701–1061) were expressed in
E. coli and purified as described [
20] and kindly supplied by Prof. A. Wittinghofer (Max-Planck-Institute of Molecular Physiology, Dortmund, Germany).
2.3. Cell Culture and Intoxication and Immunostaining
The human colon carcinoma cell line Caco2 was obtained from CLS (Cell Lines Service, Germany, independent cell repository). The cells are available from ATCC (ATCC number: HTB-37; human origin with ethnicity Caucasian. Donor age 72 years; male and from colon). These epithelial-like cells were grown and maintained at 37 °C and 5% CO
2 in plastic flasks in DMEM (Dulbecco modified Eagle medium) supplemented with 10% FCS. Since Caco2 cells express the CDTb receptor lipolysis-stimulated lipoprotein receptor (LSR) intoxication of the CDTa ART was achieved by simultaneous exposure of Caco2 cells grown on sterile glass coverslips to 500 ng/mL CDTb and 200 ng/mL CDTa under cell culture conditions as detailed previously [
18]. After increasing incubation periods, the cells were fixed by addition of 4% paraformaldehyde for 20 min.
Colon pieces were taken from male wild type mice of the C57BL/6 genetic background. The mice were maintained in the local animal house of the Ruhr-University observing the regulations of the German Animal Protection Law and sacrificed by cervical dislocation following the recommendations of the Animal Care and Use Committee of the Ruhr-University, Bochum, in compliance with the German guidelines for animal care and procedures. The colon pieces were exposed to CDTa and CDTb, fixed by 4% paraformaldehyde and immunostained as further detailed below.
For immunostaining the cells and the colon pieces were permeabilized with 0.2% Triton X-100 in PBS for 5–10 min, washed three times in PBS, incubated with primary antibodies: mouse monoclonal anti-Arp2 (FMS96; Abcam, Cambridge, UK; kindly provided by Prof. S. Linder, Hamburg, Germany) or affinity-purified rabbit anti-ArpC1 (40 kDa subunit; Abcam) at dilutions of 1:50 or 1:100, respectively, at 4 °C overnight. Subsequently the slides were incubated with Alexa Fluor
®-568-labelled phalloidin and FITC-labelled secondary antibodies (Sigma-Aldrich, Munich, Germany) for 1h at RT as detailed [
6,
9]. The nuclei were visualized with Hoechst 33342 (Riedel-de-Haen; Schwerte, Germany). Finally, the coverslips were mounted with Dako Cytomatic fluorescent mounting medium. Immunocytochemical stainings were analyzed using a Zeiss LSM 800 confocal laser scanning microscope.
2.4. Analytical Procedures
Protein concentrations were determined by the colorimetric assay [
21]. SDS-PAGE was performed using 7.5% or 10% (
w/
v) polyacrylamide gels unless stated otherwise. Trichloroethanol at 0.5% was included in the separation gel to fluorescently visualize the separated protein bands before Western blotting.
For immune-precipitation of Arp2/3 complex, cell or tissue homogenates were prepared in RIPA-buffer (250 mM NaCl, 5 mM EDTA, 50 mM NaF, 1% NP-40, 0.1% NaN3, and 50 mM Tris-HCl, pH 7.4) and frozen at −20 °C until use. After two cycles of freezing and thawing, the samples were centrifuged at 14,000 rpm for 30 min. by an Eppendorf bench centrifuge and the supernatants were collected. Then, about 50 µg (in about 100 µL) of the respective supernatant was supplemented with 2 µL of the monoclonal anti-Arp2 antibody (FMS96; Abcam, Cambridge, UK) and incubated for 1 h at 25 °C. Subsequently, 20 µL of Protein A insolubilized to Sepharose 4B (Sigma, Munich, Germany) was added and after an incubation for 1 h at 25 °C centrifuged for 10 min at 14,000 rpm. The pellet was washed 5 times with 0.5 mL HEPES-buffer. The supernatants of each washing step were carefully removed and either discharged or stored for SDS-PAGE.
Western blots were performed as detailed previously [
22]. Autoradiography was used to identify ADP-ribosylated components of the Arp2/3 complex by supplementing NAD with
32P-labelled NAD (obtained from Perkin Elmer, Rodgau, Germany) following the procedure described in [
19,
23].
For measuring the polymerization kinetics by the pyrene-assay, G-actin was modified at Cys374 by pyrenyl-iodoacetamide (pyrene-actin) as described [
24]. Pyrene-labelled actin was added to 5% of the total actin concentration. Polymerization was initiated by addition of 2 mM MgCl
2 and 50 mM KCL and the increase of pyrene-actin fluorescence was determined using a Shimadzu RF-5001-PC spectrofluorometer at wavelength settings for excitation and emission of 365 nm and 385 nm, respectively.
2.5. Electron Microscopy
Protein samples were diluted to 0.1 mg/mL in HEPES-buffer, pH 7.4, with 2 mM MgCl
2 to trigger polymerization. For negative staining, 4 µL of each sample were adsorbed to freshly glow-discharged carbon-coated copper grids (200 mesh) for 45 sec. After washing with buffer, the grids were incubated for 45 s on a drop of 1% uranylacetate [
24]. Excess staining solution was removed with filter paper and then the grids were air-dried. Three different grids were prepared for each sample. Digital micrographs were then recorded with a Zeiss transmission electron microscope EM923 run at 120 kV fitted with a TemCamF416 camera (Tietz Video and Image Processing Systems, Gauting, Germany). The number of filaments longer than 0.1 micrometer and the number of branches were counted manually on the EM micrographs.
2.6. Determination of Filament Branching by Fluorescence Microscopy
Freshly purified skeletal muscle actin at 4.8 µM (0.2 mg/mL) was mixed with 50 nM native or ADP-ribosylated Arp2/3 complex from Acanthaemoeba castellani or pig brain. After addition of 0.1 mg/mL VCA peptide it was polymerized by 10 mM HEPES-HCl buffer, pH 7.4 (containing 50 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, and 0.2 mM ATP; buffer A). After dilution to 1 µM and the F-actin samples were stained with 2 µM TRITC-phalloidin (SIGMA, Munich, Germany) and further incubated for 60 min. Then, the TRITC-phalloidin stained actins were diluted to 10 nM in buffer A and 3 μL were placed on a glass-slide, mixed with 3 μL of DAKO fluorescence mounting medium (Agilent DAKO, Santa Clara, CA, USA/Glostrup, Denmark) and covered with a coverslip.
Subsequent fluorescence microscopy was performed using Zeiss AxioImager Z2m microscope equipped with a Zeiss LD LCI Plan-Apochromat 63×/1.2 multi-immersion objective and Zeiss Axiocam 503 color camera. Glycerol was used as immersion medium. Rhodamine fluorescence was excited using the LED 555 of the solid-state light source Colibri 7 and the quadruple bandpass filter set 90 HE, both from Zeiss. Images were recorded as gray-scale pictures with the microscope-associated ZEN software. The image size was 1936 × 1460 pixels, and the pixel size was 0.116 µm/pixel. Due to prolonged snap time and slight drift of the sample double images were collected which became visible only at higher magnifications but allowed to unequivocally differentiate between filament branching or crossing.
2.7. Analytical Tools
Recorded images from fluorescence microscopy were analyzed to obtain the number of the filaments and junction points, using ImageJ and the available plugin Ridge Detection (URL:
https://imageJ.net/plugins/ridge-detection, accessed on 5 August 2022). This plugin is based on the detection algorithm described by Steger [
25] for detecting ridges and lines. The parameter was selected to indicate all visible filaments above 1 micrometer and their junction points. The final setting was line width: 4.0, sigma: 1.65, lower threshold: 2.72, upper threshold: 7.31, minimum line length: 8.60 (=1µm), and maximum line length was not defined. For some images with weak signal the upper threshold was reduced to 4.5. The locations of the indicated junction points were checked manually by zooming in on each one on the original image to determine whether they were branches, crossings or simply two filaments approaching each other, and only branches were counted. The number of identified branches and the number of filaments obtained by Ridge Detection were transcribed to Microsoft Excel [
26], and we determined for each image the frequency of branching relative to the total number of filaments. Only the results of the analysis of images with maximally 300 filaments per image were used, as higher filament density increased the frequency of filament overlap rendering it difficult to distinguish between crossing and branching.
2.8. Mass-Spectrometric (MS) Analysis
For in-gel digestion the excised gel bands were destained with 30% acetonitrile, shrunk with 100% acetonitrile, and dried in a Vacuum Concentrator (Concentrator 5301, Eppendorf, Hamburg, Germany). Digestions with trypsin (trypsin gold, mass spectrometry grade; Promega, Walldorf, Germany) were performed overnight at 37 °C in 0.05 M NH4HCO3 (pH 8). About 0.1 µg of protease was used for one gel band. Peptides were extracted from the gel slices with 5% formic acid. All LC-MS/MS analyses were performed with the 1200 Agilent Chip-HPLC system, either coupled to a Q-TOF (Agilent 6520) or an ion trap (Agilent 6340) mass spectrometer. Peptides were separated on an HPLC-Chip with an analytical column of 75-µm i.d. and 150 mm length and a 40-nL trap column, both packed with Zorbax 300SB C-18 (5 µm particle size). Peptides were eluted with a linear acetonitrile gradient with 1%/min at a flow rate of 300 nL/min (starting with 3% acetonitrile). The Q-TOF was operated in the 2 Ghz extended dynamic range mode. MS/MS analyses were performed using data-dependent acquisition mode. After a MS scan (2 spectra/s), a maximum of three peptides were selected for MS/MS (2 spectra/s). Singly charged precursor ions were excluded from selection. Internal calibration was applied using one reference mass.
ETD analyses on the ion trap were performed using data-dependent acquisition mode. After a MS scan (standard enhanced mode), a maximum of three peptides were selected for ETD-MS/MS (standard enhanced mode). The automated gain control (ICC) for MS scans was set to 350,000. The maximum accumulation time was set to 300 ms. The following ETD parameters were used. ICC target: 400,000, reaction time: 100 ms, cut-off: 140, resonance excitation (Smart Decomp) was used for doubly charged peptides.
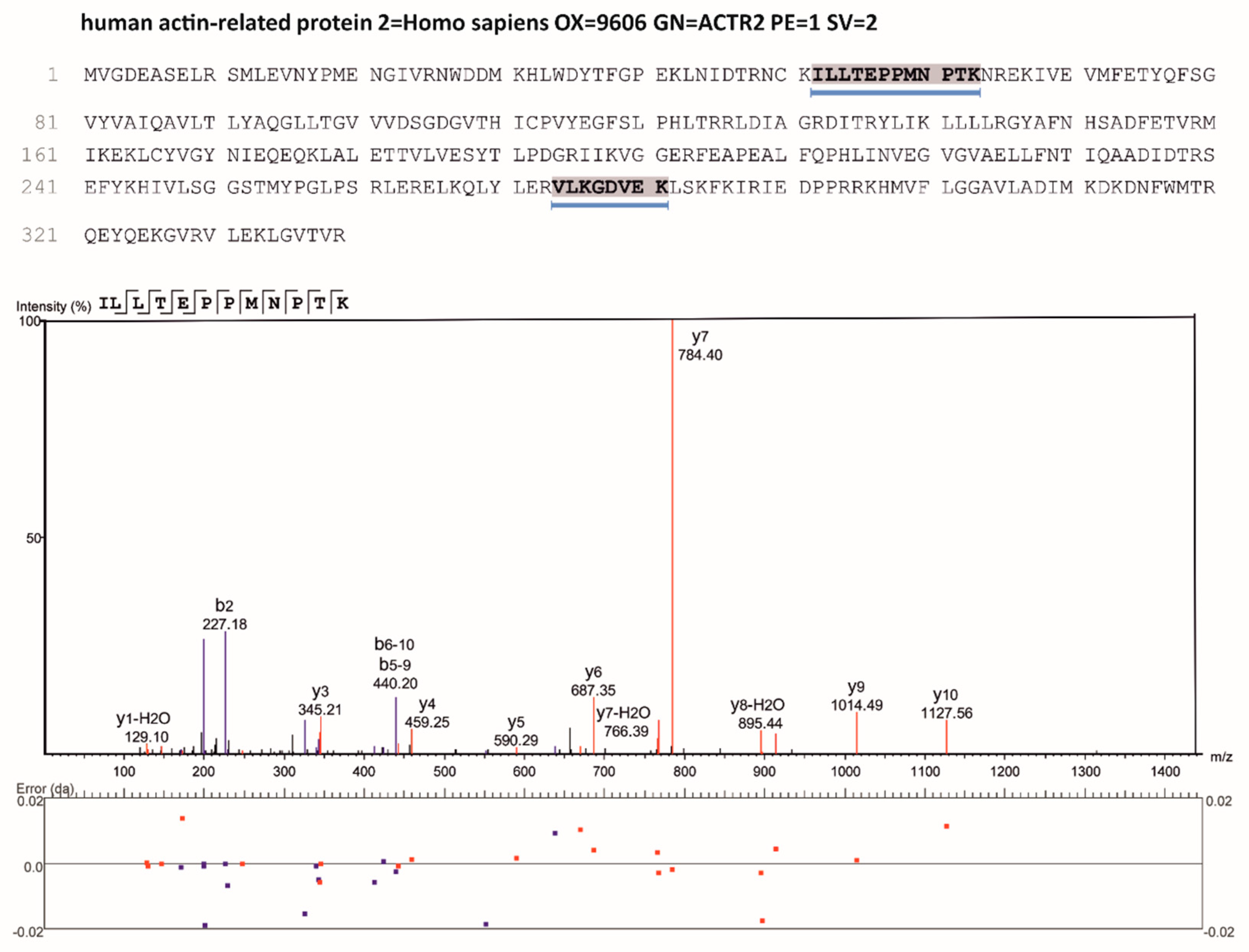
Mascot Distiller 2.3 was used for raw data processing and for generating peak lists, essentially with standard settings for the Agilent Q-Tof and ion trap. Mascot Server 2.3 was used for database searching with the following parameters: peptide mass tolerance: 20 ppm (Q-Tof) 1.1 Da (ion trap), MS/MS mass tolerance: 0.05 Da (Q-Tof), 0.3 Da (ion trap), enzyme: “trypsin” with 2 uncleaved sites allowed for trypsin, variable modifications: Carbamidomethyl), Gln-pyroGlu (N-term. Q), and oxidation (M), ADP-ribosylat (R). For protein and peptide identification a small custom database containing the protein sequence of Arp2 was used. All MS/MS spectra identified as ADP-ribosylated were validated by manual spectra interpretation. Three peptides were identified containing ADP-ribosylated arginine residues of which Arg179 was most prominently modified (
Figure 1; for details see also Figure 3).
2.9. Mass spectrometric Identification of Peptides Unique for Human Arp2
Fluorescent gel band was excised, destained with 30% acetonitrile in 0.1 M NH4HCO3 (pH 8), shrunk with 100% acetonitrile, and dried in a vacuum concentrator (Concentrator 5301, Eppendorf, Germany). Digest was performed with 0.1 µg trypsin overnight at 37 °C in 0.1 M NH4HCO3 (pH 8). After removing the supernatant, peptides were extracted from the gel slice with 5% formic acid, and extracted peptides were pooled with the supernatant.
NanoLC-MS/MS analysis was performed on an Orbitrap Fusion (Thermo Scientific, Waltham, MA, USA) equipped with a PicoView Ion Source (New Objective, Frederik, MD, USA) and coupled to an EASY-nLC 1000 (Thermo Scientific). Peptides were loaded on capillary columns (PicoFrit, 30 cm × 150 µm ID, New Objective) self-packed with ReproSil-Pur 120 C18-AQ, 1.9 µm (Dr. Maisch) and separated with a 30 min linear gradient from 3% to 30% acetonitrile and 0.1% formic acid and a flow rate of 500 nL/min.
Both MS and MS/MS scans were acquired in the Orbitrap analyzer with a resolution of 60,000 for MS scans and 7500 for MS/MS scans. HCD fragmentation with 35% normalized collision energy was applied. A Top Speed data-dependent MS/MS method with a fixed cycle time of 3 s was used. Dynamic exclusion was applied with a repeat count of 1 and an exclusion duration of 30 s; singly charged precursors were excluded from selection. Minimum signal threshold for precursor selection was set to 50,000. Predictive AGC was used with AGC a target value of 2 × 105 or MS scans and 5 × 104 for MS/MS scans. EASY-IC was used for internal calibration.
MS data was analyzed with PEAKS Studio X+ (Bioinformatics Solutions Inc., Waterloo, ON, Canada). Raw data refinement was performed with the following settings: Merge Options: no merge, Precursor Options: corrected, Charge Options: 1–6, Filter Options: no filter, Process: true, Default: true, Associate Chimera: yes. De novo sequencing and database searching were performed with a Parent Mass Error Tolerance of 10 ppm. Fragment Mass Error Tolerance was set to 0.02 Da, and Enzyme was set to trypsin with a maximum of 3 missed cleavages allowed. The following variable modifications have been used: Oxidation (M), pyro-Glu from Q (N-term Q), acetylation (protein N-terminal). A maximum of 6 variable PTMs were allowed per peptide. Database searching was performed against the human reference proteome (proteome ID UP000005640). Database search result was filtered to 1% PSM-FDR and protein –10lgP > 20.
4. Discussion
Previously it has been shown that the Arp2/3 complex is targeted by a number of bacterial pathogens. For instance,
Listeria monocytogenes bacteria express ActA—a bacterial surface protein that activates Arp2/3 complex and uses it for comet tail formation and their intra- and intercellular transport [
34]. Our data show for the first time that clostridial toxins with ART-activity also target the Arp2/3 complex by ADP-ribosylating the Arp2 subunit and some accessory proteins of the Arp2/3 complex. Indeed, Arp2 possesses about 50% sequence identity and high structural homology to actin [
28,
31]. Therefore, Arp2 is a well-suited second substrate for bacterial ARTs, since it also possesses an arginine at their target site (Arg179), whereas this residue is a histidine in Arp3 [
29].
This Arp2 modification affects the whole Arp2/3 complex and leads to inhibition of its stimulatory activity on actin polymerization and filament branching. Since it has been shown that during branch formation the first actin molecule of the growing daughter filament attaches to the barbed end area of Arp2, the ADP-ribosylation of Arp2 at Arg179 could block the attachment of this first actin subunit similar to the inhibition of actin subunit addition to the plus end of F-actin by the capping activity of ADP-ribosylated actin [
35].
When comparing the time dependence of ADP-ribosylation of Arp2 of the Arp2/3 complex with that of G-actin, it was obvious that G-actin is much faster modified that Arp2 (
Figure 1H). Within F-actin the Arg177 residue is located at the interstrand interface [
36,
37] and therefore is not accessible for the ARTs although they may bind to F-actin. Structural studies have shown that
C. perfrigens Iota-a toxin binds to a large target area of monomeric (G-) actin covering subdomains 1,3, and 4 [
38], of which subdomains 3 and 4 are not fully exposed in F-actin [
38]. Therefore, the ARTs will bind to F-actin with reduced affinity, but be unable to ADP-ribosylate Arg177 of actin, thus explaining the inhibitory effect of F-actin on ART´s ADP-ribosylation activity.
Though CDTa, C2I and Iota-a toxin modify only monomeric but not filamentous actin (see also
Figure 1A–C), this result does not necessarily indicate a negligible in vivo effect of Arp2 ADP-ribosylation. Only a small fraction of the total actin is in monomeric state in established cell lines, tissue epithelial or migrating white blood cells. Instead, most of the intracellular actin is polymerized to filamentous structures forming static or dynamic supramolecular organizations by interacting with other actin binding proteins, which may further inhibit ADP-ribosylation of actin as shown also for the TccC3 toxin [
23]. It has been estimated that the intracellular concentration of actin is about 40- to 100-fold higher than that of Arp2/3 complex [
28]. Our data suggest that actin and Arp2 were almost equally strongly modified in treated Caco2 cells and mouse colon. This observation will most probably be due to the fact that intracellularly only a small amount of the total actin is in monomeric state (5 to 10%), of which only a smaller fraction will not be complexed to G-actin binding proteins like thymosin ß4 or profilin, which might further reduce the accessibility of bacterial ARTs [
23]. Additionally, capping of the plus-ends of F-actin by ADP-ribosylated actin [
36] will reduce the rate of depolymerization of intracellular actin filaments, since then actin subunit dissociation occurs only slower from their minus ends.
In epithelial cells the F-actin is additionally stabilized by interactions with a large number of actin binding proteins. The Arp2/3 complex is of paramount importance for the connection of F-actin to for instance to adherence or tight junctions, which firmly attach neighboring cells to each other, or the stabilization of the cortical F-actin beneath the plasma membrane. Both structures appear to be affected by clostridial toxins possessing ART activity as shown by the effects of CDTa on the contacts of Caco2 cells and the dissolution of the epithelial layer of the mouse colon mucosa. The disruption of the cortical F-actin will reduce the resistance of the plasma membrane and lead to the observed outgrowth of microtubule bundles, a process that further increases pathogen attachment [
18]. Infection of Caco2 cells by CDTa showed a translocation of Arp2 immunofluorescence away from the plasma membrane into the cell interior and the formation of large aggregates leading to disruption of the cell–cell and possibly also of cell-substratum contacts. This process will be aggravated by the simultaneous ADP-ribosylation of actin leading to the disassembly of the actin cytoskeleton [
1,
2,
3,
4]. The immunofluorescence data indicated 45 min after intoxication a partial colocalisation of Arp2 and actin in these aggregates. The preferential staining of F-actin by TRITC-phalloidin might, however, miss the presence of G-actin in these aggregates. Nevertheless, the intracellular formation of Arp2 and presumably also of Arp2/3 complex aggregations and the disassembly of the actin cytoskeleton will have arrested lamellipodial activity and intracellular vesicle transport terminating in cell death [
4]. Our data suggest that an similar process occurs within the colon enterocytes after exposure to CDTa,b finally resulting in their detachment from the basal lamina.
Furthermore, the CDTa component might perform a dual toxic effect when inhibiting the function of Arp2/3 complex as it will also lead to an interruption of essential signaling pathways to the actin cytoskeleton affecting also processes like morphogenesis, cell motility, intracellular vesicle transport, and phagocytosis. In this respect, the action of toxins like CDT appears to have similar consequences as the lack of the Arp2/3 complex activating factor WASP in patients with Wiskott-Aldrich syndrome, which suffer from thrombocytopenia, an insufficiency of the immune cells to migrate and form contacts (as seen in CDTa-treated Caco2 cells), and gastrointestinal hemorrhage [
6,
7,
8].
Finally, it has been shown that the Arp2/3 complex attaches via its additional subunits ArpC2, ArpC3 and ArpC4/or5 to the mother filament [
27]. A recent cryo-electron study has shown that the main contacts of the Arp2/3 complex with the growing daughter filament are established by both Arp2 and Arp3 [
39,
40]. Therefore, it appears plausible that ADP-ribosylation of Arp2 will inhibit the addition of actin subunits and thereby the growth of a daughter filament. Furthermore, a recent study also showed that the additional ArpC2 and ArpC4 subunits form an extensive contact area with the mother filament [
40]. Our data show that exactly these subunits were also ADP-ribosylated by particularly the C2I and Iota-a toxins (
Figure 1). It is tempting to assume that their ADP-ribosylation (of ArpC2 and ArpC4) might inhibit the attachment of Arp2/3 complex to a mother filament [
40] and thereby additionally contribute to the failure of the modified Arp2/3 complex to nucleate actin polymerization and branch formation. This aspect will, however, necessitate future investigations.
In summary, our findings show that the Arp2/3 complex is an additional target of toxins with ART-activity like CDT, C2I and Iota-a. ADP-ribosylation of Arp2 of the Arp2/3 complex inhibits its stimulatory activity on actin polymerization. Moreover, our studies employing culture cells and an intestinal tissue model suggest that the modification of the Arp2/3 complex together with that of actin induce complete disassembly of the actin cytoskeleton in Caco2 cells and most likely lead to the morphological alterations of colon tissue organization, especially of its epithelial layer that appear reminiscent of pseudomembranous colitis. These results may open new approaches and targets for the treatment for the severe diarrhea and pseudomembranous colitis caused by C. difficile and diseases induced by other clostridial bacteria.