Stem Cell Factor SOX9 Interacts with a Cell Death Regulator RIPK1 and Results in Escape of Cancer Stem Cell Death
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines, Reagents, and Chemicals
2.2. Immunohistochemistry and Quantification
2.3. Immunofluorescence (IF)
2.4. Quantitative Real-Time PCR
2.5. Immunoblotting and Co-Immunoprecipitation
2.6. Cell Cycle Assay
2.7. Apoptosis
2.8. Viability Assay/Proliferation Assay
2.9. Xenograft Experiments
2.10. Sphere Formation Assay
2.11. Invasion Assay
2.12. ALDEFLUOR Assay
2.13. Yeast Two-Hybrid (Y2H) Screening and Quantitation of Interaction
2.14. Statistical Analysis
3. Results
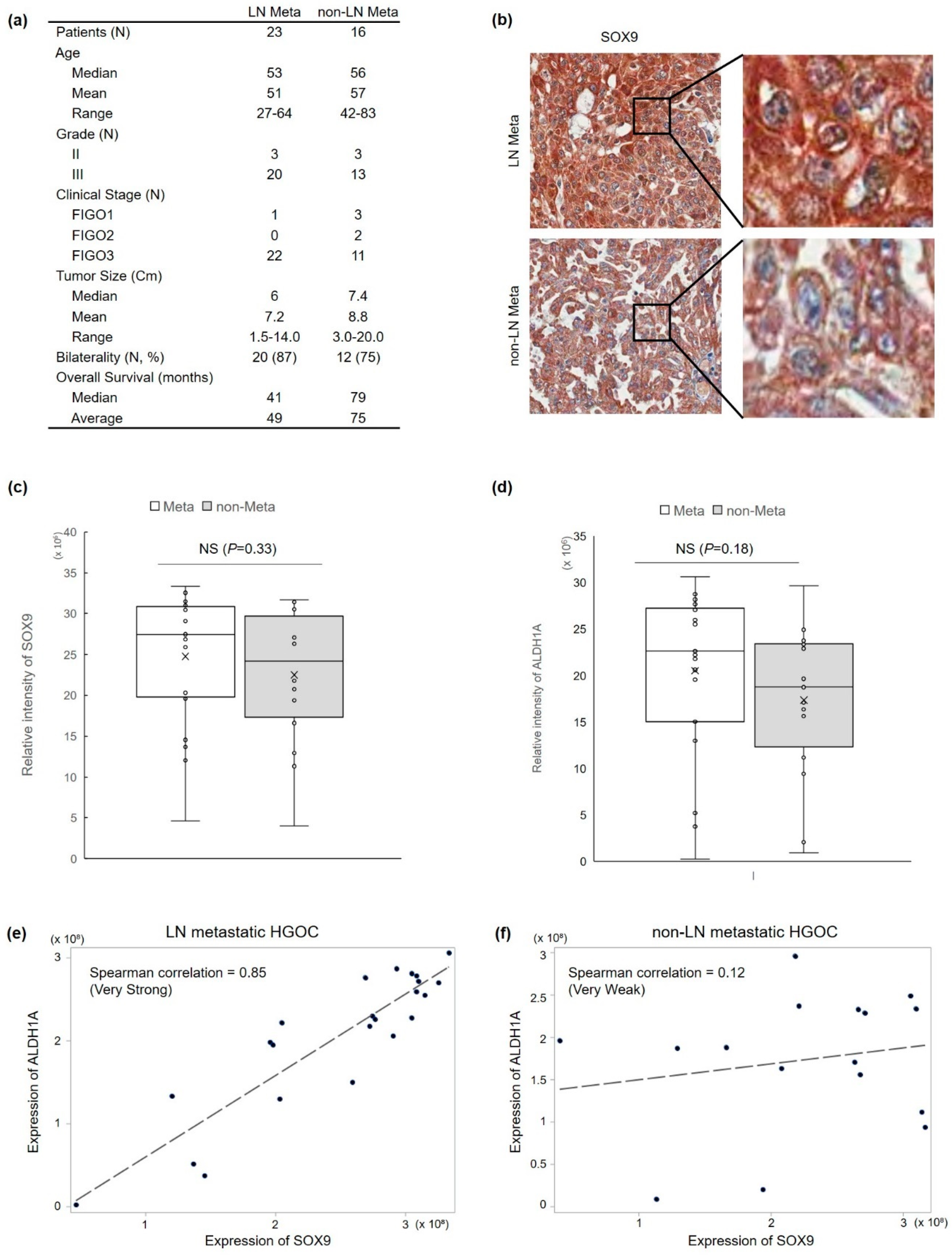
3.1. SOX9 Protein Expression Is Correlated with Lymph Node Metastasis and ALDH1 Protein Expression in HGOC
3.2. SOX9 Silencing Abrogates Platinum Resistance and Tumorigenesis in HGOC Cells
3.3. Silencing of Sox9 Abrogated the Cancer Stemness Properties in Ovarian Cancer Cells
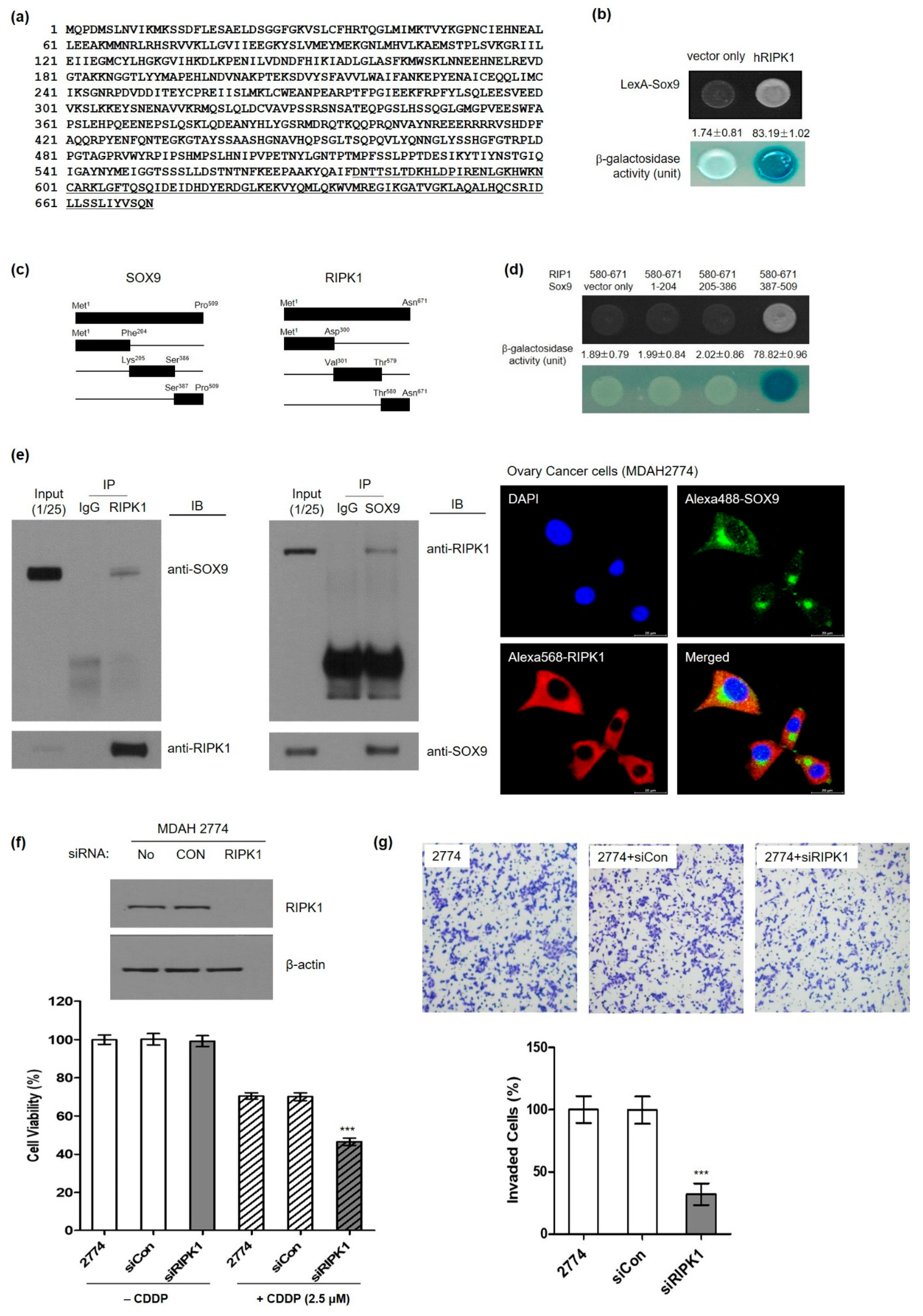
3.4. Identification of Serine/Threonine-Protein Kinase 1 (RIPK1) as a Novel SOX9-Interaction Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Binju, M.; Amaya-Padilla, M.A.; Wan, G.; Gunosewoyo, H.; Suryo Rahmanto, Y.; Yu, Y. Therapeutic inducers of apoptosis in ovarian cancer. Cancers 2019, 11, 1786. [Google Scholar] [CrossRef]
- Eisenhauer, E.L.; Abu-Rustum, N.R.; Sonoda, Y.; Aghajanian, C.; Barakat, R.R.; Chi, D.S. The effect of maximal surgical cytoreduction on sensitivity to platinum-taxane chemotherapy and subsequent survival in patients with advanced ovarian cancer. Gynecol. Oncol. 2008, 108, 276–281. [Google Scholar] [CrossRef]
- Motohara, T.; Yoshida, G.J.; Katabuchi, H. The hallmarks of ovarian cancer stem cells and niches: Exploring their harmonious interplay in therapy resistance. Semin. Cancer Biol. 2021, 77, 182–193. [Google Scholar] [CrossRef]
- Beck, B.; Blanpain, C. Unravelling cancer stem cell potential. Nat. Rev. Cancer 2013, 13, 727–738. [Google Scholar] [CrossRef]
- O’Connell, E.; Reynolds, I.S.; McNamara, D.A.; Burke, J.P.; Prehn, J.H.M. Resistance to cell death in mucinous colorectal cancer—A review. Cancers 2021, 13, 1389. [Google Scholar] [CrossRef]
- Della Torre, L.; Nebbioso, A.; Stunnenberg, H.G.; Martens, J.H.A.; Carafa, V.; Altucci, L. The role of necroptosis: Biological relevance and its involvement in cancer. Cancers 2021, 13, 684. [Google Scholar] [CrossRef]
- Chefetz, I.; Grimley, E.; Yang, K.; Hong, L.; Vinogradova, E.V.; Suciu, R.; Kovalenko, I.; Karnak, D.; Morgan, C.A.; Chtcherbinine, M.; et al. A Pan-ALDH1A inhibitor induces necroptosis in ovarian cancer stem-like cells. Cell Rep. 2019, 26, 3061–3075.e6. [Google Scholar] [CrossRef]
- McMullen, M.; Madariaga, A.; Lheureux, S. New approaches for targeting platinum-resistant ovarian cancer. Semin. Cancer Biol. 2020, 77, 167–181. [Google Scholar] [CrossRef]
- Mohammad, R.M.; Muqbil, I.; Lowe, L.; Yedjou, C.; Hsu, H.Y.; Lin, L.T.; Siegelin, M.D.; Fimognari, C.; Kumar, N.B.; Dou, Q.P.; et al. Broad targeting of resistance to apoptosis in cancer. Semin. Cancer Biol. 2015, 35, S78–S103. [Google Scholar] [CrossRef]
- Chen, J.; Kos, R.; Garssen, J.; Redegeld, F. Molecular insights into the mechanism of necroptosis: The necrosome as a potential therapeutic target. Cells 2019, 8, 1486. [Google Scholar] [CrossRef]
- Gong, Y.; Fan, Z.; Luo, G.; Yang, C.; Huang, Q.; Fan, K.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. The role of necroptosis in cancer biology and therapy. Mol. Cancer 2019, 18, 100. [Google Scholar] [CrossRef]
- Sekido, R.; Lovell-Badge, R. Sex determination and SRY: Down to a wink and a nudge? Trends Genet. 2009, 25, 19–29. [Google Scholar] [CrossRef]
- Matheu, A.; Collado, M.; Wise, C.; Manterola, L.; Cekaite, L.; Tye, A.J.; Canamero, M.; Bujanda, L.; Schedl, A.; Cheah, K.S.; et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012, 72, 1301–1315. [Google Scholar] [CrossRef]
- Aguilar-Medina, M.; Avendano-Felix, M.; Lizarraga-Verdugo, E.; Bermudez, M.; Romero-Quintana, J.G.; Ramos-Payan, R.; Ruiz-Garcia, E.; Lopez-Camarillo, C. SOX9 stem-cell factor: Clinical and functional relevance in cancer. J. Oncol. 2019, 2019, 6754040. [Google Scholar] [CrossRef]
- Sherman-Samis, M.; Onallah, H.; Holth, A.; Reich, R.; Davidson, B. SOX2 and SOX9 are markers of clinically aggressive disease in metastatic high-grade serous carcinoma. Gynecol. Oncol. 2019, 153, 651–660. [Google Scholar] [CrossRef]
- Sun, L.; Mathews, L.A.; Cabarcas, S.M.; Zhang, X.; Yang, A.; Zhang, Y.; Young, M.R.; Klarmann, K.D.; Keller, J.R.; Farrar, W.L. Epigenetic regulation of SOX9 by the NF-κB signaling pathway in pancreatic cancer stem cells. Stem Cells 2013, 31, 1454–1466. [Google Scholar] [CrossRef]
- Park, K.; Kim, K.; Rho, S.B.; Choi, K.; Kim, D.; Oh, S.H.; Park, J.; Lee, S.H.; Lee, J.H. Homeobox Msx1 interacts with p53 tumor suppressor and inhibits tumor growth by inducing apoptosis. Cancer Res. 2005, 65, 749–757. [Google Scholar]
- Rho, S.B.; Lee, K.H.; Kim, J.W.; Shiba, K.; Jo, Y.J.; Kim, S. Interaction between human tRNA synthetases involves repeated sequence elements. Proc. Natl. Acad. Sci. USA 1996, 93, 10128–10133. [Google Scholar] [CrossRef]
- Voronkova, M.A.; Rojanasakul, L.W.; Kiratipaiboon, C.; Rojanasakul, Y. The SOX9-aldehyde dehydrogenase axis determines resistance to chemotherapy in Non-Small-Cell lung cancer. Mol. Cell Biol. 2020, 40, e00307-19. [Google Scholar] [CrossRef]
- Panda, M.; Tripathi, S.K.; Biswal, B.K. SOX9: An emerging driving factor from cancer progression to drug resistance. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188517. [Google Scholar] [CrossRef]
- Toledo-Guzmán, M.E.; Hernández, M.I.; Gómez-Gallegos, Á.A.; Ortiz-Sánchez, E. ALDH as a stem cell marker in solid tumors. Curr. Stem. Cell Res. Ther. 2019, 14, 375–388. [Google Scholar] [CrossRef]
- Chakravarty, G.; Moroz, K.; Makridakis, N.M.; Lloyd, S.A.; Galvez, S.E.; Canavello, P.R.; Lacey, M.R.; Agrawal, K.; Mondal, D. Prognostic significance of cytoplasmic SOX9 in invasive ductal carcinoma and metastatic breast cancer. Exp. Biol. Med. 2011, 236, 145–155. [Google Scholar] [CrossRef]
- Chakravarty, G.; Rider, B.; Mondal, D. Cytoplasmic compartmentalization of SOX9 abrogates the growth arrest response of breast cancer cells that can be rescued by trichostatin A treatment. Cancer Biol. Ther. 2011, 11, 71–83. [Google Scholar] [CrossRef][Green Version]
- Ma, Y.; Shepherd, J.; Zhao, D.; Bollu, L.R.; Tahaney, W.M.; Hill, J.; Zhang, Y.; Mazumdar, A.; Brown, P.H. SOX9 Is Essential for Triple-Negative Breast Cancer Cell Survival and Metastasis. Mol. Cancer Res. 2020, 18, 1825–1838. [Google Scholar] [CrossRef]
- Liu, L.; Lalaoui, N. 25 years of research put RIPK1 in the clinic. Semin Cell Dev. Biol. 2021, 109, 86–95. [Google Scholar] [CrossRef]
- Woo, Y.; Lee, H.J.; Jung, Y.M.; Jung, Y.J. Regulated necrotic cell death in alternative tumor therapeutic strategies. Cells 2020, 9, 2709. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, Q.; Li, J.; Zhang, K.; Qin, J.; Zhao, J.; Sun, Q.; Wang, Z.; Wartmann, T.; Jauch, K.W.; et al. Targeting cancer stem cells and their niche: Perspectives for future therapeutic targets and strategies. Semin. Cancer Biol. 2018, 53, 139–155. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, M.; Son, C.; Rho, S.B.; Kim, M.; Park, K.; Song, S.Y. Stem Cell Factor SOX9 Interacts with a Cell Death Regulator RIPK1 and Results in Escape of Cancer Stem Cell Death. Cells 2022, 11, 363. https://doi.org/10.3390/cells11030363
Oh M, Son C, Rho SB, Kim M, Park K, Song SY. Stem Cell Factor SOX9 Interacts with a Cell Death Regulator RIPK1 and Results in Escape of Cancer Stem Cell Death. Cells. 2022; 11(3):363. https://doi.org/10.3390/cells11030363
Chicago/Turabian StyleOh, Mijung, Chaeyeon Son, Seung Bae Rho, Minjeong Kim, Kyoungsook Park, and Sang Yong Song. 2022. "Stem Cell Factor SOX9 Interacts with a Cell Death Regulator RIPK1 and Results in Escape of Cancer Stem Cell Death" Cells 11, no. 3: 363. https://doi.org/10.3390/cells11030363
APA StyleOh, M., Son, C., Rho, S. B., Kim, M., Park, K., & Song, S. Y. (2022). Stem Cell Factor SOX9 Interacts with a Cell Death Regulator RIPK1 and Results in Escape of Cancer Stem Cell Death. Cells, 11(3), 363. https://doi.org/10.3390/cells11030363






