Overexpression of RCAN1, a Gene on Human Chromosome 21, Alters Cell Redox and Mitochondrial Function in Enamel Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Use
2.2. Overexpression of RCAN1 in LS8 Cells
2.3. Western Blot
2.4. Immunofluorescence
2.5. RNA Extraction and Quantitative PCR
2.6. GSH/GSSG Assay
2.7. Mitochondrial Superoxide and OCR Measurements
2.8. Mitochondrial NADH Autofluorescence Measurements
2.9. Mitochondrial Ca2+ Uptake
2.10. Statistical Analysis
3. Results
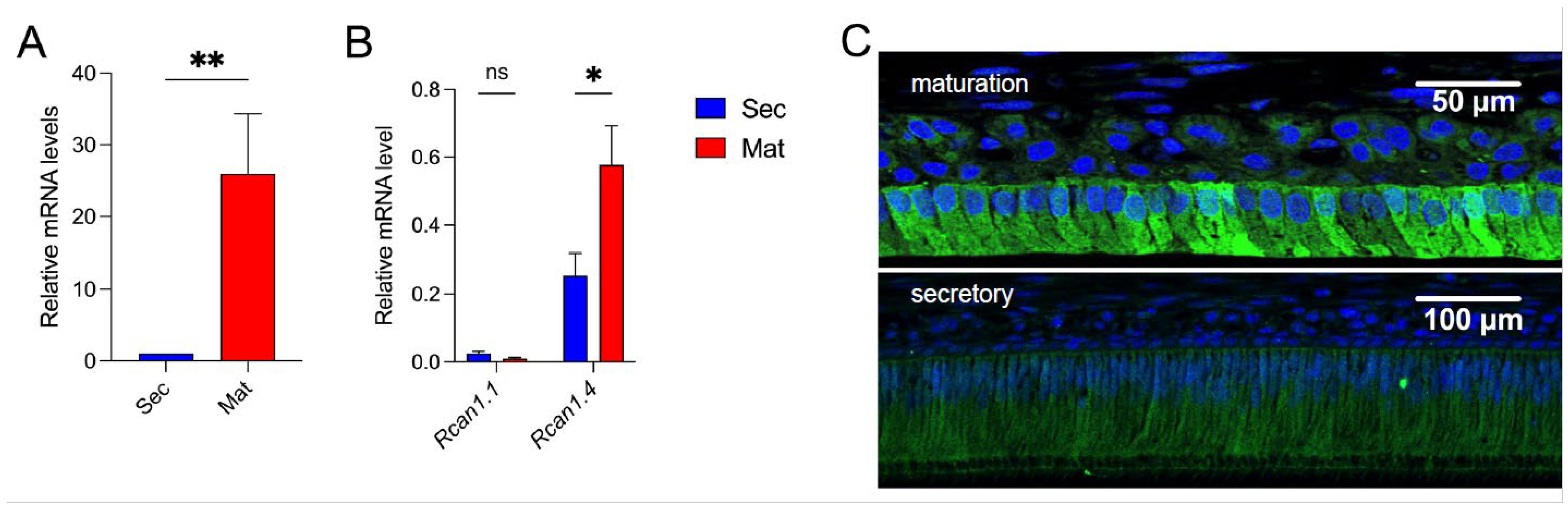
3.1. RCAN1 Is Highly Expressed in Rat Maturation Stage Ameloblasts
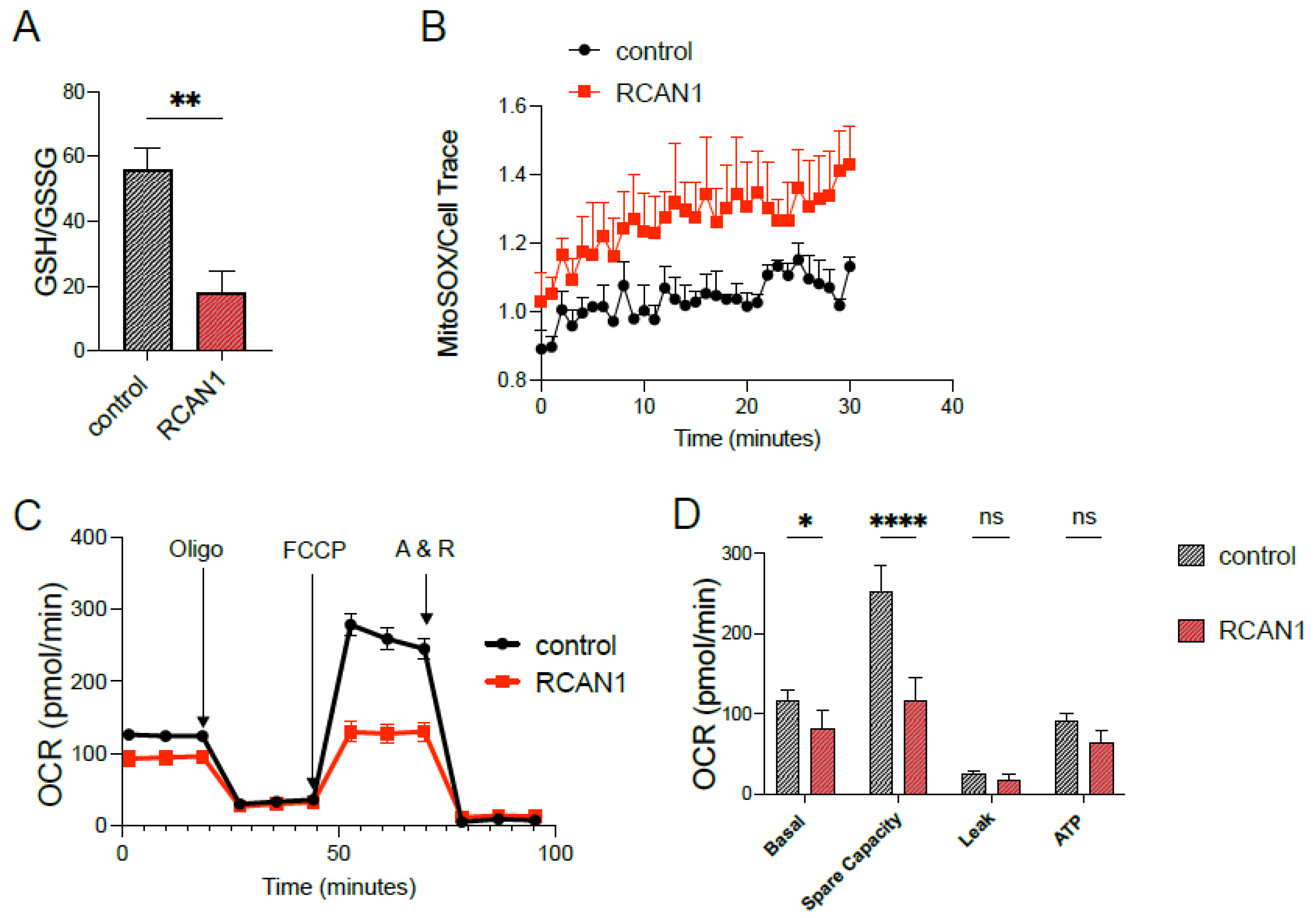
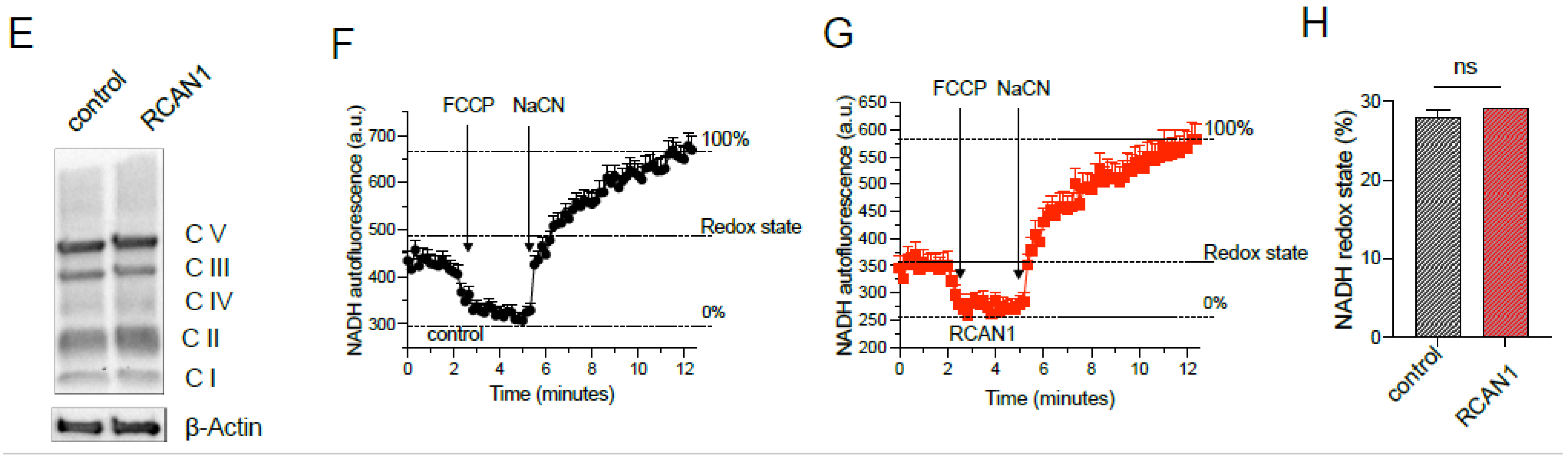
3.2. RCAN1 Overexpression in LS8 Cells Alters Redox and Mitochondrial Function
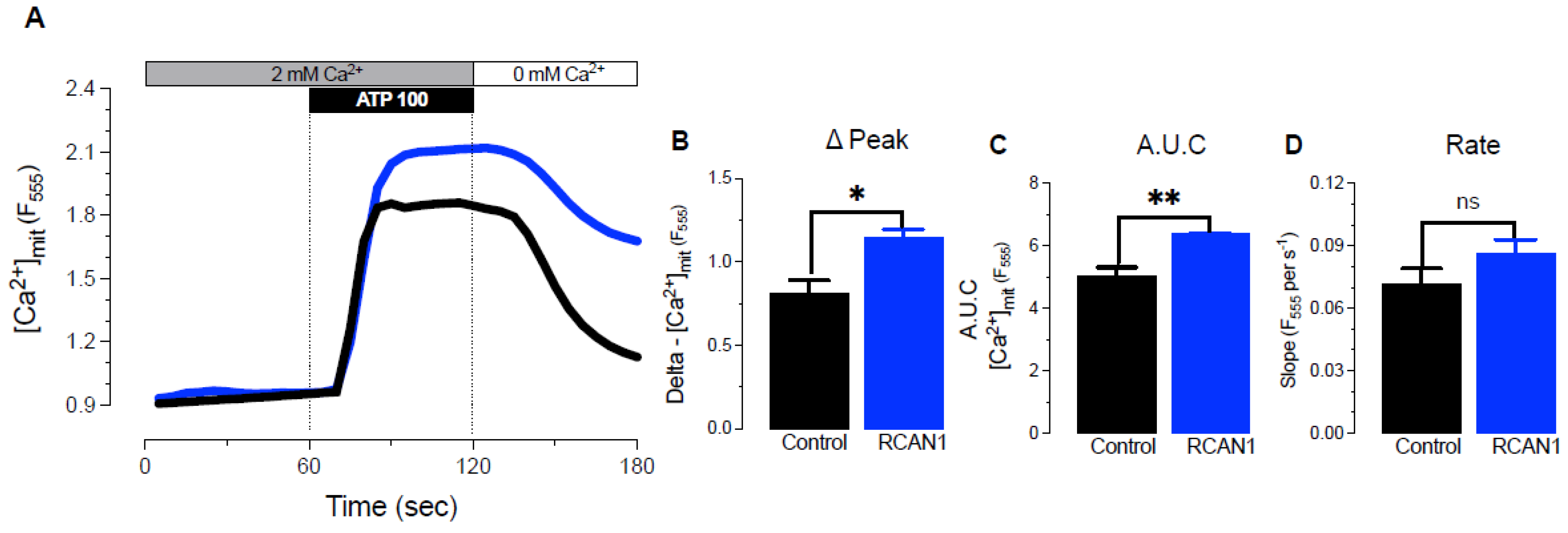
3.3. RCAN1 Overexpression Elevated Mitochondrial Ca2+ Uptake
3.4. Enamel Gene Expression Is Reduced by Overexpression of Rcan1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Smith, C.E. Cellular and chemical events during enamel maturation. Crit. Rev. Oral. Biol. Med. 1998, 9, 128–161. [Google Scholar] [CrossRef] [PubMed]
- Lacruz, R.S.; Smith, C.E.; Bringas, P., Jr.; Chen, Y.B.; Smith, S.M.; Snead, M.L.; Kurtz, I.; Hacia, J.G.; Hubbard, M.J.; Paine, M.L. Identification of novel candidate genes involved in mineralization of dental enamel by genome-wide transcript profiling. J. Cell Physiol. 2012, 227, 2264–2275. [Google Scholar] [CrossRef]
- Lacruz, R.S.; Habelitz, S.; Wright, J.T.; Paine, M.L. Dental Enamel Formation and Implications for Oral Health and Disease. Physiol. Rev. 2017, 97, 939–993. [Google Scholar] [CrossRef] [PubMed]
- Rothermel, B.; Vega, R.B.; Yang, J.; Wu, H.; Bassel-Duby, R.; Williams, R.S. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J. Biol. Chem. 2000, 275, 8719–8725. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.; Levenga, J.; Cain, P.; Rothermel, B.; Klann, E.; Hoeffer, C. RCAN1 overexpression promotes age-dependent mitochondrial dysregulation related to neurodegeneration in Alzheimer’s disease. Acta Neuropathol. 2015, 130, 829–843. [Google Scholar] [CrossRef]
- Fuentes, J.J.; Genescà, L.; Kingsbury, T.J.; Cunningham, K.W.; Pérez-Riba, M.; Estivill, X.; Luna, S.D.L. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum. Mol. Genet. 2000, 9, 1681–1690. [Google Scholar] [CrossRef]
- Patterson, D. Molecular genetic analysis of Down syndrome. Hum. Genet. 2009, 126, 195–214. [Google Scholar] [CrossRef]
- Bell, E.; Townsend, G.; Wilson, D.; Kieser, J.; Hughes, T. Effect of Down syndrome on the dimensions of dental crowns and tissues. Am. J. Hum. Biol. 2001, 13, 690–698. [Google Scholar] [CrossRef]
- Bell, E.J.; Kaidonis, J.; Townsend, G.C. Tooth wear in children with Down syndrome. Aust. Dent. J. 2002, 47, 30–35. [Google Scholar] [CrossRef]
- Moraes, M.E.L.D.; Moraes, L.C.D.; Dotto, G.N.; Dotto, P.P.; Santos, L.R.D.A.D. Dental anomalies in patients with Down syndrome. Braz. Dent. J. 2007, 18, 346–350. [Google Scholar] [CrossRef]
- Costiniti, V.; Bomfim, G.H.S.; Neginskaya, M.; Son, G.Y.; Mitaishvili, E.; Giacomello, M.; Pavlov, E.; Lacruz, R.S. Mitochondria modulate ameloblast Ca2+ signaling. FASEB J. 2022, 36, e22169. [Google Scholar] [CrossRef]
- Costiniti, V.; Bomfim, G.H.; Li, Y.; Mitaishvili, E.; Ye, Z.W.; Zhang, J.; Townsend, D.M.; Giacomello, M.; Lacruz, R.S. Mitochondrial Function in Enamel Development. Front. Physiol. 2020, 11, 538. [Google Scholar] [CrossRef]
- Ermak, G.; Sojitra, S.; Yin, F.; Cadenas, E.; Cuervo, A.M.; Davies, K.J. Chronic expression of RCAN1-1L protein induces mitochondrial autophagy and metabolic shift from oxidative phosphorylation to glycolysis in neuronal cells. J. Biol. Chem. 2012, 287, 14088–14098. [Google Scholar] [CrossRef]
- Sarkar, J.; Simanian, E.J.; Tuggy, S.Y.; Bartlett, J.D.; Snead, M.L.; Sugiyama, T.; Paine, M.L. Comparison of two mouse ameloblast-like cell lines for enamel-specific gene expression. Front. Physiol. 2014, 5, 277. [Google Scholar] [CrossRef]
- Chen, L.S.; Couwenhoven, R.I.; Hsu, D.; Luo, W.; Snead, M.L. Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ. Arch. Oral Biol. 1992, 37, 771–778. [Google Scholar] [CrossRef]
- Eckstein, M.; Vaeth, M.; Fornai, C.; Vinu, M.; Bromage, T.G.; Nurbaeva, M.K.; Sorge, J.L.; Coelho, P.G.; Idaghdour, Y.; Feske, S. Store-operated Ca(2+) entry controls ameloblast cell function and enamel development. JCI Insight 2017, 2, e91166. [Google Scholar] [CrossRef]
- Eckstein, M.; Vaeth, M.; Aulestia, F.J.; Costiniti, V.; Kassam, S.N.; Bromage, T.G.; Pedersen, P.; Issekutz, T.; Idaghdour, Y.; Moursi, A.M. Differential regulation of Ca(2+) influx by ORAI channels mediates enamel mineralization. Sci. Signal. 2019, 12, eaav4663. [Google Scholar] [CrossRef]
- Aulestia, F.J.; Groeling, J.; Bomfim, G.H.S.; Costiniti, V.; Manikandan, V.; Chaloemtoem, A.; Concepcion, A.R.; Li, Y.; Wagner, L.E., 2nd; Idaghdour, Y.; et al. Fluoride exposure alters Ca(2+) signaling and mitochondrial function in enamel cells. Sci. Signal. 2020, 13, eaay0086. [Google Scholar] [CrossRef]
- Blacker, T.S.; Duchen, M.R. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic. Biol. Med. 2016, 100, 53–65. [Google Scholar] [CrossRef]
- Harris, C.D.; Ermak, G.; Davies, K.J. Multiple roles of the DSCR1 (Adapt78 or RCAN1) gene and its protein product calcipressin 1 (or RCAN1) in disease. Cell Mol. Life Sci. 2005, 62, 2477–2486. [Google Scholar] [CrossRef]
- Hubbard, M.J. Calbindin28kDa and calmodulin are hyperabundant in rat dental enamel cells. Identification of the protein phosphatase calcineurin as a principal calmodulin target and of a secretion-related role for calbindin28kDa. Eur. J. Biochem. 1995, 230, 68–79. [Google Scholar] [CrossRef]
- Peiris, H.; Dubach, D.; Jessup, C.F.; Unterweger, P.; Raghupathi, R.; Muyderman, H.; Zanin, M.P.; Mackenzie, K.; Pritchard, M.A.; Keating, D.J. RCAN1 regulates mitochondrial function and increases susceptibility to oxidative stress in mammalian cells. Oxid. Med. Cell Longev. 2014, 2014, 520316. [Google Scholar] [CrossRef] [PubMed]
- Rebrin, I.; Sohal, R.S. Pro-oxidant shift in glutathione redox state during aging. Adv. Drug Deliv. Rev. 2008, 60, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Glancy, B.; Balaban, R.S. Role of mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry 2012, 51, 2959–2973. [Google Scholar] [CrossRef] [PubMed]
- Nurbaeva, M.K.; Eckstein, M.; Snead, M.L.; Feske, S.; Lacruz, R.S. Store-operated Ca2+ Entry Modulates the Expression of Enamel Genes. J. Dent. Res. 2015, 94, 1471–1477. [Google Scholar] [CrossRef]
- Yang, J.; Rothermel, B.; Vega, R.B.; Frey, N.; McKinsey, T.A.; Olson, E.N.; Bassel-Duby, R.; Williams, R.S. Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circ. Res. 2000, 87, E61–E68. [Google Scholar] [CrossRef]
- Lacruz, R.S.; Smith, C.E.; Kurtz, I.; Hubbard, M.J.; Paine, M.L. New paradigms on the transport functions of maturation-stage ameloblasts. J. Dent. Res. 2013, 92, 122–129. [Google Scholar] [CrossRef]
- Porta, S.; Serra, S.A.; Huch, M.; Valverde, M.A.; Llorens, F.; Estivill, X.; Arbones, M.L.; Martí, E. RCAN1 (DSCR1) increases neuronal susceptibility to oxidative stress: A potential pathogenic process in neurodegeneration. Hum. Mol. Genet. 2007, 16, 1039–1050. [Google Scholar] [CrossRef]
- Parra, V.; Altamirano, F.; Hernández-Fuentes, C.P.; Tong, D.; Kyrychenko, V.; Rotter, D.; Pedrozo, Z.; Hill, J.A.; Eisner, V.; Lavandero, S.; et al. Down Syndrome Critical Region 1 Gene, Rcan1, Helps Maintain a More Fused Mitochondrial Network. Circ. Res. 2018, 122, e20–e33. [Google Scholar] [CrossRef]
- Parekh, A.B. Mitochondrial regulation of store-operated CRAC channels. Cell Calcium 2008, 44, 6–13. [Google Scholar] [CrossRef]
- Drago, I.; De Stefani, D.; Rizzuto, R.; Pozzan, T. Mitochondrial Ca2+ uptake contributes to buffering cytoplasmic Ca2+ peaks in cardiomyocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 12986–12991. [Google Scholar] [CrossRef]
- Rizzuto, R.; Pinton, P.; Carrington, W.; Fay, F.S.; Fogarty, K.E.; Lifshitz, L.M.; Tuft, R.A.; Pozzan, T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 1998, 280, 1763–1766. [Google Scholar] [CrossRef]
- Denton, R.M. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim. Biophys. Acta 2009, 1787, 1309–1316. [Google Scholar] [CrossRef]
- Rutter, G.A.; Burnett, P.; Rizzuto, R.; Brini, M.; Murgia, M.; Pozzan, T.; Tavaré, J.M.; Denton, R.M. Subcellular imaging of intramitochondrial Ca2+ with recombinant targeted aequorin: Significance for the regulation of pyruvate dehydrogenase activity. Proc. Natl. Acad. Sci. USA 1996, 93, 5489–5494. [Google Scholar] [CrossRef]
- Finkel, T.; Menazza, S.; Holmström, K.M.; Parks, R.J.; Liu, J.; Sun, J.; Liu, J.; Pan, X.; Murphy, E. The ins and outs of mitochondrial calcium. Circ. Res. 2015, 116, 1810–1819. [Google Scholar] [CrossRef]
- Pandya, J.D.; Nukala, V.N.; Sullivan, P.G. Concentration dependent effect of calcium on brain mitochondrial bioenergetics and oxidative stress parameters. Front. Neuroenergetics 2013, 5, 10. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Costiniti, V.; Souza Bomfim, G.H.; Neginskaya, M.; Son, G.-Y.; Rothermel, B.; Pavlov, E.; Lacruz, R.S. Overexpression of RCAN1, a Gene on Human Chromosome 21, Alters Cell Redox and Mitochondrial Function in Enamel Cells. Cells 2022, 11, 3576. https://doi.org/10.3390/cells11223576
Li Y, Costiniti V, Souza Bomfim GH, Neginskaya M, Son G-Y, Rothermel B, Pavlov E, Lacruz RS. Overexpression of RCAN1, a Gene on Human Chromosome 21, Alters Cell Redox and Mitochondrial Function in Enamel Cells. Cells. 2022; 11(22):3576. https://doi.org/10.3390/cells11223576
Chicago/Turabian StyleLi, Yi, Veronica Costiniti, Guilherme H. Souza Bomfim, Maria Neginskaya, Ga-Yeon Son, Beverly Rothermel, Evgeny Pavlov, and Rodrigo S. Lacruz. 2022. "Overexpression of RCAN1, a Gene on Human Chromosome 21, Alters Cell Redox and Mitochondrial Function in Enamel Cells" Cells 11, no. 22: 3576. https://doi.org/10.3390/cells11223576
APA StyleLi, Y., Costiniti, V., Souza Bomfim, G. H., Neginskaya, M., Son, G.-Y., Rothermel, B., Pavlov, E., & Lacruz, R. S. (2022). Overexpression of RCAN1, a Gene on Human Chromosome 21, Alters Cell Redox and Mitochondrial Function in Enamel Cells. Cells, 11(22), 3576. https://doi.org/10.3390/cells11223576






