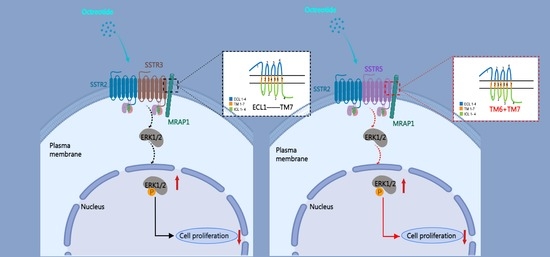
Selective Interactions of Mouse Melanocortin Receptor Accessory Proteins with Somatostatin Receptors
Abstract
1. Introduction
2. Method and Materials
2.1. Plasmids
2.2. Cell Culture and Transfection
2.3. Western-Blot and Co-Immunoprecipitation (CoIP)
2.4. Immunofluorescence
2.5. FACS Analysis
2.6. Surface Expression Measurement
2.7. Annexin/Propidium Iodide (PI) Staining
2.8. MTT Assay
2.9. In Silico and Statistical Analysis
3. Results
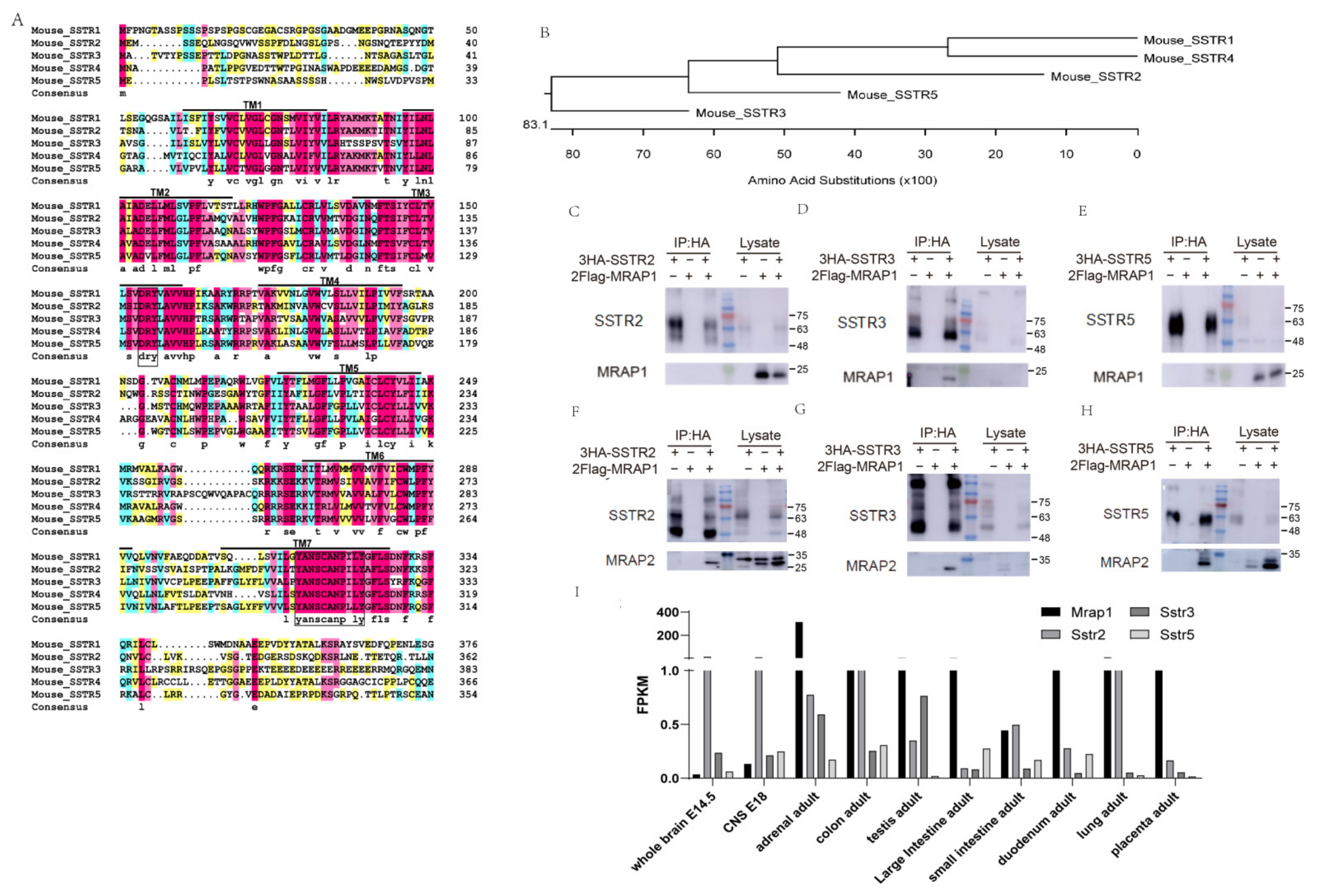
3.1. Homology of SSTRs and Selective Interactions with MRAP Proteins
3.2. Identification of the Binding Regions of MRAP1 with SSTR3 and SSTR5
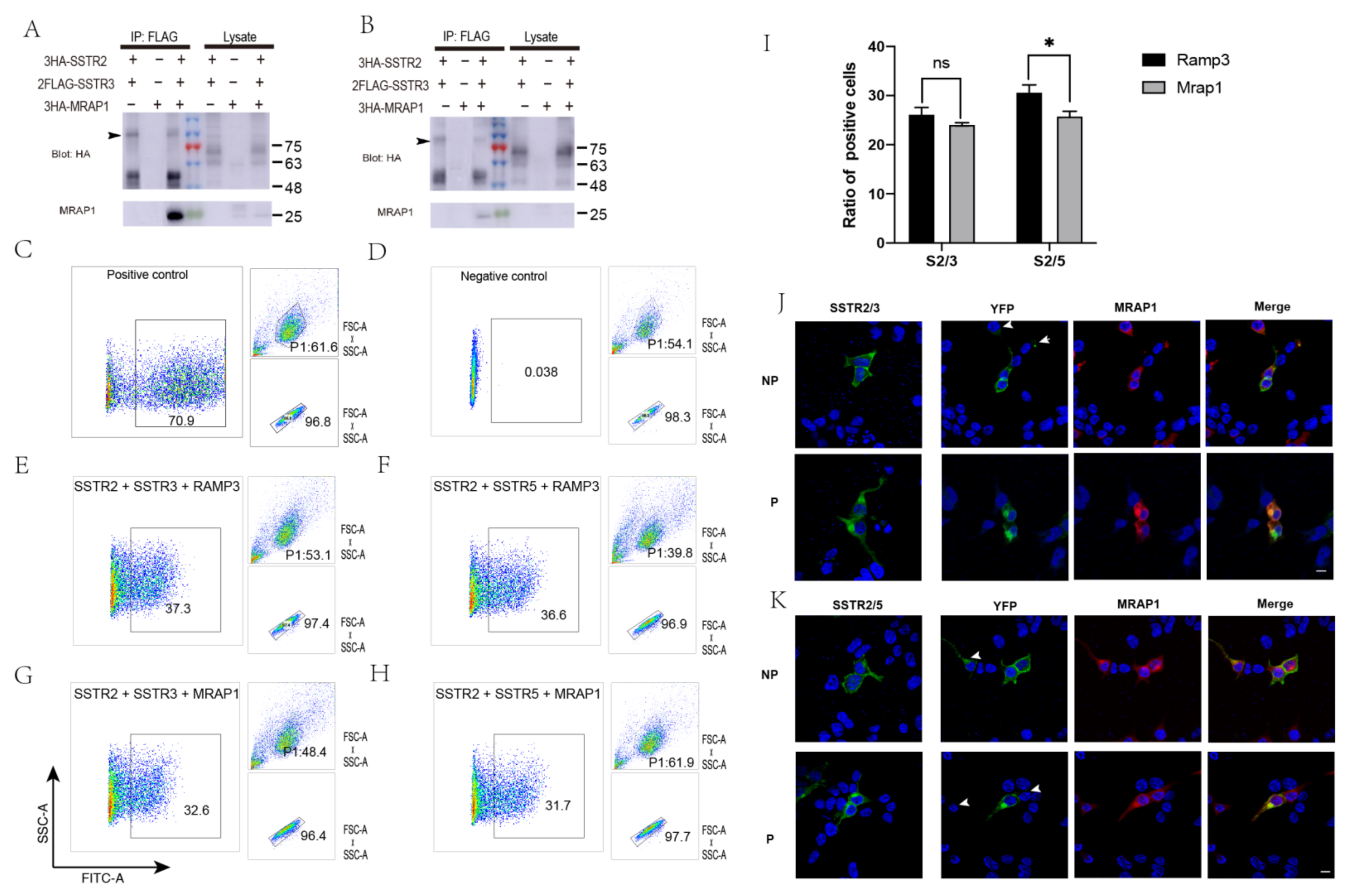
3.3. Inhibition of the Formation of SSTR2/SSTR3, SSTR2/SSTR5 Heterodimers by MRAP1
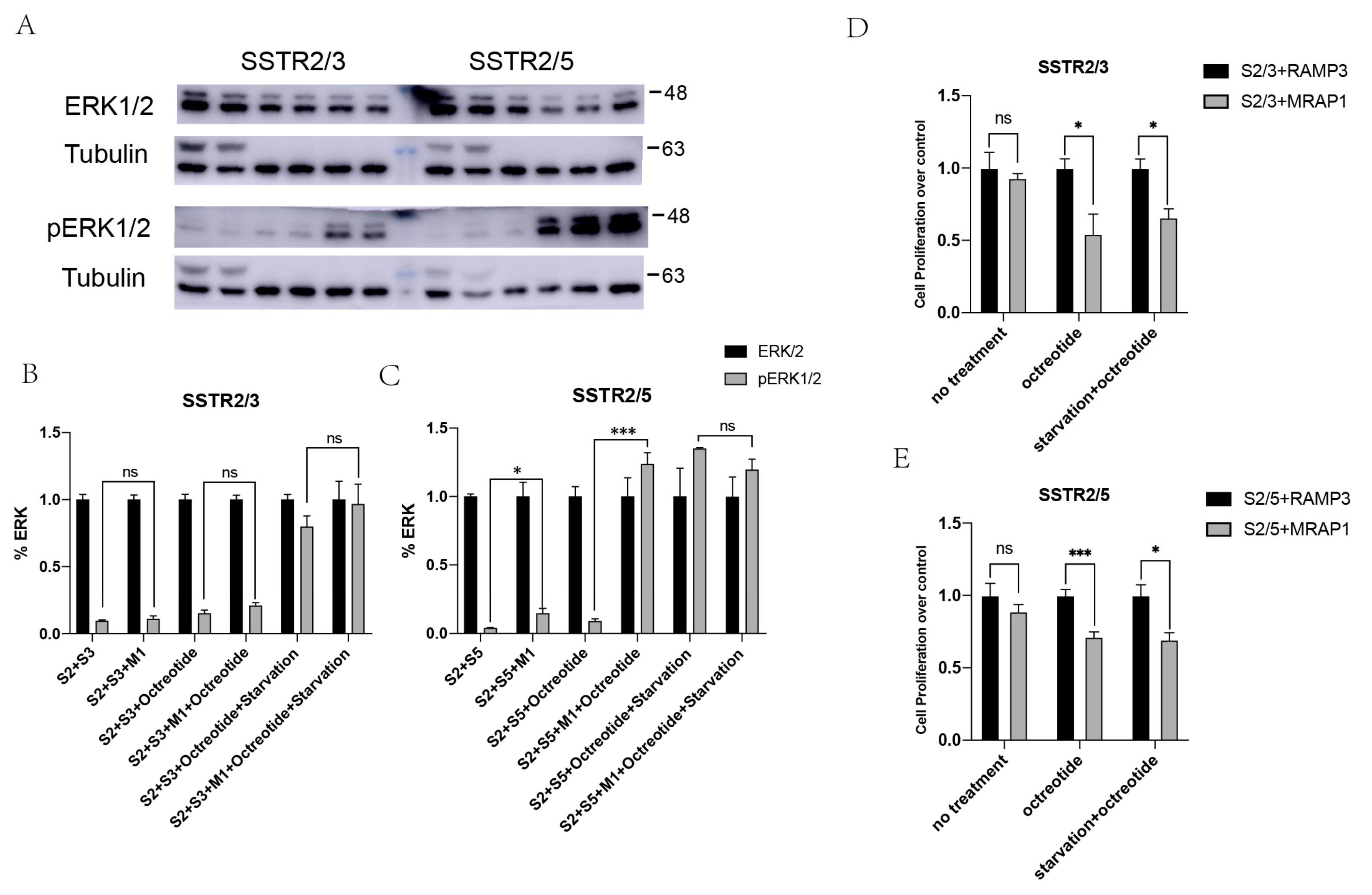
3.4. Enhancement of MAPK Signaling of SSTR2/3 and SSTR2/5 Heterodimers by MRAP1 Protein
3.5. The Role of SSTR2/3 and SSTR2/5 Heterodimers in Cell Apoptosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tzotzas, T.; Papazisis, K.; Perros, P.; Krassas, G.E. Use of Somatostatin Analogues in Obesity. Drugs 2008, 68, 1963–1973. [Google Scholar] [CrossRef]
- Burgos-Ramos, E.; Hervas-Aguilar, A.; Aguado-Llera, D.; Puebla-Jimenez, L.; Hernandez-Pinto, A.M.; Barrios, V.; Arilla-Ferreiro, E. Somatostatin and Alzheimer’s disease. Mol. Cell. Endocrinol. 2008, 286, 104–111. [Google Scholar] [CrossRef]
- Rorsman, P.; Huising, M.O. The somatostatin-secreting pancreatic delta-cell in health and disease. Nat. Rev. Endocrinol. 2018, 14, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Shahbaz, M.; Fang, R.L.; Zhang, X.; Liang, B.J.; Wang, C.; He, Z.B.; Niu, J. mRNA expression of somatostatin receptor subtypes SSTR-2, SSTR-3, and SSTR-5 and its significance in pancreatic cancer. World J. Surg. Oncol. 2015, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Moller, L.N.; Stidsen, C.E.; Hartmann, B.; Holst, J.J. Somatostatin receptors. Biochim. Biophys. Acta-Biomembr. 2003, 1616, 1–84. [Google Scholar] [CrossRef]
- Weckbecker, G.; Lewis, I.; Albert, R.; Schmid, H.A.; Hoyer, D.; Bruns, C. Opportunities in somatostatin research: Biological, chemical and therapeutic aspects. Nat. Rev. Drug Discov. 2003, 2, 999–1017. [Google Scholar] [CrossRef] [PubMed]
- Patel, Y.C. Somatostatin and its receptor family. Front. Neuroendocrinol. 1999, 20, 157–198. [Google Scholar] [CrossRef] [PubMed]
- Olias, G.; Viollet, C.; Kusserow, H.; Epelbaum, J.; Meyerhof, W. Regulation and function of somatostatin receptors. J. Neurochem. 2004, 89, 1057–1091. [Google Scholar] [CrossRef]
- Dal Monte, M.; Petrucci, C.; Cozzi, A.; Allen, J.P.; Bagnoli, P. Somatostatin inhibits potassium-evoked glutamate release by activation of the sst2 somatostatin receptor in the mouse retina. Naunyn-Schmiedebergs Arch. Pharmacol. 2003, 367, 188–192. [Google Scholar] [CrossRef]
- Dutar, P.; Vaillend, C.; Viollet, C.; Billard, J.M.; Potier, B.; Carlo, A.S.; Ungerer, A.; Epelbaum, J. Spatial learning and synaptic hippocampal plasticity in type 2 somatostatin receptor knock-out mice. Neuroscience 2002, 112, 455–466. [Google Scholar] [CrossRef]
- Moneta, D.; Richichi, C.; Aliprandi, M.; Dournaud, P.; Dutar, P.; Billard, J.M.; Carlo, A.S.; Viollet, C.; Hannon, J.P.; Fehlmann, D.; et al. Somatostatin receptor subtypes 2 and 4 affect seizure susceptibility and hippocampal excitatory neurotransmission in mice. Eur. J. Neurosci. 2002, 16, 843–849. [Google Scholar] [CrossRef]
- Grant, M.; Alturaihi, H.; Jaquet, P.; Collier, B.; Kumar, U. Cell growth inhibition and functioning of human somatostatin receptor type 2 are modulated by receptor heterodimerization. Mol. Endocrinol. 2008, 22, 2278–2292. [Google Scholar] [CrossRef] [PubMed]
- Baragli, A.; Alturaihi, H.; Watt, H.L.; Abdallah, A.; Kumar, U. Heterooligomerization of human dopamine receptor 2 and somatostatin receptor 2—Co-immunoprecipitation and fluorescence resonance energy transfer analysis. Cell. Signal. 2007, 19, 2304–2316. [Google Scholar] [CrossRef] [PubMed]
- Duran-Prado, M.; Bucharles, C.; Gonzalez, B.J.; Vazquez-Martinez, R.; Martinez-Fuentes, A.J.; Garcia-Navarro, S.; Rhodes, S.J.; Vaudry, H.; Malagon, M.M.; Castano, J.P. Porcine somatostatin receptor 2 displays typical pharmacological sst2 features but unique dynamics of homodimerization and internalization. Endocrinology 2007, 148, 411–421. [Google Scholar] [CrossRef]
- Grant, M.; Collier, B.; Kumar, U. Agonist-dependent dissociation of human somatostatin receptor 2 dimers—A role in receptor trafficking. J. Biol. Chem. 2004, 279, 36179–36183. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.; Patel, R.C.; Kumar, U. The role of subtype-specific ligand binding and the C-tail domain in dimer formation of human somatostatin receptors. J. Biol. Chem. 2004, 279, 38636–38643. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.C.; Kumar, U.; Lamb, D.C.; Eid, J.S.; Rocheville, M.; Grant, M.; Rani, A.; Hazlett, T.; Patel, S.C.; Gratton, E.; et al. Ligand binding to somatostatin receptors induces receptor-specific oligomer formation in live cells. Proc. Natl. Acad. Sci. USA 2002, 99, 3294–3299. [Google Scholar] [CrossRef]
- Rocheville, M.; Lange, D.C.; Kumar, U.; Sasi, R.; Patel, R.C.; Patel, Y.C. Subtypes of the somatostatin receptor assemble as functional homo- and heterodimers. J. Biol. Chem. 2000, 275, 7862–7869. [Google Scholar] [CrossRef]
- Pfeiffer, M.; Koch, T.; Schroder, H.; Klutzny, M.; Kirscht, S.; Kreienkamp, H.J.; Hollt, V.; Schulz, S. Homo- and heterodimerization of somatostatin receptor subtypes—Inactivation of sst3 receptor function by heterodimerization with sst2A. J. Biol. Chem. 2001, 276, 14027–14036. [Google Scholar] [CrossRef]
- Sharif, N.; Gendron, L.; Wowchuk, J.; Sarret, P.; Mazella, J.; Beaudet, A.; Stroh, T. Coexpression of somatostatin receptor subtype 5 affects internalization and trafficking of somatostatin receptor subtype 2. Endocrinology 2007, 148, 2095–2105. [Google Scholar] [CrossRef][Green Version]
- Ben-Shlomo, A.; Wawrowsky, K.A.; Proekt, I.; Wolkenfeld, N.M.; Ren, S.G.; Taylor, J.; Culler, M.D.; Melmed, S. Somatostatin receptor type 5 modulates somatostatin receptor type 2 regulation of adrenocorticotropin secretion. J. Biol. Chem. 2005, 280, 24011–24021. [Google Scholar] [CrossRef]
- Ren, S.G.; Kim, S.; Taylor, J.; Dong, J.S.; Moreau, J.P.; Culler, M.D.; Melmed, S. Suppression of rat and human growth hormone and prolactin secretion by a novel somatostatin/dopaminergic chimeric ligand. J. Clin. Endocrinol. Metab. 2003, 88, 5414–5421. [Google Scholar] [CrossRef][Green Version]
- Gruszka, A.; Ren, S.G.; Dong, J.; Culler, M.D.; Melmed, S. Regulation of growth hormone and prolactin gene expression and secretion by chimeric somatostatin-dopamine molecules. Endocrinology 2007, 148, 6107–6114. [Google Scholar] [CrossRef][Green Version]
- Saveanu, A.; Lavaque, E.; Gunz, G.; Barlier, A.; Kim, S.; Taylor, J.E.; Culler, M.D.; Enjalbert, A.; Jaquet, P. Demonstration of enhanced potency of a chimeric somatostatin-dopamine molecule, BIM-23A387, in suppressing growth hormone and prolactin secretion from human pituitary somatotroph adenoma cells. J. Clin. Endocrinol. Metab. 2002, 87, 5545–5552. [Google Scholar] [CrossRef] [PubMed]
- Somvanshi, R.K.; Billova, S.; Kharmate, G.; Rajput, P.S.; Kumar, U. C-tail mediated modulation of somatostatin receptor type-4 homo- and heterodimerizations and signaling. Cell. Signal. 2009, 21, 1396–1414. [Google Scholar] [CrossRef] [PubMed]
- War, S.A.; Somvanshi, R.K.; Kumar, U. Somatostatin receptor-3 mediated intracellular signaling and apoptosis is regulated by its cytoplasmic terminal. Biochim. Biophys. Acta-Mol. Cell Res. 2011, 1813, 390–402. [Google Scholar] [CrossRef]
- Watt, H.L.; Kharmate, G.D.; Kumar, U. Somatostatin receptors 1 and 5 heterodimerize with epidermal growth factor receptor: Agonist-dependent modulation of the downstream MAPK signalling pathway in breast cancer cells. Cell. Signal. 2009, 21, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.; Ji, R.L.; Huang, L.; Fan, S.Y.; Liu, T.; Liu, S.J.; Tao, Y.X. Regulation of Melanocortin-4 Receptor Pharmacology by Two Isoforms of Melanocortin Receptor Accessory Protein 2 in Topmouth Culter (Culter alburnus). Front. Endocrinol. 2020, 11, 538. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Pi, L.; Lei, X.; Li, L.; Xu, J.; Kuang, Z.; Zhang, C.; Li, L.; Zhang, C. Functional Characterization of the Internal Symmetry of MRAP2 Antiparallel Homodimer. Front. Endocrinol. 2021, 12, 750797. [Google Scholar] [CrossRef]
- Xu, Y.; Li, L.; Zheng, J.; Wang, M.; Jiang, B.; Zhai, Y.; Lu, L.; Zhang, C.; Kuang, Z.; Yang, X.; et al. Pharmacological modulation of the cAMP signaling of two isoforms of melanocortin-3 receptor by melanocortin receptor accessory proteins in the tetrapod Xenopus laevis. Endocr. Connect. 2021, 10, 1477–1488. [Google Scholar] [CrossRef]
- Wang, M.; Zhai, Y.; Lu, L.; Zhang, C.; Li, N.; Xue, S.; Cheng, D.; Fu, S.; Liu, Q.; Zhang, C. Elucidation of the dimeric interplay of dual MRAP2 proteins in the zebrafish. J. Cell. Physiol. 2021, 236, 6472–6480. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, Y.; Zhu, M.; Xu, B.; Guo, W.; Lyu, Y.; Zhang, C. Pharmacological modulation of melanocortin-4 receptor by melanocortin receptor accessory protein 2 in Nile tilapia. Gen. Comp. Endocrinol. 2019, 282, 113219. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Xu, B.; Wang, M.; Liu, S.; Zhang, Y.; Zhang, C. Pharmacological modulation of MRAP2 protein on melanocortin receptors in the sea lamprey. Endocr. Connect. 2019, 8, 378–388. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, M.; Chen, Y.; Zhang, C. Pharmacological modulation of two melanocortin-5 receptors by MRAP2 proteins in zebrafish. J. Mol. Endocrinol. 2018, 62, 27–36. [Google Scholar] [CrossRef]
- Sebag, J.A.; Hinkle, P.M. Opposite Effects of the Melanocortin-2 (MC2) Receptor Accessory Protein MRAP on MC2 and MC5 Receptor Dimerization and Trafficking. J. Biol. Chem. 2009, 284, 22641–22648. [Google Scholar] [CrossRef]
- Chaly, A.L.; Srisai, D.; Gardner, E.E.; Sebag, J.A. The Melanocortin Receptor Accessory Protein 2 promotes food intake through inhibition of the Prokineticin Receptor-1. eLife 2016, 5, e12397. [Google Scholar] [CrossRef]
- Rouault, A.A.J.; Lee, A.A.; Sebag, J.A. Regions of MRAP2 required for the inhibition of orexin and prokineticin receptor signaling. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2017, 1864, 2322–2329. [Google Scholar] [CrossRef]
- Srisai, D.; Yin, T.C.; Lee, A.A.; Rouault, A.A.J.; Pearson, N.A.; Grobe, J.L.; Sebag, J.A. MRAP2 regulates ghrelin receptor signaling and hunger sensing. Nat. Commun. 2017, 8, 713. [Google Scholar] [CrossRef]
- Rouault, A.A.J.; Rosselli-Murai, L.K.; Hernandez, C.C.; Gimenez, L.E.; Tall, G.G.; Sebag, J.A. The GPCR accessory protein MRAP2 regulates both biased signaling and constitutive activity of the ghrelin receptor GHSR1a. Sci. Signal. 2020, 13, eaax4569. [Google Scholar] [CrossRef]
- Yang, L.K.; Hou, Z.S.; Tao, Y.X. Biased signaling in naturally occurring mutations of G protein-coupled receptors associated with diverse human diseases. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2021, 1867, 165973. [Google Scholar] [CrossRef] [PubMed]
- War, S.A.; Kumar, U. Coexpression of human somatostatin receptor-2 (SSTR2) and SSTR3 modulates antiproliferative signaling and apoptosis. J. Mol. Signal. 2012, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Pages, P.; Benali, N.; Saint-Laurent, N.; Esteve, J.P.; Schally, A.V.; Tkaczuk, J.; Vaysse, N.; Susini, C.; Buscail, L. sst2 somatostatin receptor mediates cell cycle arrest and induction of p27Kip1—Evidence for the role of SHP-1. J. Biol. Chem. 1999, 274, 15186–15193. [Google Scholar] [CrossRef] [PubMed]
- Lahlou, H.; Saint-Laurent, N.; Esteve, J.P.; Eychene, A.; Pradayrol, L.; Pyronnet, S.; Susini, C. sst2 somatostatin receptor inhibits cell proliferation through Ras-, Rap1-, and B-Raf-dependent ERK2 activation. J. Biol. Chem. 2003, 278, 39356–39371. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Lu, L.; Wang, M.; Zhai, Y.; Tai, X.; Dilimulati, D.; Lei, X.; Xu, J.; Zhang, C.; et al. Identification of novel GPCR partners of the central melanocortin signaling. Mol. Metab. 2021, 53, 101317. [Google Scholar] [CrossRef]
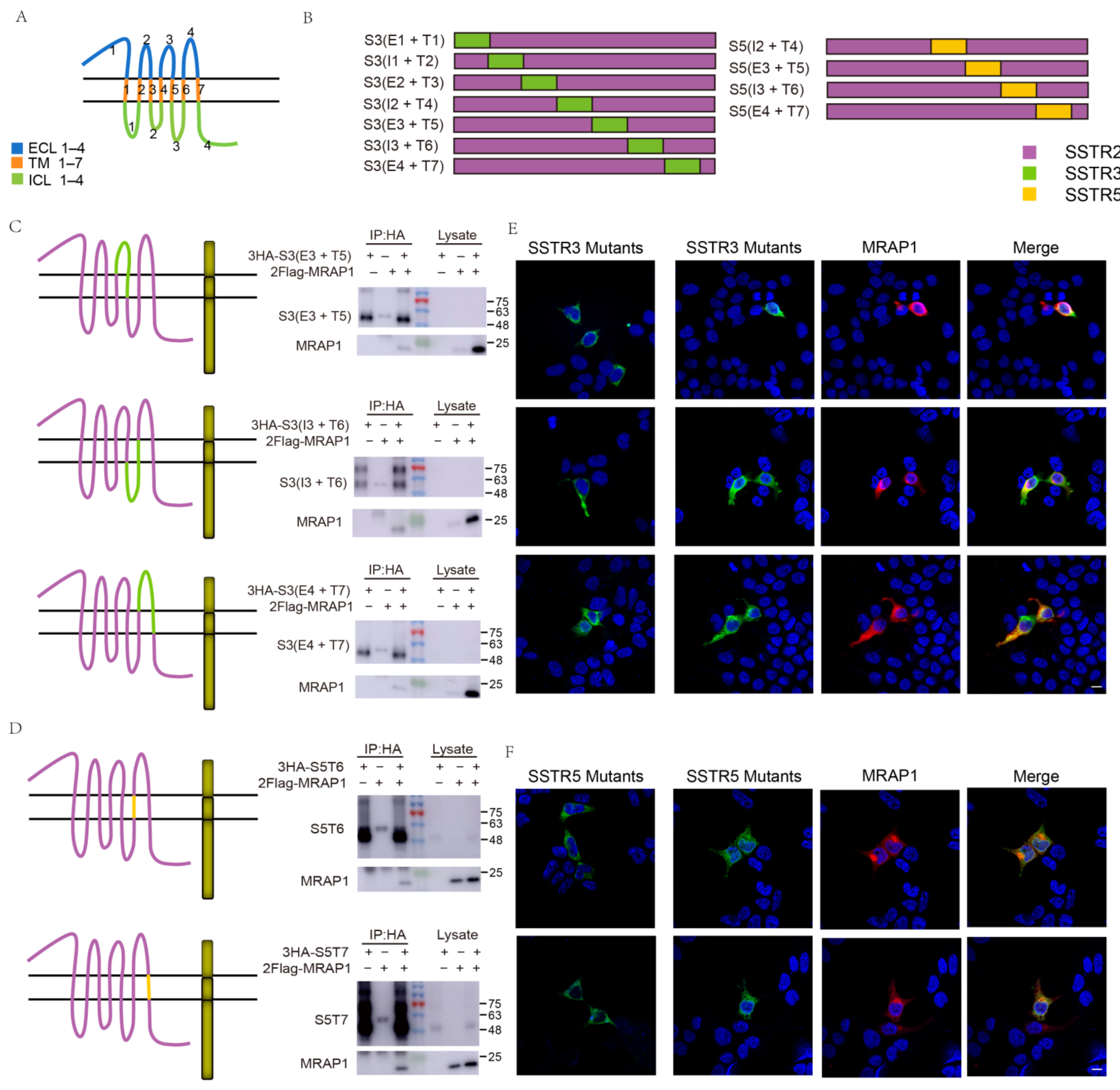
| E1 | T1 | I1 | T2 | E2 | T3 | I2 | T4 | E3 | T5 | I3 | T6 | E4 | T7 | I4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SSTR1 | + | ||||||||||||||
| + | + | ||||||||||||||
| − | + | + | + | + | + | + | |||||||||
| SSTR3 | + | ||||||||||||||
| + | + | ||||||||||||||
| + (weak) | + | + (weak) | − | + | + | + | |||||||||
| SSTR5 | + | ||||||||||||||
| − | + | ||||||||||||||
| − | − | + | + | ||||||||||||
| − | + | + | |||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Xu, J.; Lei, X.-W.; Zhang, C.; Liu, S.-Y.; Jin, L.-N.; Zhang, C. Selective Interactions of Mouse Melanocortin Receptor Accessory Proteins with Somatostatin Receptors. Cells 2022, 11, 267. https://doi.org/10.3390/cells11020267
Wang M, Xu J, Lei X-W, Zhang C, Liu S-Y, Jin L-N, Zhang C. Selective Interactions of Mouse Melanocortin Receptor Accessory Proteins with Somatostatin Receptors. Cells. 2022; 11(2):267. https://doi.org/10.3390/cells11020267
Chicago/Turabian StyleWang, Meng, Jing Xu, Xiao-Wei Lei, Cong Zhang, Shang-Yun Liu, Li-Na Jin, and Chao Zhang. 2022. "Selective Interactions of Mouse Melanocortin Receptor Accessory Proteins with Somatostatin Receptors" Cells 11, no. 2: 267. https://doi.org/10.3390/cells11020267
APA StyleWang, M., Xu, J., Lei, X.-W., Zhang, C., Liu, S.-Y., Jin, L.-N., & Zhang, C. (2022). Selective Interactions of Mouse Melanocortin Receptor Accessory Proteins with Somatostatin Receptors. Cells, 11(2), 267. https://doi.org/10.3390/cells11020267