Cuprotosis Programmed-Cell-Death-Related lncRNA Signature Predicts Prognosis and Immune Landscape in PAAD Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Data Sources
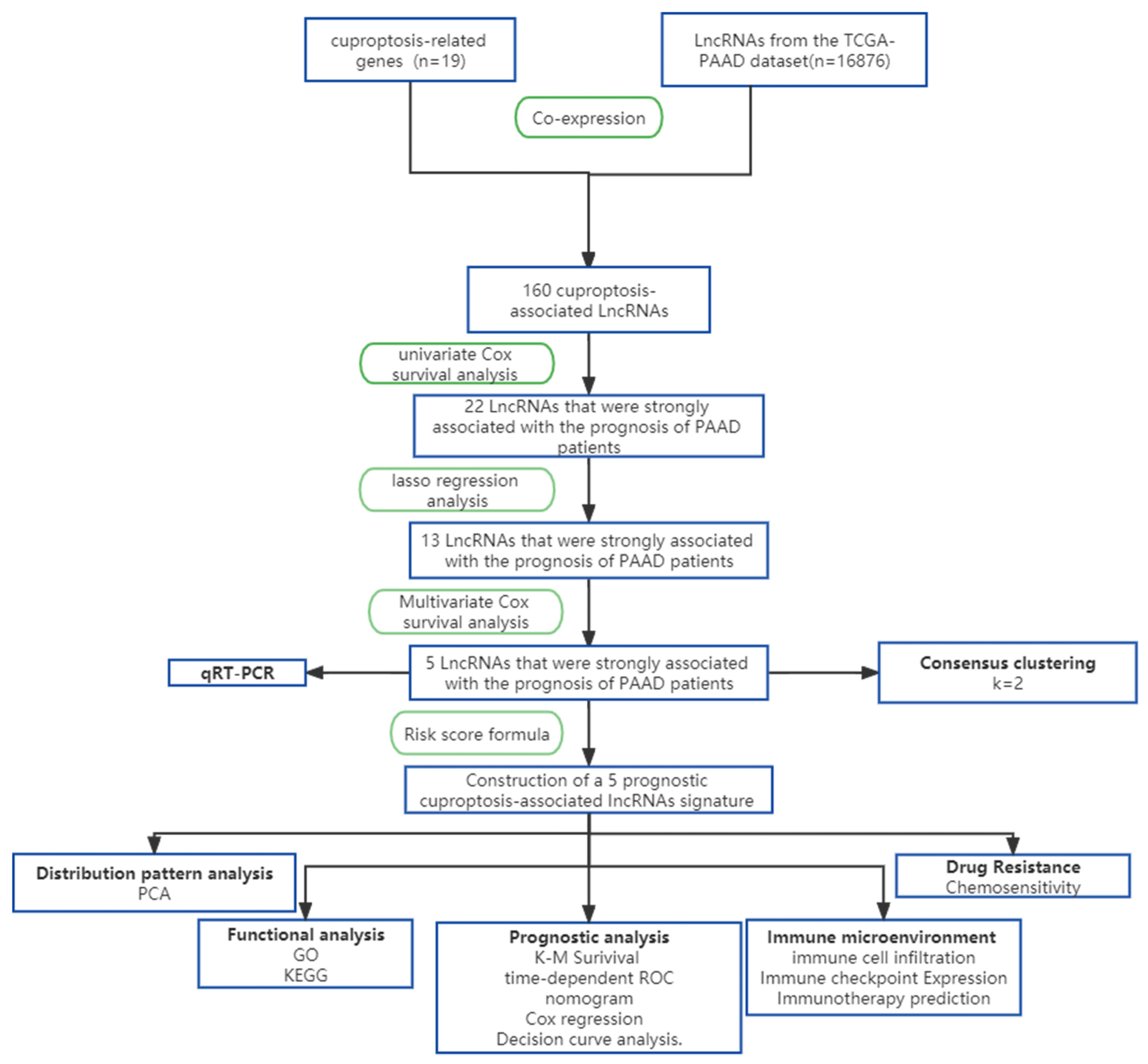
2.2. Model Construction and Validation
2.3. Consensus Clustering Analysis
2.4. Model Formulae
2.5. Independent Prognostic Analysis and Nomogram Construction
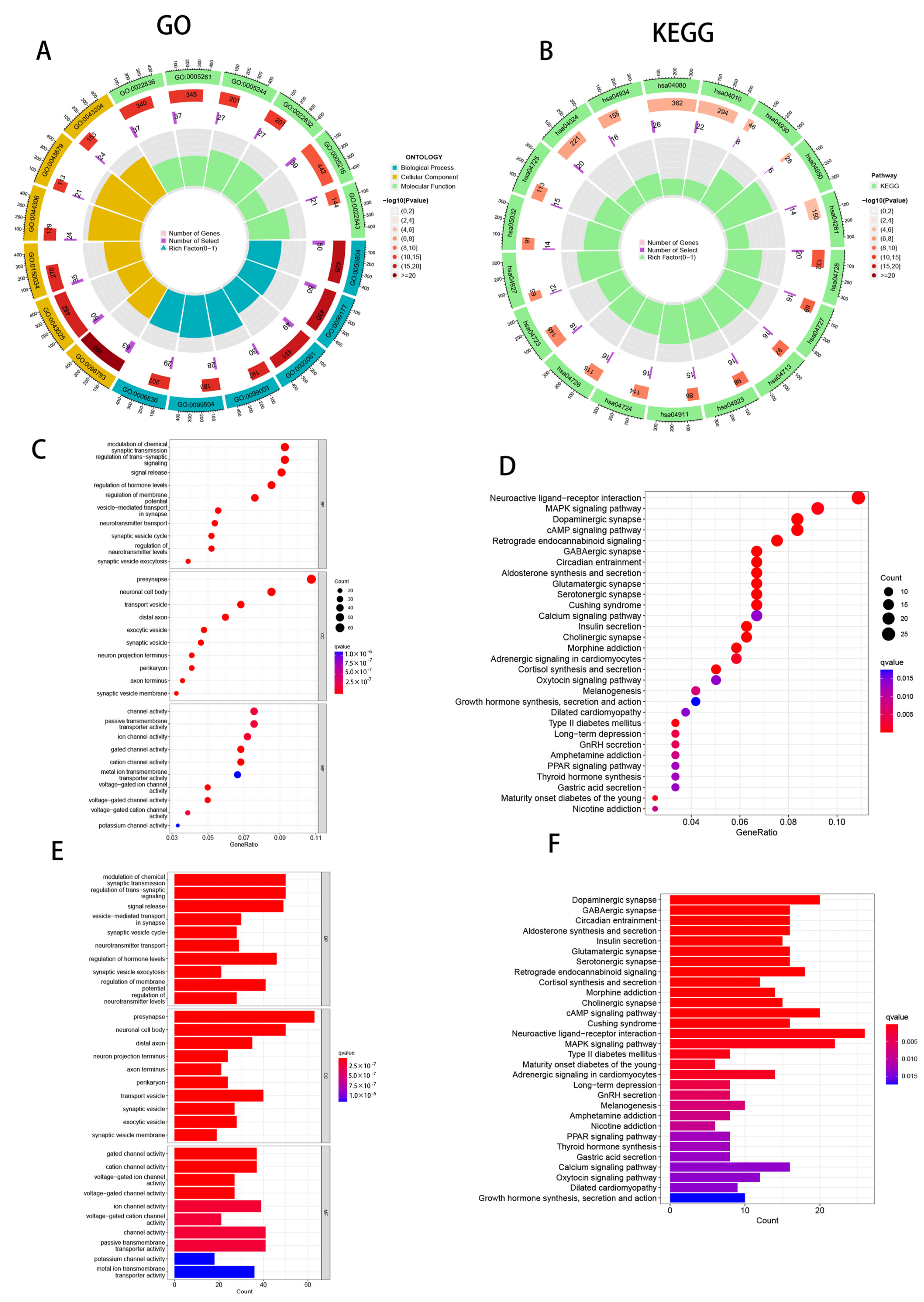
2.6. Functional Enrichment Analysis
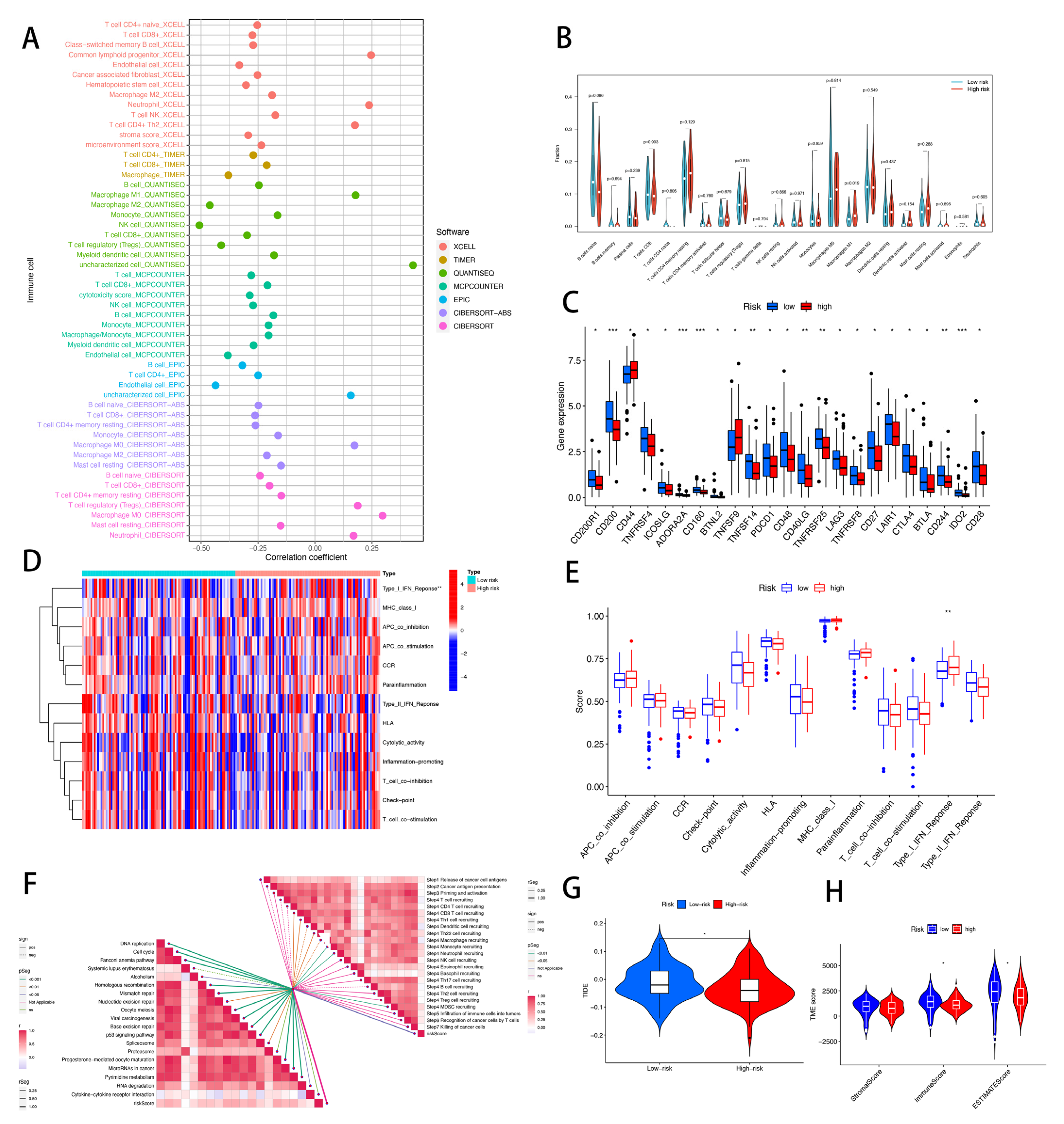
2.7. Immunity Analysis of the Risk Signature
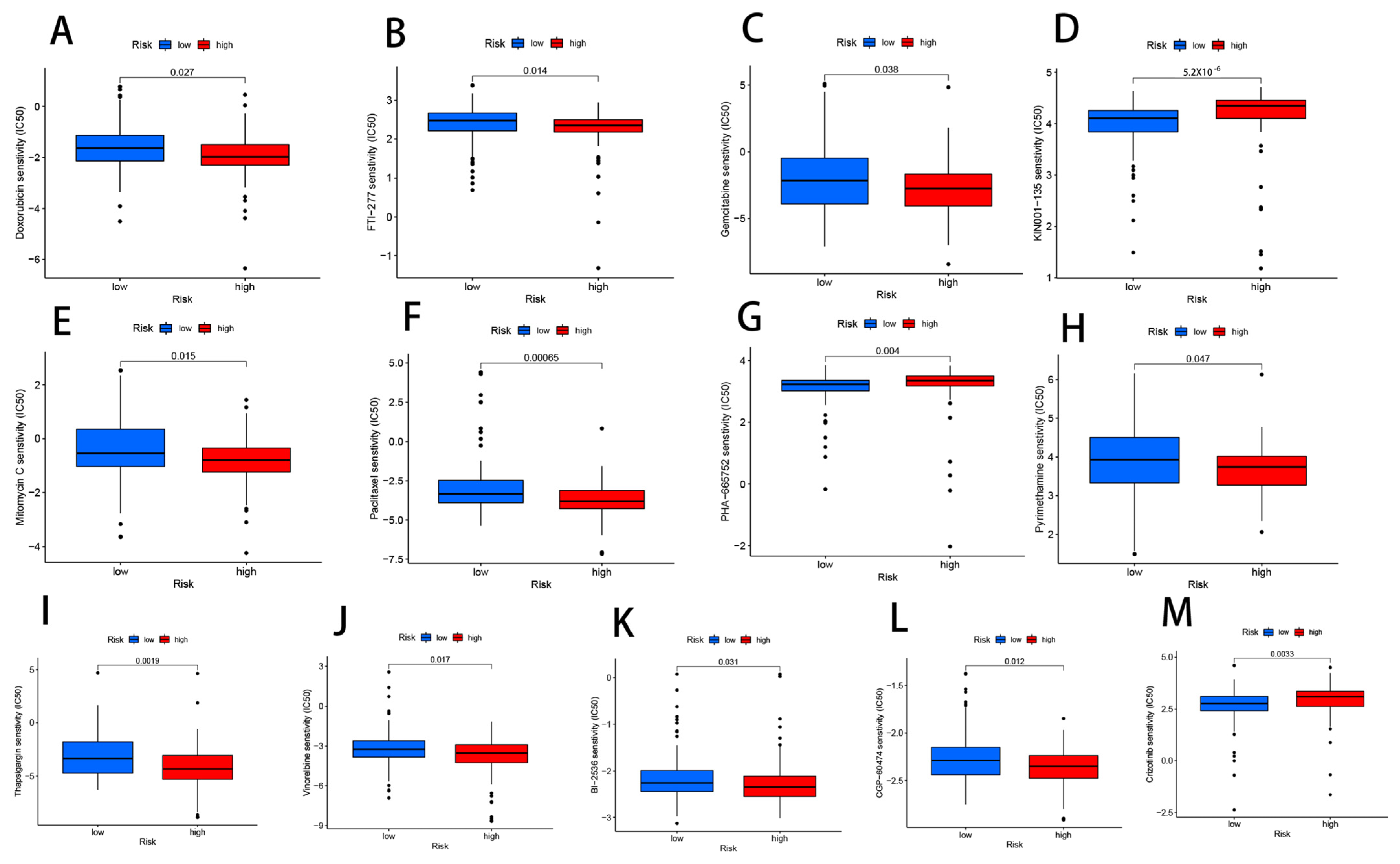
2.8. Drug Sensitivity
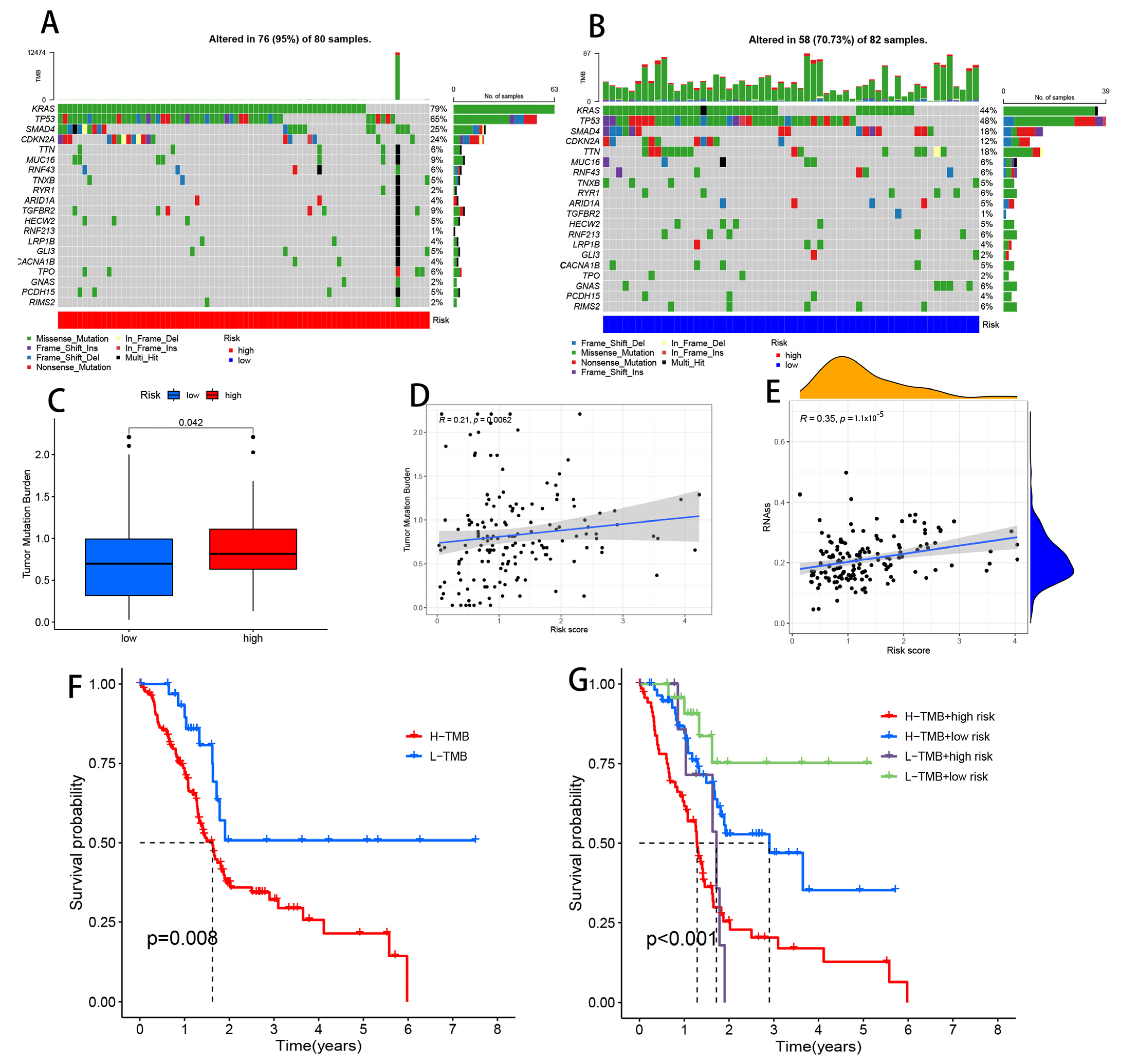
2.9. Somatic Mutation Analysis
2.10. Construction of Cuprotosis Cell Model and qPCR Assay
2.11. Statistical Analysis
3. Result
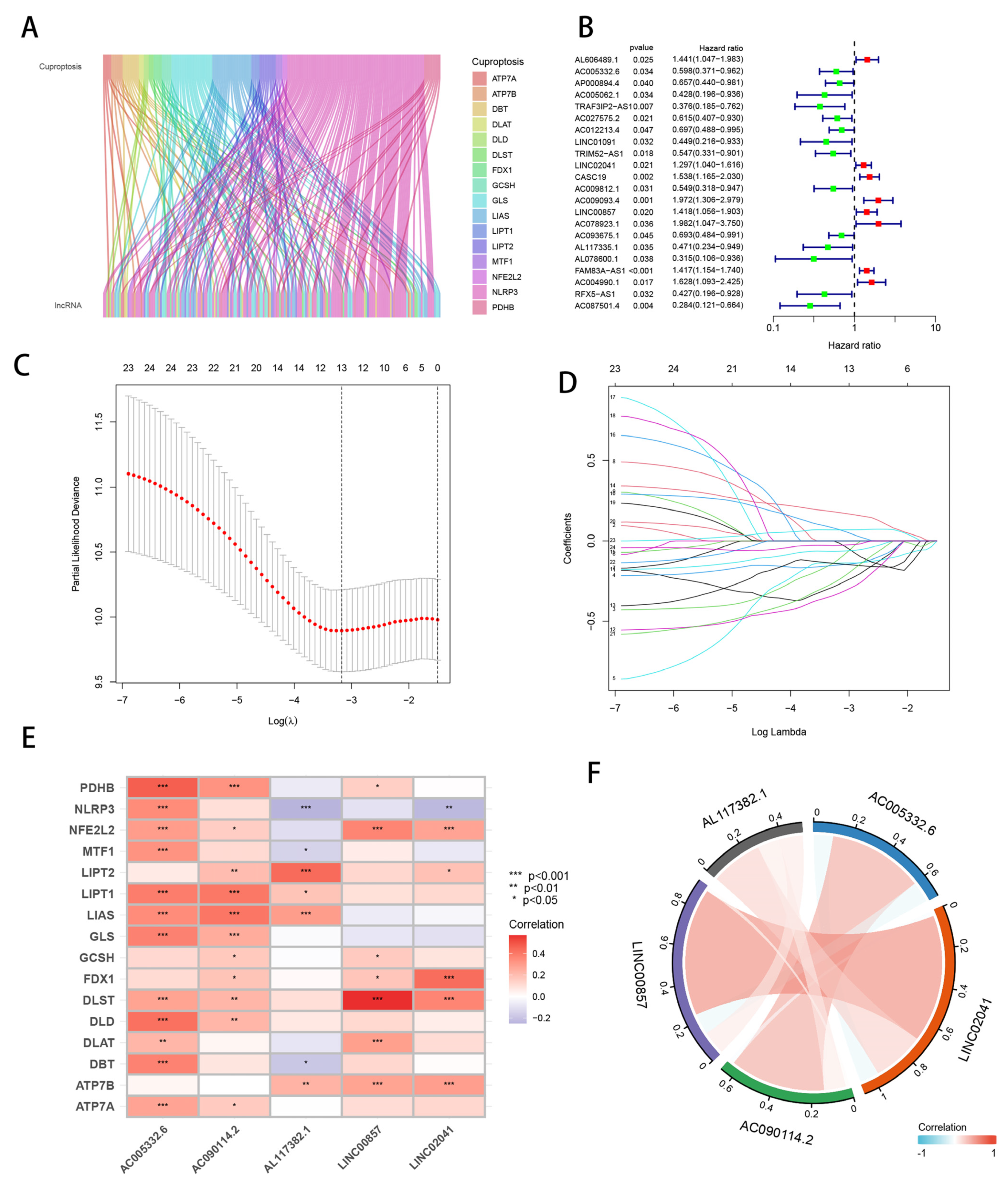
3.1. Identification of Candidate Cuprotosis-Related lncRNAs
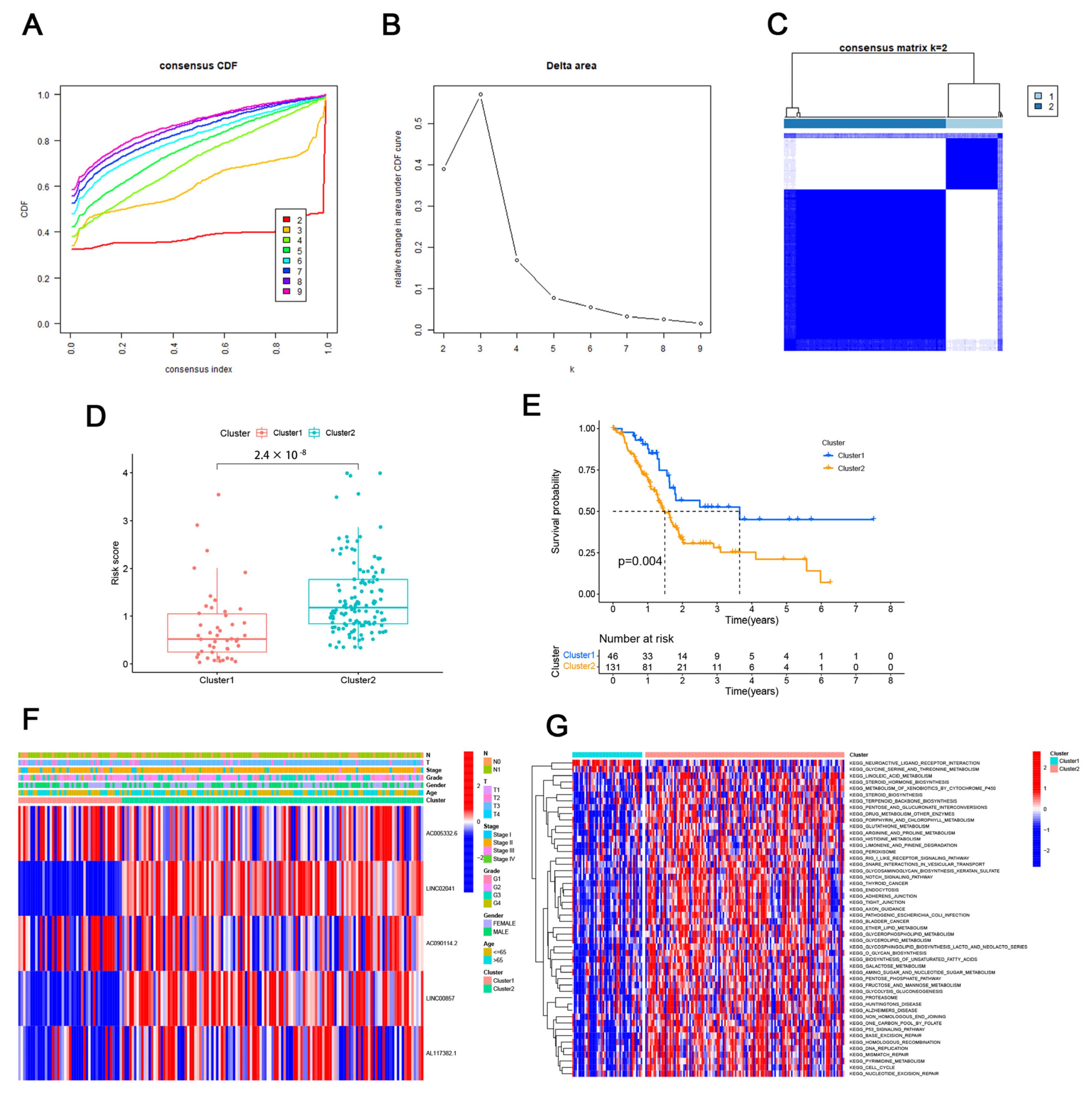
3.2. Consensus Clustering Identified the Molecular Subtypes of PAAD
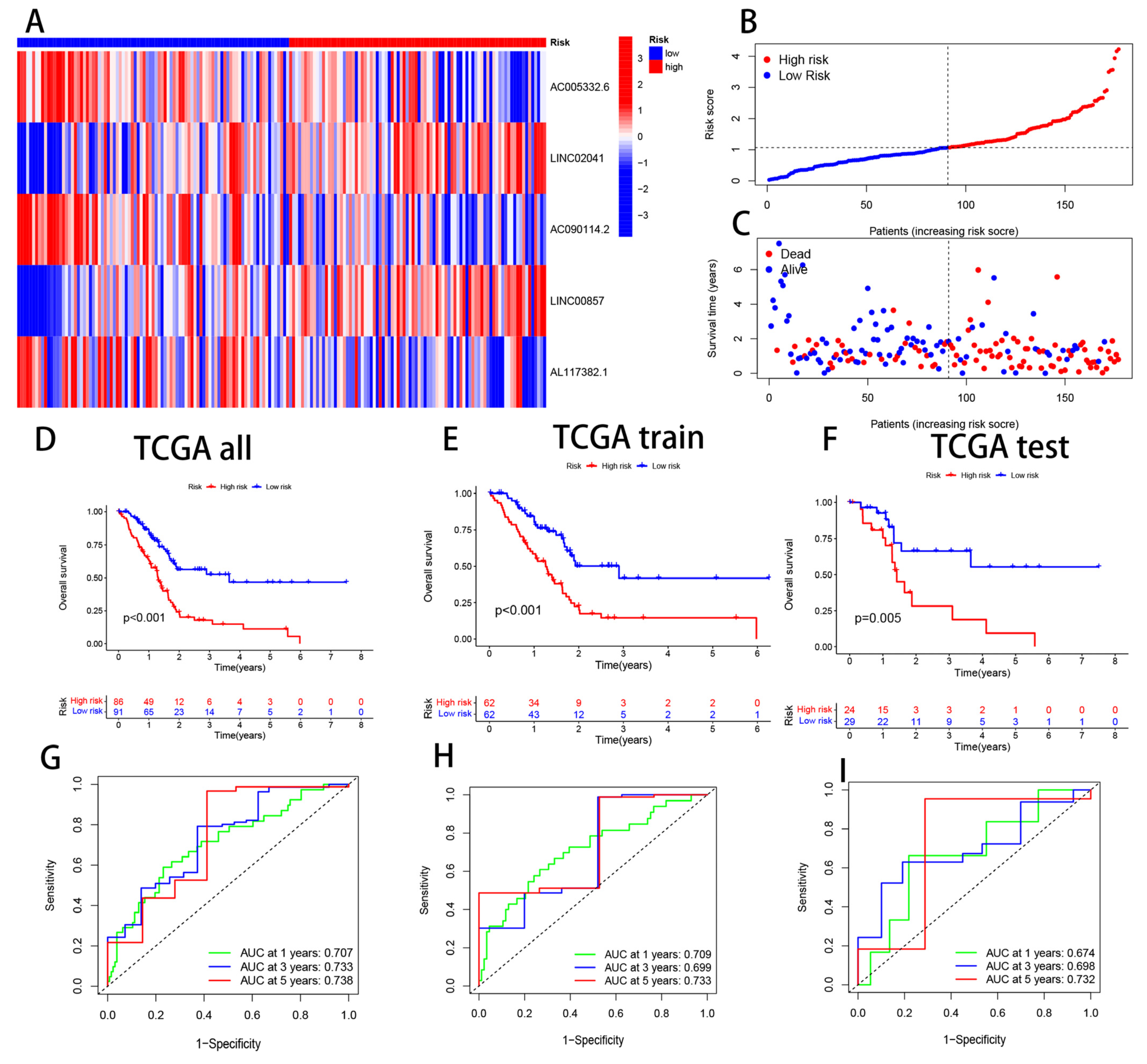
3.3. Validation of CRL Signature and its Prognostic Value
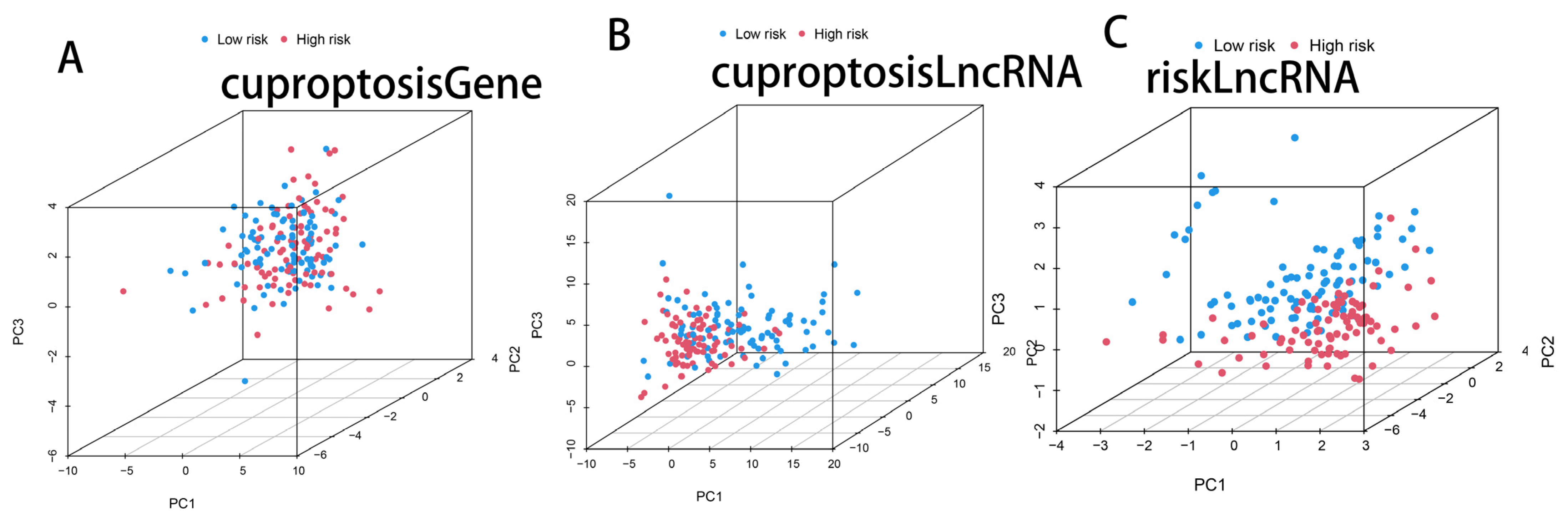
3.4. PCA Analysis of Cuprotosis-Related Genes, Cuprotosis-Related lncRNAs, and CRL Model lncRNAs
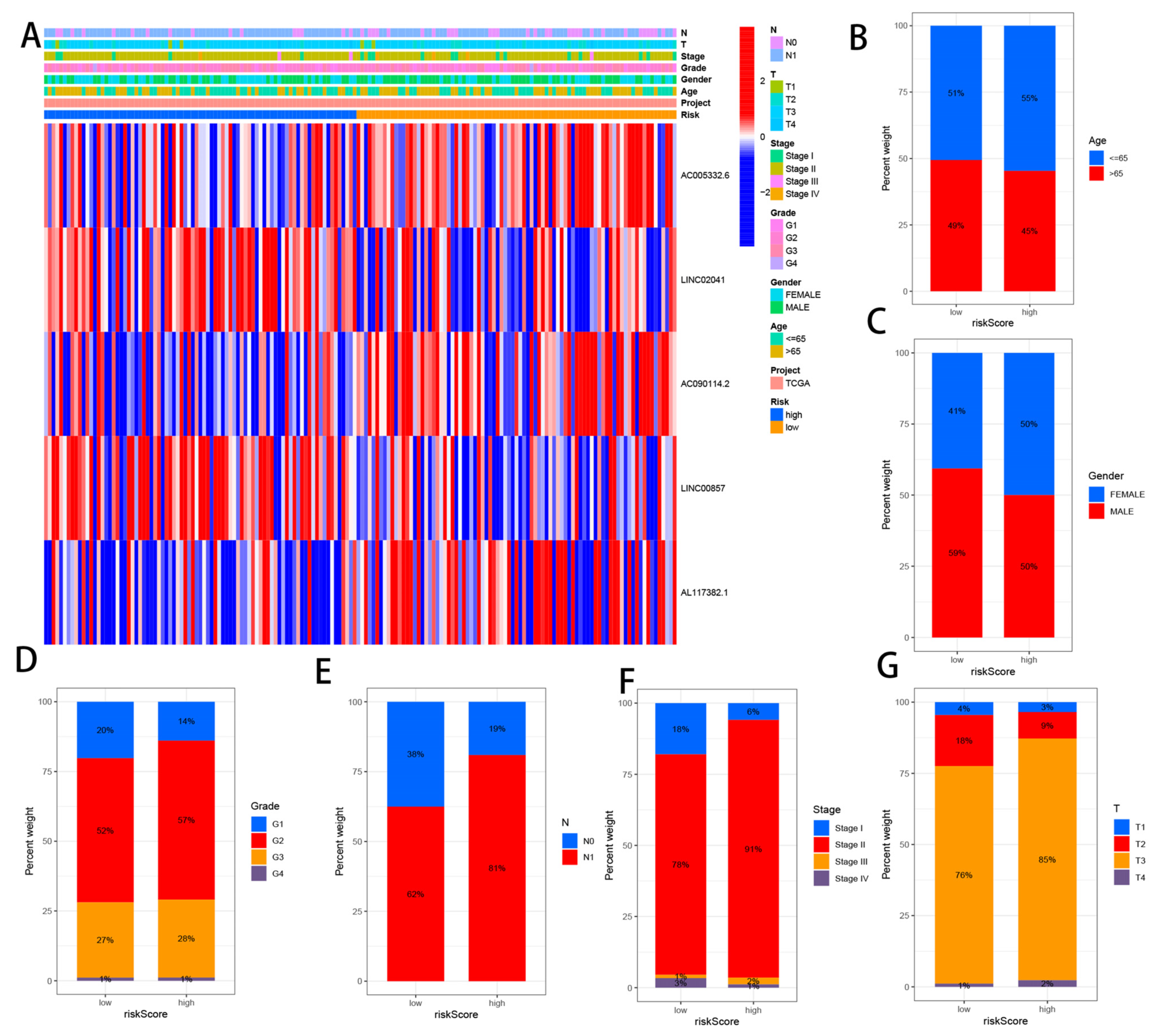
3.5. Correlation Analysis between CRLs and Clinicopathological Features
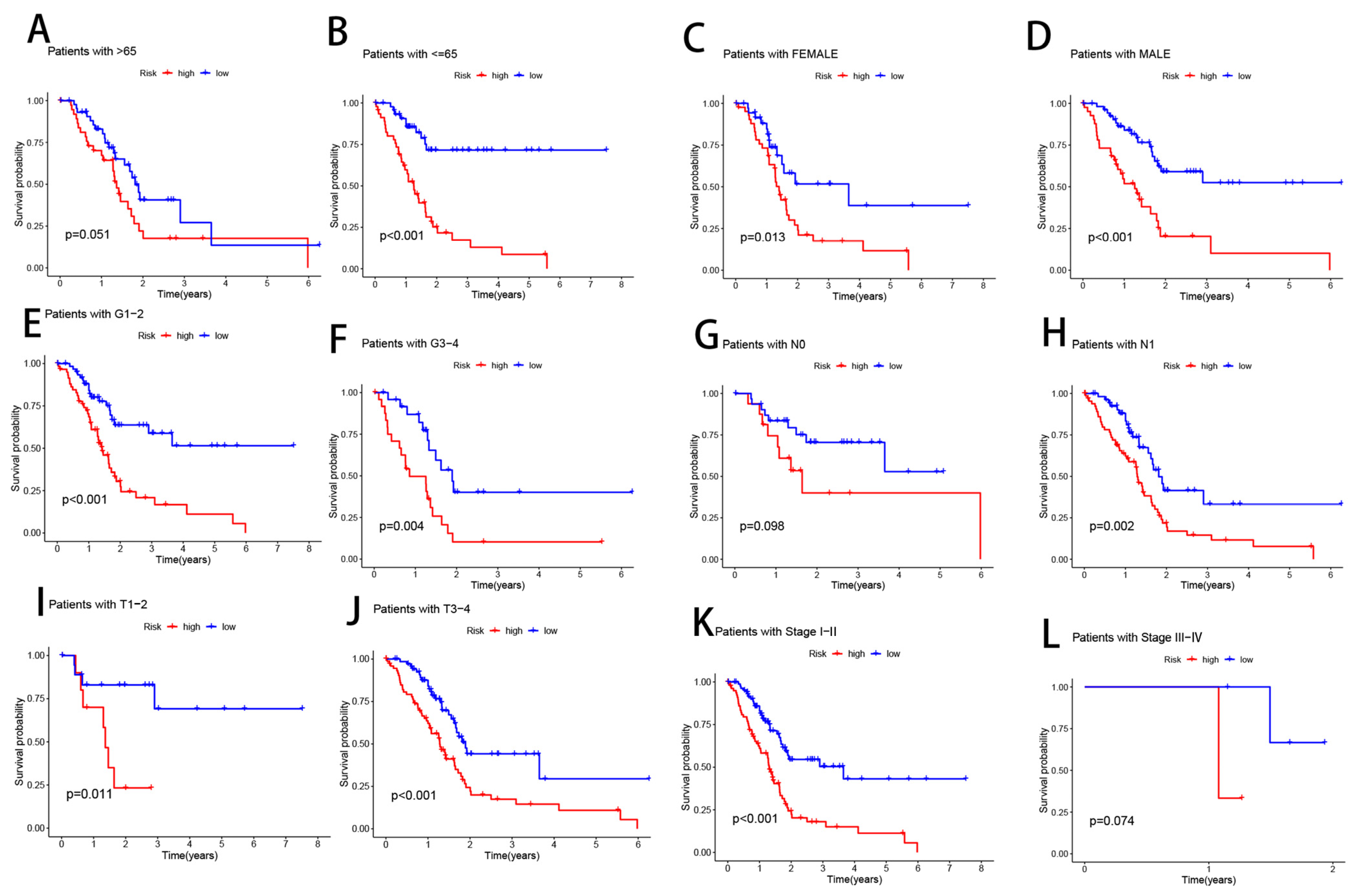
3.6. Clinical Subgroup Analysis of Risk Models for CRLs
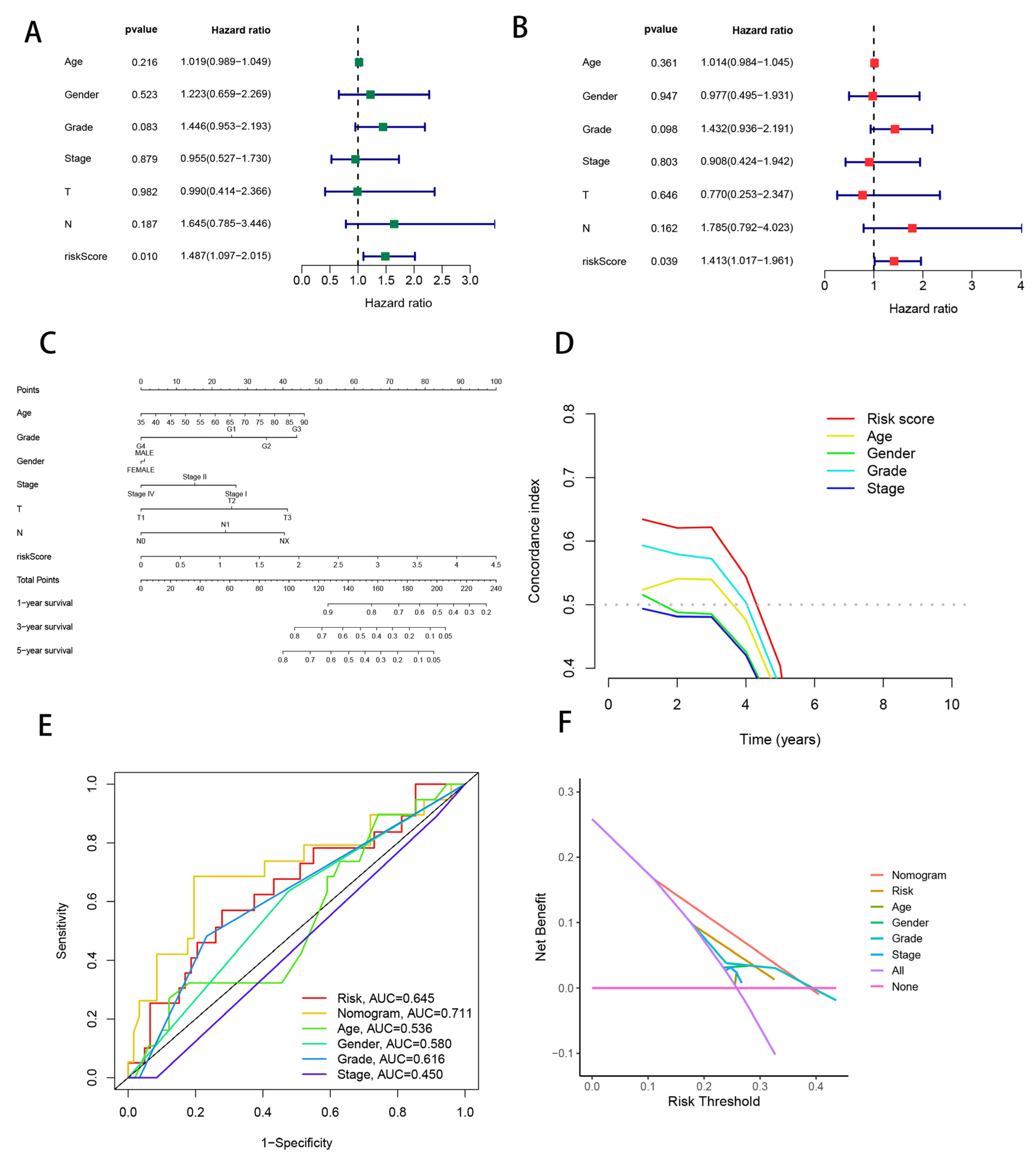
3.7. Combining Clinical Characteristics to Build Nomograms
3.8. Enrichment Analysis of PAAD Patients Based on Prognostic Markers
3.9. Risk Score of CRLs Predicts Tumor Microenvironment and Immune Cell Infiltration
3.10. Drug Sensitivity Analysis Related to CRLs
3.11. Comparison of Somatic Mutations between Low- and High-Risk Groups
3.12. qRT-PCR Assay in Cuprotosis Cell Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ansari, D.; Tingstedt, B.; Andersson, B.; Holmquist, F.; Sturesson, C.; Williamsson, C.; Sasor, A.; Borg, D.; Bauden, M.; Andersson, R. Pancreatic cancer: Yesterday, today and tomorrow. Future Oncol. 2016, 12, 1929–1946. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Bilimoria, K.Y.; Bentrem, D.J.; Ko, C.Y.; Ritchey, J.; Stewart, A.K.; Winchester, D.P.; Talamonti, M.S. Validation of the 6th edition AJCC Pancreatic Cancer Staging System: Report from the National Cancer Database. Cancer 2007, 110, 738–744. [Google Scholar] [CrossRef]
- Aroldi, F.; Bertocchi, P.; Rosso, E.; Prochilo, T.; Zaniboni, A. Pancreatic Cancer: Promises and Failures of Target Therapies. Rev. Recent Clin. Trials 2016, 11, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.; Donahue, T. Pancreatic Cancer. Jama 2019, 322, 1426. [Google Scholar] [CrossRef]
- Huang, Z.D.; Liu, Z.Z.; Liu, Y.Y.; Fu, Y.C.; Lin, L.L.; Hu, C.; Gu, H.Y.; Wei, R.X. Molecular Subtypes Based on Cell Differentiation Trajectories in Head and Neck Squamous Cell Carcinoma: Differential Prognosis and Immunotherapeutic Responses. Front. Immunol. 2021, 12, 791621. [Google Scholar] [CrossRef]
- Ruiz, L.M.; Libedinsky, A.; Elorza, A.A. Role of Copper on Mitochondrial Function and Metabolism. Front. Mol. Biosci. 2021, 8, 711227. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Lelièvre, P.; Sancey, L.; Coll, J.L.; Deniaud, A.; Busser, B. The Multifaceted Roles of Copper in Cancer: A Trace Metal Element with Dysregulated Metabolism, but Also a Target or a Bullet for Therapy. Cancers 2020, 12, 3594. [Google Scholar] [CrossRef]
- Blockhuys, S.; Celauro, E.; Hildesjö, C.; Feizi, A.; Stål, O.; Fierro-González, J.C.; Wittung-Stafshede, P. Defining the human copper proteome and analysis of its expression variation in cancers. Met. Integr. Biometal Sci. 2017, 9, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Andreux, P.; Poitry-Yamate, C.; Auwerx, J.; Hanahan, D. Bioavailable copper modulates oxidative phosphorylation and growth of tumors. Proc. Natl. Acad. Sci. United States Am. 2013, 110, 19507–19512. [Google Scholar] [CrossRef]
- Lener, M.R.; Scott, R.J.; Wiechowska-Kozłowska, A.; Serrano-Fernández, P.; Baszuk, P.; Jaworska-Bieniek, K.; Sukiennicki, G.; Marciniak, W.; Muszyńska, M.; Kładny, J.; et al. Serum Concentrations of Selenium and Copper in Patients Diagnosed with Pancreatic Cancer. Cancer Res. Treat. 2016, 48, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhou, R.; Zhao, Y.; Pan, Y.; Liang, H.; Zhang, J.S.; Tai, S.; Jin, L.; Teng, C.B. Blockage of SLC31A1-dependent copper absorption increases pancreatic cancer cell autophagy to resist cell death. Cell Prolif. 2019, 52, e12568. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Jandura, A.; Krause, H.M. The New RNA World: Growing Evidence for Long Noncoding RNA Functionality. Trends Genet. TIG 2017, 33, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Koyama, K.; Ota, Y.; Iwamoto, H.; Yamakita, K.; Fujii, S.; Kitano, Y. The Interaction Between Long Non-coding RNA HULC and MicroRNA-622 via Transfer by Extracellular Vesicles Regulates Cell Invasion and Migration in Human Pancreatic Cancer. Front. Oncol. 2020, 10, 1013. [Google Scholar] [CrossRef]
- Takahashi, K.; Taniue, K.; Ono, Y.; Fujiya, M.; Mizukami, Y.; Okumura, T. Long Non-Coding RNAs in Epithelial-Mesenchymal Transition of Pancreatic Cancer. Front. Mol. Biosci. 2021, 8, 717890. [Google Scholar] [CrossRef]
- Kishikawa, T.; Otsuka, M.; Ohno, M.; Yoshikawa, T.; Takata, A.; Koike, K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J. Gastroenterol. 2015, 21, 8527–8540. [Google Scholar] [CrossRef]
- Peng, W.; Jiang, A. Long noncoding RNA CCDC26 as a potential predictor biomarker contributes to tumorigenesis in pancreatic cancer. Biomed. Pharmacother. 2016, 83, 712–717. [Google Scholar] [CrossRef]
- Luo, Y.; Yan, B.; Liu, L.; Yin, L.; Ji, H.; An, X.; Gladkich, J.; Qi, Z.; De La Torre, C.; Herr, I. Sulforaphane Inhibits the Expression of Long Noncoding RNA H19 and Its Target APOBEC3G and Thereby Pancreatic Cancer Progression. Cancers 2021, 13, 827. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Aran, D. Cell-Type Enrichment Analysis of Bulk Transcriptomes Using xCell. Methods Mol. Biol. 2020, 2120, 263–276. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Dienstmann, R.; Villacampa, G.; Sveen, A.; Mason, M.J.; Niedzwiecki, D.; Nesbakken, A.; Moreno, V.; Warren, R.S.; Lothe, R.A.; Guinney, J. Relative contribution of clinicopathological variables, genomic markers, transcriptomic subtyping and microenvironment features for outcome prediction in stage II/III colorectal cancer. Ann. Oncol. 2019, 30, 1622–1629. [Google Scholar] [CrossRef]
- Racle, J.; de Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 2017, 6, e26476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, R.; Cao, Y.; Gu, Y.; Lin, C.; Liu, X.; Lv, K.; He, X.; Fang, H.; Jin, K.; et al. Poor Clinical Outcomes and Immunoevasive Contexture in Intratumoral IL-10-Producing Macrophages Enriched Gastric Cancer Patients. Ann. Surg. 2022, 275, e626–e635. [Google Scholar] [CrossRef]
- Tamminga, M.; Hiltermann, T.J.N.; Schuuring, E.; Timens, W.; Fehrmann, R.S.; Groen, H.J. Immune microenvironment composition in non-small cell lung cancer and its association with survival. Clin. Transl. Immunol. 2020, 9, e1142. [Google Scholar] [CrossRef] [PubMed]
- Auslander, N.; Zhang, G.; Lee, J.S.; Frederick, D.T.; Miao, B.; Moll, T.; Tian, T.; Wei, Z.; Madan, S.; Sullivan, R.J.; et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 2018, 24, 1545–1549. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Deng, C.; Pang, B.; Zhang, X.; Liu, W.; Liao, G.; Yuan, H.; Cheng, P.; Li, F.; Long, Z.; et al. TIP: A Web Server for Resolving Tumor Immunophenotype Profiling. Cancer Res. 2018, 78, 6575–6580. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Geeleher, P.; Cox, N.J.; Huang, R.S. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014, 15, R47. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Lonigro, R.J.; Vats, P.; Cobain, E.; Everett, J.; Cao, X.; Rabban, E.; Kumar-Sinha, C.; Raymond, V.; et al. Integrative clinical genomics of metastatic cancer. Nature 2017, 548, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Li, B.; Li, Z.; Sun, L. Identification of Autophagy Related circRNA-miRNA-mRNA-Subtypes Network With Radiotherapy Responses and Tumor Immune Microenvironment in Non-small Cell Lung Cancer. Front. Genet. 2021, 12, 730003. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Zheng, J.; Mou, W.; Lu, G.; Chen, S.; Du, J.; Zheng, Y.; Chen, S.; Shen, B.; Li, J.; et al. Development and validation of a hypoxia-stemness-based prognostic signature in pancreatic adenocarcinoma. Front. Pharmacol. 2022, 13, 939542. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Y.; Jiang, Z. Immune induction identified by TMT proteomics analysis in Fusobacterium nucleatum autoinducer-2 treated macrophages. Expert Rev. Proteom. 2020, 17, 175–185. [Google Scholar] [CrossRef]
- Ye, Y.; Zheng, S. Successful Immunotherapy for Pancreatic Cancer in a Patient With TSC2 and SMAD4 Mutations: A Case Report. Front. Immunol. 2021, 12, 785400. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, X.; Shi, Y.; Lu, Y.; Qiu, P.; Deng, Z.; Yao, W.; Wang, J. A Comprehensive Analysis of Pyroptosis-Related lncRNAs Signature Associated With Prognosis and Tumor Immune Microenvironment of Pancreatic Adenocarcinoma. Front. Genet. 2022, 13, 899496. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Y.; Xing, C.; Ding, C.; Zhang, H.; Chen, L.; You, L.; Dai, M.; Zhao, Y. Tumor microenvironment in chemoresistance, metastasis and immunotherapy of pancreatic cancer. Am. J. Cancer Res. 2020, 10, 1937–1953. [Google Scholar] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Tuveson, D.A.; Neoptolemos, J.P. Understanding metastasis in pancreatic cancer: A call for new clinical approaches. Cell 2012, 148, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.; Zeng, T.; Liu, S.; Cheng, H.; Ma, L.; Wang, Y. A novel 10 glycolysis-related genes signature could predict overall su rvival for clear cell renal cell carcinoma. BMC Cancer 2021, 21, 381. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zheng, M.; Zhang, D.; Gou, R.; Liu, O.; Wang, S.; Lin, B. Identification of the prognostic value of a 2-gene signature of the WN T gene family in UCEC using bioinformatics and real-world data. Cancer Cell Int. 2021, 21, 516. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, V.C.; Gudekar, N.; Jasmer, K.; Papageorgiou, C.; Singh, K.; Petris, M.J. Copper metabolism as a unique vulnerability in cancer. Biochim. Biophys. Acta. Mol. Cell Res. 2021, 1868, 118893. [Google Scholar] [CrossRef] [PubMed]
- Kahlson, M.A.; Dixon, S.J. Copper-induced cell death. Science 2022, 375, 1231–1232. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Ge, C. LncRNA ZFAS1 promotes pancreatic adenocarcinoma metastasis via the RHOA/ROCK2 pathway by sponging miR-3924. Cancer Cell Int. 2020, 20, 249. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhao, H.; Zhou, H.; Geng, T. LncRNA LINC00857 strengthens the malignancy behaviors of pancreatic adenocarcinoma cells by serving as a competing endogenous RNA for miR-340-5p to upregulate TGFA expression. PLoS ONE 2021, 16, e0247817. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lei, C.; Lu, C.; Wang, J.; Gao, M.; Gao, W. LINC01232 exerts oncogenic activities in pancreatic adenocarcinoma via regulation of TM9SF2. Cell Death Dis. 2019, 10, 698. [Google Scholar] [CrossRef]
- Deng, Z.; Li, X.; Shi, Y.; Lu, Y.; Yao, W.; Wang, J. A Novel Autophagy-Related IncRNAs Signature for Prognostic Prediction and Clinical Value in Patients With Pancreatic Cancer. Front. Cell Dev. Biol. 2020, 8, 606817. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.; Castro, E.; Belfort, M.A.; Shamsirshaz, A.A.; Nassr, A.A.; Espinoza, J. Massive Extra-Abdominal Umbilical Vein Varix: A Case Report. Fetal Diagn. Ther. 2021, 48, 158–162. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, J.; Cai, B.; Chen, Q.; Wang, G.; Lu, Z.; Jiang, K.; Miao, Y. Development of epithelial-mesenchymal transition-related lncRNA signature for predicting survival and immune microenvironment in pancreatic cancerwithexperiment validation. Bioengineered 2021, 12, 10553–10567. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Yang, M.; Wang, Q.; Zhao, L.; Zhu, S.; Zhu, L.; Xu, T.; Cao, R.; Li, C.; Liu, Q.; et al. A Novel Prognostic Prediction Model for Colorectal Cancer Based on Nine Autophagy-Related Long Noncoding RNAs. Front. Oncol. 2021, 11, 613949. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zeng, Z.; Wang, J.; Cao, W.; Song, C.; Lei, S.; Li, Y.; Ren, Z. Long noncoding RNA LINC00857 promotes pancreatic cancer proliferation and metastasis by regulating the miR-130b/RHOA axis. Cell Death Discov. 2022, 8, 198. [Google Scholar] [CrossRef]
- Spill, F.; Reynolds, D.S.; Kamm, R.D.; Zaman, M.H. Impact of the physical microenvironment on tumor progression and metastasis. Curr. Opin. Biotechnol. 2016, 40, 41–48. [Google Scholar] [CrossRef]
- Del Prete, A.; Schioppa, T.; Tiberio, L.; Stabile, H.; Sozzani, S. Leukocyte trafficking in tumor microenvironment. Curr. Opin. Pharmacol. 2017, 35, 40–47. [Google Scholar] [CrossRef]
- Zhang, Z.; Qiu, X.; Yan, Y.; Liang, Q.; Cai, Y.; Peng, B.; Xu, Z.; Xia, F. Evaluation of Ferroptosis-related Gene AKR1C1 as a Novel Biomarker Associated with the Immune Microenvironment and Prognosis in Breast Cancer. Int. J. Gen. Med. 2021, 14, 6189–6200. [Google Scholar] [CrossRef]
- Xiao, J.; Lv, C.; Xiao, C.; Ma, J.; Liao, J.; Liu, T.; Du, J.; Zuo, S.; Li, H.; Gu, H. Construction of a ceRNA Network and Analysis of Tumor Immune Infiltration in Pancreatic Adenocarcinoma. Front. Mol. Biosci. 2021, 8, 745409. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Cui, L.; Yang, L.; Wang, B.; Zhuo, Y.; Zhang, L.; Wang, X.; Zhang, Q.; Zhang, S. Pancreatic Stellate Cells Promote Tumor Progression by Promoting an Immunosuppressive Microenvironment in Murine Models of Pancreatic Cancer. Pancreas 2020, 49, 120–127. [Google Scholar] [CrossRef]
- Yan, Y.; Gao, R.; Trinh, T.L.P.; Grant, M.B. Immunodeficiency in Pancreatic Adenocarcinoma with Diabetes Revealed by Comparative Genomics. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 6363–6373. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Yang, Y.; Liu, F.; Jiang, Z.; Jiang, Z. TNFSF9 promotes metastasis of pancreatic cancer by regulating M2 polarization of macrophages through Src/FAK/p-Akt/IL-1β signaling. Int. Immunopharmacol. 2022, 102, 108429. [Google Scholar] [CrossRef]
- Lakshman, M.; Subramaniam, V.; Wong, S.; Jothy, S. CD44 promotes resistance to apoptosis in murine colonic epithelium. J. Cell. Physiol. 2005, 203, 583–588. [Google Scholar] [CrossRef]
- Lakshman, M.; Subramaniam, V.; Rubenthiran, U.; Jothy, S. CD44 promotes resistance to apoptosis in human colon cancer cells. Exp. Mol. Pathol. 2004, 77, 18–25. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Singleton, P.A.; Zhu, H.; Diedrich, F. Hyaluronan-mediated CD44 interaction with RhoGEF and Rho kinase promotes Grb2-associated binder-1 phosphorylation and phosphatidylinositol 3-kinase signaling leading to cytokine (macrophage-colony stimulating factor) production and breast tumor progression. J. Biol. Chem. 2003, 278, 29420–29434. [Google Scholar] [CrossRef]
- Hu, Z.; Gao, J.; Zhang, D.; Liu, Q.; Yan, L.; Gao, L.; Liu, J.; Liu, D.; Zhang, S.; Lin, B. High expression of Lewis y antigen and CD44 is correlated with resistance to chemotherapy in epithelial ovarian cancers. PLoS ONE 2013, 8, e57250. [Google Scholar] [CrossRef]
- Choueiry, F.; Torok, M.; Shakya, R.; Agrawal, K.; Deems, A.; Benner, B.; Hinton, A.; Shaffer, J.; Blaser, B.W.; Noonan, A.M.; et al. CD200 promotes immunosuppression in the pancreatic tumor microenvironment. J. Immunother. Cancer 2020, 8, e000189. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Garcia-Marques, F.; Zhang, C.A.; Lee, J.J.; Nolley, R.; Shen, M.; Hsu, E.C.; Aslan, M.; Koul, K.; Pitteri, S.J.; et al. Discovery of CASP8 as a potential biomarker for high-risk prostate cancer through a high-multiplex immunoassay. Sci. Rep. 2021, 11, 7612. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Peng, Q.; Cheng, D.; Pan, T.; Sun, W.; Wang, H.; Ma, X.; He, R.; Zhang, H.; Cui, Z.; et al. Cancer cell-expressed BTNL2 facilitates tumour immune escape via engagement with IL-17A-producing γδ T cells. Nat. Commun. 2022, 13, 231. [Google Scholar] [CrossRef]
- Foley, K.; Kim, V.; Jaffee, E.; Zheng, L. Current progress in immunotherapy for pancreatic cancer. Cancer Lett. 2016, 381, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.; Hwu, W.J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. New Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.R.; Drake, C.G.; Wollner, I.; Powderly, J.D.; Picus, J.; Sharfman, W.H.; Stankevich, E.; Pons, A.; Salay, T.M.; McMiller, T.L.; et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: Safety, clinical activity, pharmacodynamics, and immunologic correlates. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 3167–3175. [Google Scholar] [CrossRef] [PubMed]
- Seifert, L.; Plesca, I.; Müller, L.; Sommer, U.; Heiduk, M.; von Renesse, J.; Digomann, D.; Glück, J.; Klimova, A.; Weitz, J.; et al. LAG-3-Expressing Tumor-Infiltrating T Cells Are Associated with Reduced Disease-Free Survival in Pancreatic Cancer. Cancers 2021, 13, 1297. [Google Scholar] [CrossRef]
- Andrews, L.P.; Marciscano, A.E.; Drake, C.G.; Vignali, D.A. LAG3 (CD223) as a cancer immunotherapy target. Immunol. Rev. 2017, 276, 80–96. [Google Scholar] [CrossRef]
- Witkiewicz, A.K.; Costantino, C.L.; Metz, R.; Muller, A.J.; Prendergast, G.C.; Yeo, C.J.; Brody, J.R. Genotyping and expression analysis of IDO2 in human pancreatic cancer: A novel, active target. J. Am. Coll. Surg. 2009, 208, 781–787. [Google Scholar] [CrossRef]
- Zhang, J.; Wolfgang, C.L.; Zheng, L. Precision Immuno-Oncology: Prospects of Individualized Immunotherapy for Pancreatic Cancer. Cancers 2018, 10, 39. [Google Scholar] [CrossRef]
- Mace, T.A.; Shakya, R.; Pitarresi, J.R.; Swanson, B.; McQuinn, C.W.; Loftus, S.; Nordquist, E.; Cruz-Monserrate, Z.; Yu, L.; Young, G.; et al. IL-6 and PD-L1 antibody blockade combination therapy reduces tumour progression in murine models of pancreatic cancer. Gut 2018, 67, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Motz, G.T.; Coukos, G. Deciphering and reversing tumor immune suppression. Immunity 2013, 39, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Yu, A.; Othmane, B.; Qiu, D.; Li, H.; Li, C.; Liu, P.; Ren, W.; Chen, M.; Gong, G.; et al. Siglec15 shapes a non-inflamed tumor microenvironment and predicts the molecular subtype in bladder cancer. Theranostics 2021, 11, 3089–3108. [Google Scholar] [CrossRef] [PubMed]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer cell 2021, 39, 154–173. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef]
- Patel, S.P.; Kurzrock, R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015, 14, 847–856. [Google Scholar] [CrossRef]
- Yarchoan, M.; Albacker, L.A.; Hopkins, A.C.; Montesion, M.; Murugesan, K.; Vithayathil, T.T.; Zaidi, N.; Azad, N.S.; Laheru, D.A.; Frampton, G.M.; et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight 2019, 4, e126908. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, H.; Peng, G.; Wang, R.; Yang, F.; Xie, X.; Zhang, J.; Xu, K.; Gu, T.; Yang, X.; Tian, G. Cuprotosis Programmed-Cell-Death-Related lncRNA Signature Predicts Prognosis and Immune Landscape in PAAD Patients. Cells 2022, 11, 3436. https://doi.org/10.3390/cells11213436
Chi H, Peng G, Wang R, Yang F, Xie X, Zhang J, Xu K, Gu T, Yang X, Tian G. Cuprotosis Programmed-Cell-Death-Related lncRNA Signature Predicts Prognosis and Immune Landscape in PAAD Patients. Cells. 2022; 11(21):3436. https://doi.org/10.3390/cells11213436
Chicago/Turabian StyleChi, Hao, Gaoge Peng, Rui Wang, Fengyi Yang, Xixi Xie, Jinhao Zhang, Ke Xu, Tao Gu, Xiaoli Yang, and Gang Tian. 2022. "Cuprotosis Programmed-Cell-Death-Related lncRNA Signature Predicts Prognosis and Immune Landscape in PAAD Patients" Cells 11, no. 21: 3436. https://doi.org/10.3390/cells11213436
APA StyleChi, H., Peng, G., Wang, R., Yang, F., Xie, X., Zhang, J., Xu, K., Gu, T., Yang, X., & Tian, G. (2022). Cuprotosis Programmed-Cell-Death-Related lncRNA Signature Predicts Prognosis and Immune Landscape in PAAD Patients. Cells, 11(21), 3436. https://doi.org/10.3390/cells11213436







