Of Flies and Men—The Discovery of TLRs
Abstract
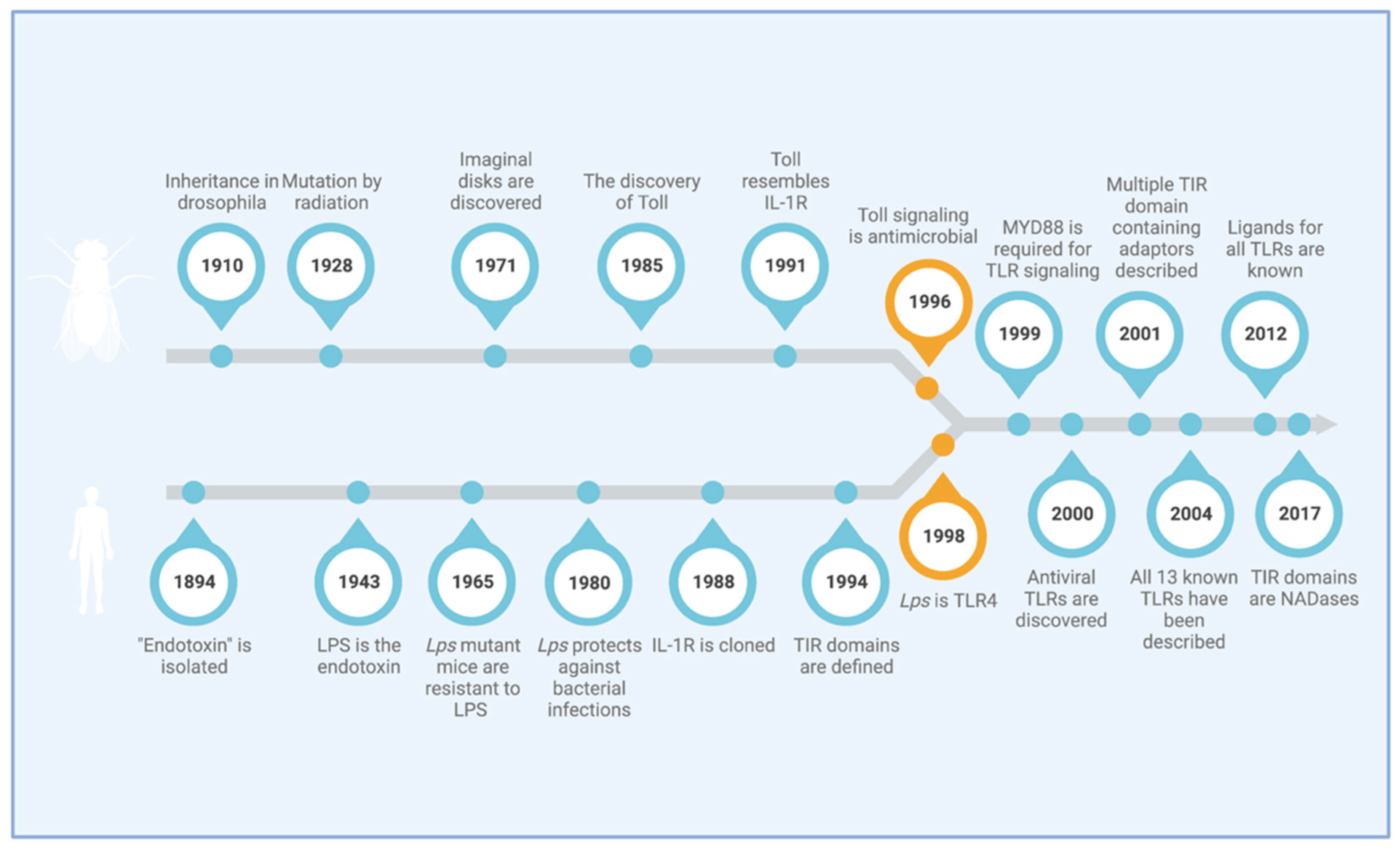
1. Introduction
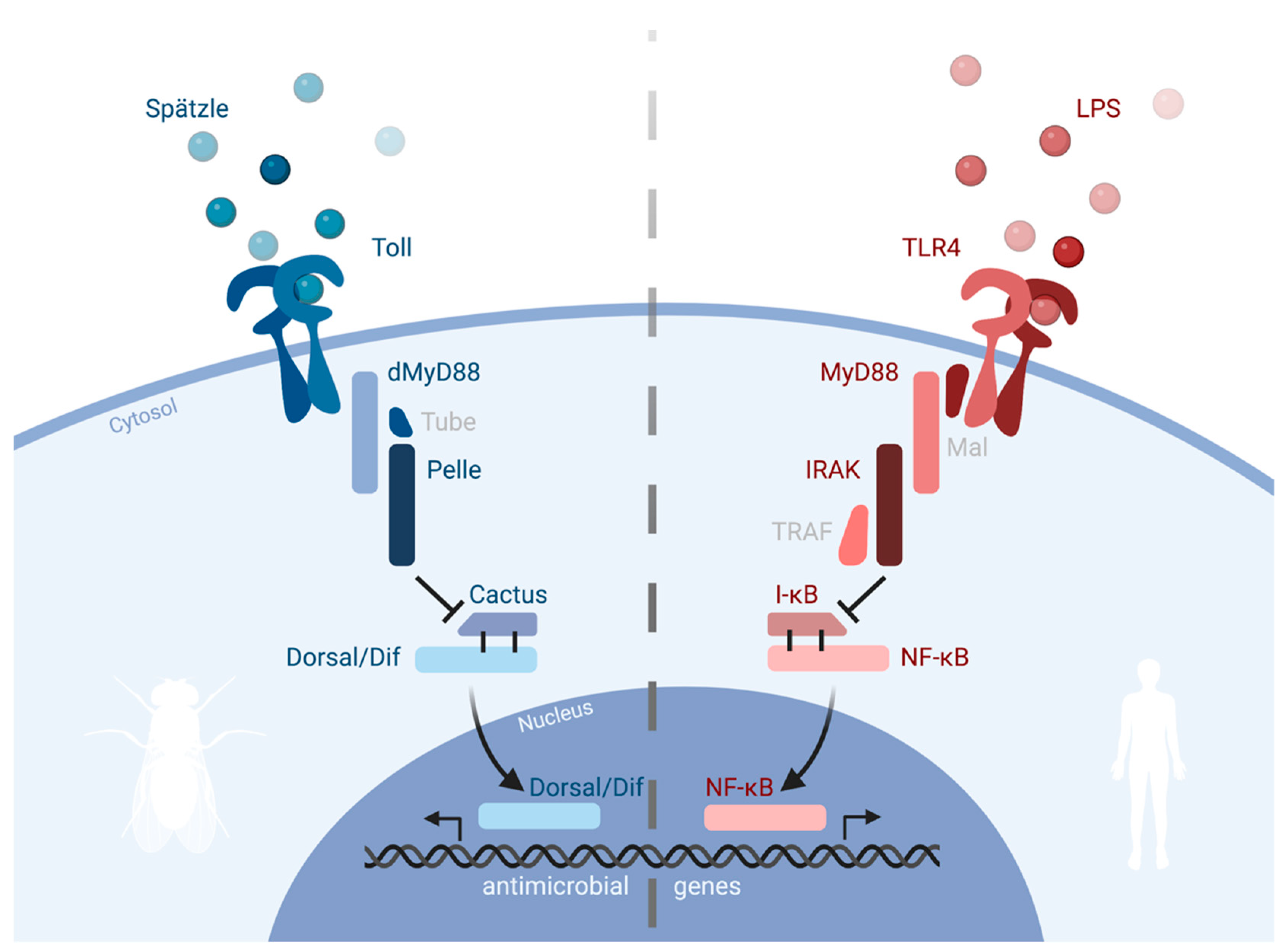
2. Toll and Pathways in Drosophila
3. Toll Has Antimicrobial Functions
4. Innate Immune Signaling and LPS
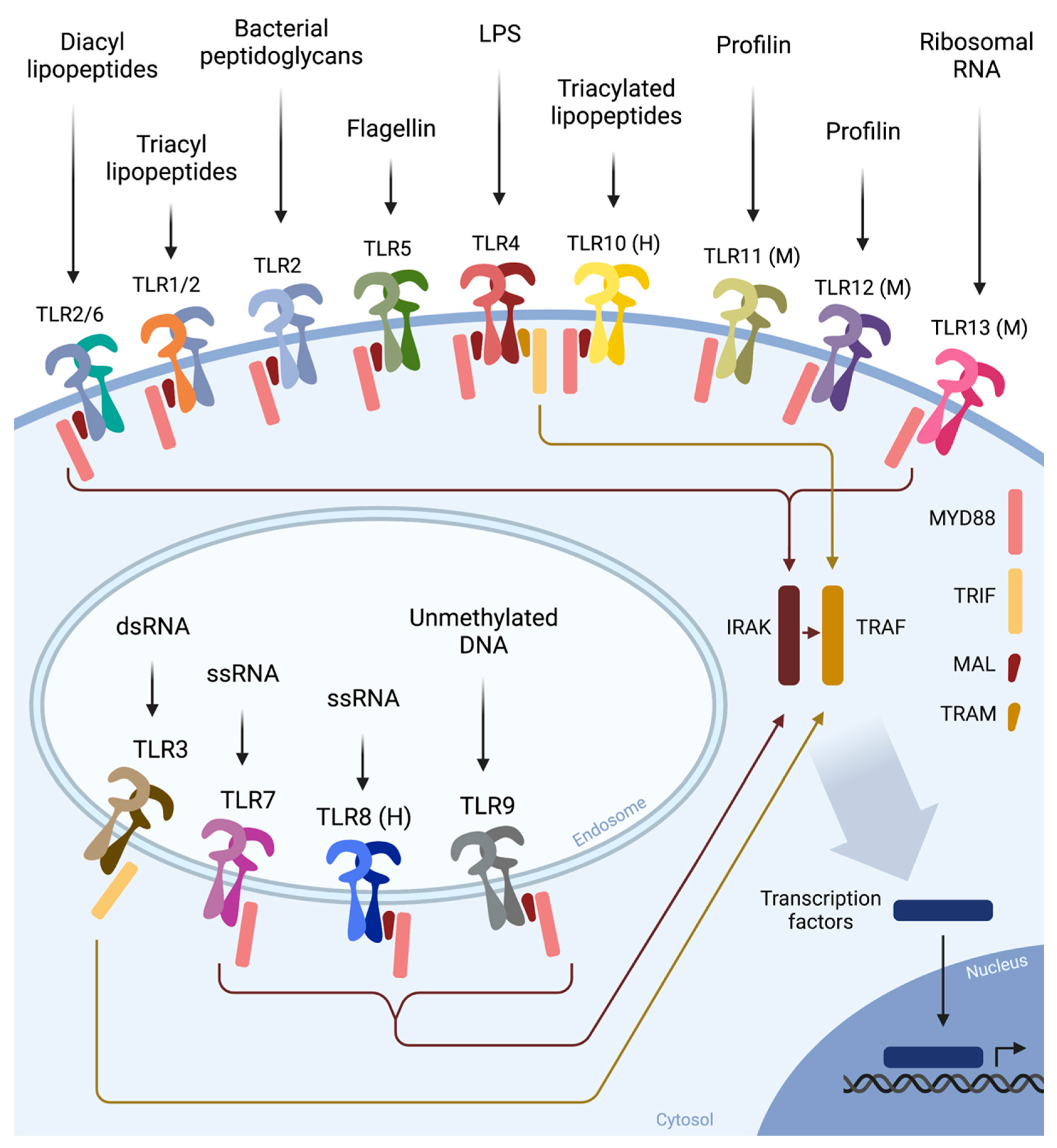
5. TLR4 Is the Receptor for Bacterial LPS
6. Scientific Impact
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steinman, R.M.; Cohn, Z.A. Identification of a Novel Cell Type in Peripheral Lymphoid Organs of Mice. J. Exp. Med. 1973, 137, 1142–1162. [Google Scholar] [CrossRef] [PubMed]
- Steinman, R.M.; Witmer, M.D. Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. Proc. Natl. Acad. Sci. USA 1978, 75, 5132–5136. [Google Scholar] [CrossRef] [PubMed]
- Schuler, G.; Steinman, R.M. Murine epidermal Langerhans cells mature into potent immunostimulatory dendritic cells in vitro. J. Exp. Med. 1985, 161, 526–546. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.-M.; Hoffmann, J.A. The Dorsoventral Regulatory Gene Cassette spätzle/Toll/cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef]
- Poltorak, A.; He, X.; Smirnova, I.; Liu, M.-Y.; Huffel, C.V.; Du, X.; Birdwell, D.; Alejos, E.; Silva, M.; Galanos, C.; et al. Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 Gene. Science 1998, 282, 2085–2088. [Google Scholar] [CrossRef]
- Jennings, B.H. Drosophila—A versatile model in biology & medicine. Mater. Today 2011, 14, 190–195. [Google Scholar] [CrossRef]
- Morgan, T.H. Sex Limited Inheritance in Drosophila. Science 1910, 32, 120–122. [Google Scholar] [CrossRef]
- Muller, H.J. The Production of Mutations by X-Rays. Proc. Natl. Acad. Sci. USA 1928, 14, 714–726. [Google Scholar] [CrossRef]
- Mandaravally Madhavan, M.; Schneiderman, H.A. Histological analysis of the dynamics of growth of imaginal discs and histoblast nests during the larval development ofDrosophila melanogaster. Wilhelm Roux’s Arch. Dev. Biol. 1977, 183, 269–305. [Google Scholar] [CrossRef]
- Garcia-Bellido, A.; Merriam, J.R. Parameters of the wing imaginal disc development ofDrosophila melanogaster. Dev. Biol. 1971, 24, 61–87. [Google Scholar] [CrossRef]
- Gehring, W. The Stability of the Determined State in Cultures of Imaginal Disks in Drosophila. Develop. Biol. 1967, 16, 438–456. [Google Scholar] [CrossRef]
- Beadle, G.W.; Ephrussi, B. The differentiation of eye pigments in drosophila as studied by transplantation. Genetics 1936, 21, 225–247. [Google Scholar] [CrossRef] [PubMed]
- Bryant, P.J. Cell lineage relationships in the imaginal wing disc of Drosophila melanogaster. Dev. Biol. 1970, 22, 389–411. [Google Scholar] [CrossRef]
- Steiner, E. Establishment of compartments in the developing leg imaginal discs ofDrosophila melanogaster. Wilhelm Roux’s Arch. Dev. Biol. 1976, 180, 9–30. [Google Scholar] [CrossRef]
- MORATA, G.; LAWRENCE, P.A. Anterior and posterior compartments in the head of Drosophila. Nature 1978, 274, 473–474. [Google Scholar] [CrossRef]
- Lawrence, P.A.; Morata, G. Compartments in the wing of Drosophila: A study of the engrailed gene. Dev. Biol. 1976, 50, 321–337. [Google Scholar] [CrossRef]
- Nüsslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef]
- Santamaria, P.; Nüsslein-Volhard, C. Partial rescue of dorsal, a maternal effect mutation affecting the dorso-ventral pattern of the Drosophila embryo, by the injection of wild-type cytoplasm. EMBO J. 1983, 2, 1695–1699. [Google Scholar] [CrossRef]
- Anderson, K.V.; Nüsslein-Volhard, C. Information for the dorsal–ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature 1984, 311, 223–227. [Google Scholar] [CrossRef]
- Schüpbach, T.; Wieschaus, E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics 1989, 121, 101–117. [Google Scholar] [CrossRef]
- Morata, G.; Lawrence, P. An exciting period of Drosophila developmental biology: Of imaginal discs, clones, compartments, parasegments and homeotic genes. Dev. Biol. 2022, 484, 12–21. [Google Scholar] [CrossRef]
- Press Release: The Nobel Prize in Physiology or Medicine. 1995. Available online: https://www.nobelprize.org/prizes/medicine/1995/press-release/ (accessed on 10 September 2022).
- Anderson, K.V.; Bokla, L.; Nüsslein-Volhard, C. Establishment of dorsal-ventral polarity in the drosophila embryo: The induction of polarity by the Toll gene product. Cell 1985, 42, 791–798. [Google Scholar] [CrossRef]
- Nüsslein-Volhard, C. The Toll gene in Drosophila pattern formation. Trends Genet. 2022, 38, 231–245. [Google Scholar] [CrossRef]
- Hashimoto, C.; Hudson, K.L.; Anderson, K.V. The Toll gene of drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell 1988, 52, 269–279. [Google Scholar] [CrossRef]
- Stein, D.; Roth, S.; Vogelsang, E.; Nu¨sslein-Volhard, C. The polarity of the dorsoventral axis in the drosophila embryo is defined by an extracellular signal. Cell 1991, 65, 725–735. [Google Scholar] [CrossRef]
- Schneider, D.S.; Jin, Y.; Morisato, D.; Anderson, K.V. A processed form of the Spatzle protein defines dorsal-ventral polarity in the Drosophila embryo. Development 1994, 120, 1243–1250. [Google Scholar] [CrossRef]
- Schneider, D.S.; Hudson, K.L.; Lin, T.Y.; Anderson, K.V. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev. 1991, 5, 797–807. [Google Scholar] [CrossRef]
- Steward, R. Dorsal, an Embryonic Polarity Gene in Drosophila, Is Homologous to the Vertebrate Proto-Oncogene, c- rel. Science 1987, 238, 692–694. [Google Scholar] [CrossRef]
- Kidd, S. Characterization of the Drosophila cactus locus and analysis of interactions between cactus and dorsal proteins. Cell 1992, 71, 623–635. [Google Scholar] [CrossRef]
- Geisler, R.; Bergmann, A.; Hiromi, Y.; Nüsslein-Volhard, C. cactus, a gene involved in dorsoventral pattern formation of Drosophila, is related to the IκB gene family of vertebrates. Cell 1992, 71, 613–621. [Google Scholar] [CrossRef]
- Shelton, C.A.; Wasserman, S.A. pelle encodes a protein kinase required to establish dorsoventral polarity in the Drosophila embryo. Cell 1993, 72, 515–525. [Google Scholar] [CrossRef]
- Cao, Z.; Henzel, W.J.; Gao, X. IRAK: A Kinase Associated with the Interleukin-1 Receptor. Science 1996, 271, 1128–1131. [Google Scholar] [CrossRef]
- Reichhart, J.M.; Georgel, P.; Meister, M.; Lemaitre, B.; Kappler, C.; Hoffmann, J.A. Expression and nuclear translocation of the rel/NF-kappa B-related morphogen dorsal during the immune response of Drosophila. C. R. Acad. Sci. III. 1993, 316, 1218–1224. [Google Scholar]
- Ip, Y.T.; Reach, M.; Engstrom, Y.; Kadalayil, L.; Cai, H.; González-Crespo, S.; Tatei, K.; Levine, M. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell 1993, 75, 753–763. [Google Scholar] [CrossRef]
- Whitham, S.; Dinesh-Kumar, S.P.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef]
- Hoffmann, D.; Brehelin, M.; Hoffmann, J.A. Modifications of the hemogram and of the hemocytopoietic tissue of male adults of Locusta migratoria (Orthoptera) after injection of Bacillus thuringiensis. J. Invertebr. Pathol. 1974, 24, 238–247. [Google Scholar] [CrossRef]
- Dimarcq, J.-L.; Keppi, E.; Dunbar, B.; Lambert, J.; Reichart, J.-M.; Hoffmann, D.; Rankine, S.M.; Fothergill, J.E.; Hoffmann, J.A. Insect immunity. Purification and characterization of a family of novel inducible antibacterial proteins from immunized larvae of the dipteran Phormia terranovae and complete amino-acid sequence of the predominant member, diptericin A. Eur. J. Biochem. 1988, 171, 17–22. [Google Scholar] [CrossRef]
- Dimarcq, J.-L.; Hoffmann, D.; Meister, M.; Bulet, P.; Lanot, R.; Reichhart, J.-M.; Hoffmann, J.A. Characterization and transcriptional profiles of a Drosophila gene encoding an insect defensin. A study in insect immunity. Eur. J. Biochem. 1994, 221, 201–209. [Google Scholar] [CrossRef]
- Morisalo, D.; Anderson, K.V. Signaling pathways that establish the dorsal-ventral pattern of the drosophila embryo. Annu. Rev. Genet. 1995, 29, 371–399. [Google Scholar] [CrossRef]
- GAY, N.J.; KEITH, F.J. Drosophila Toll and IL-1 receptor. Nature 1991, 351, 355–356. [Google Scholar] [CrossRef]
- Lemaitre, B.; Kromer-Metzger, E.; Michaut, L.; Nicolas, E.; Meister, M.; Georgel, P.; Reichhart, J.M.; Hoffmann, J.A. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl. Acad. Sci. USA 1995, 92, 9465–9469. [Google Scholar] [CrossRef]
- Janeway, C.A. Approaching the Asymptote? Evolution and Revolution in Immunology. Cold Spring Harb. Symp. Quant. Biol. 1989, 54, 1–13. [Google Scholar] [CrossRef]
- Pfeiffer, R. Untersuchungen über das Choleragift. Z. Hyg. Infekt. 1892, 11, 393–412. [Google Scholar] [CrossRef]
- Centanni, E.; Bruschettini, A. Untersuchungen über das Infectionsfieber. DMW-Dtsch. Med. Wochenschr. 1894, 20, 270–272. [Google Scholar] [CrossRef]
- Coley, W.B. The Treatment of Inoperable Sarcoma by Bacterial Toxins (the Mixed Toxins of the Streptococcus erysipelas and the Bacillus prodigiosus). Proc. R. Soc. Med. 1910, 3, 1–48. [Google Scholar] [CrossRef]
- Starnes, C.O. Coley’s toxins in perspective. Nature 1992, 357, 11–12. [Google Scholar] [CrossRef]
- Shear, M.J.; Turner, F.C.; Perrault, A.; Shovelton, T. Chemical Treatment of Tumors. V. Isolation of the Hemorrhage-Producing Fraction from Serratia marcescens (Bacillus prodigiosus) Culture Filtrate. JNCI J. Natl. Cancer Inst. 1943, 4, 81–97. [Google Scholar] [CrossRef]
- Strain, S.M.; Fesik, S.W.; Armitage, I.M. Characterization of lipopolysaccharide from a heptoseless mutant of Escherichia coli by carbon 13 nuclear magnetic resonance. J. Biol. Chem. 1983, 258, 2906–2910. [Google Scholar] [CrossRef]
- Beutler, B.; Rietschel, E.T. Innate immune sensing and its roots: The story of endotoxin. Nat. Rev. Immunol. 2003, 3, 169–176. [Google Scholar] [CrossRef]
- Heppner, G.; Weiss, D.W. High Susceptibility of Strain A Mice to Endotoxin and Endotoxin-Red Blood Cell Mixtures. J. Bacteriol. 1965, 90, 696–703. [Google Scholar] [CrossRef]
- Watson, J.; Riblet, R. Genetic control of responses to bacterial lipopolysaccharides in mice. J. Exp. Med. 1974, 140, 1147–1161. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.; Riblet, R. Genetic control of responses to bacterial lipopolysaccharides in mice. II. A gene that influences a membrane component involved in the activation of bone marrow-derived lymphocytes by lipipolysaccharides. J. Immunol. 1975, 114, 1462–1468. [Google Scholar] [PubMed]
- Coutinho, A.; Forni, L.; Melchers, F.; Watanabe, T. Genetic defect in responsiveness to the B cell mitogen lipopolysaccharide. Eur. J. Immunol. 1977, 7, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.N.; Goodrum, K.J.; Berry, L.J. Mediation of an endotoxic effect by macrophages. J. Reticuloendothel. Soc. 1976, 19, 187–197. [Google Scholar]
- SIBAL, L.R. Effect of endotoxin on antibody production by chicken spleen cells transferred to chick chorioallantois. J. Immunol. 1961, 87, 362–366. [Google Scholar]
- Davies, A.M.; Gery, I.; Rosenmann, E.; Laufer, A. Endotoxin as Adjuvant in Autoimmunity to Cardiac Tissue. Exp. Biol. Med. 1963, 114, 520–523. [Google Scholar] [CrossRef]
- Neter, E. Endotoxins and the Immune Response. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 1969; pp. 82–124. [Google Scholar]
- O’Brien, A.D.; Rosenstreich, D.L.; Scher, I.; Campbell, G.H.; MacDermott, R.P.; Formal, S.B. Genetic control of susceptibility to Salmonella typhimurium in mice: Role of the LPS gene. J. Immunol. 1980, 124, 20–24. [Google Scholar]
- Hagberg, L.; Hull, R.; Hull, S.; McGhee, J.R.; Michalek, S.M.; Svanborg Edén, C. Difference in susceptibility to gram-negative urinary tract infection between C3H/HeJ and C3H/HeN mice. Infect. Immun. 1984, 46, 839–844. [Google Scholar] [CrossRef]
- Beutler, B.; Greenwald, D.; Hulmes, J.D.; Chang, M.; Pan, Y.-C.E.; Mathison, J.; Ulevitch, R.; Cerami, A. Identity of tumour necrosis factor and the macrophage-secreted factor cachectin. Nature 1985, 316, 552–554. [Google Scholar] [CrossRef]
- Beutler, B.; Milsark, I.W.; Cerami, A.C. Passive Immunization Against Cachectin/Tumor Necrosis Factor Protects Mice from Lethal Effect of Endotoxin. Science 1985, 229, 869–871. [Google Scholar] [CrossRef]
- Havell, E.A. Production of tumor necrosis factor during murine listeriosis. J. Immunol. 1987, 139, 4225–4231. [Google Scholar] [PubMed]
- Kindler, V.; Sappino, A.-P.; Grau, G.E.; Piguet, P.-F.; Vassalli, P. The inducing role of tumor necrosis factor in the development of bactericidal granulomas during BCG infection. Cell 1989, 56, 731–740. [Google Scholar] [CrossRef]
- Shakhov, A.N.; Collart, M.A.; Vassalli, P.; Nedospasov, S.A.; Jongeneel, C. V Kappa B-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor alpha gene in primary macrophages. J. Exp. Med. 1990, 171, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Tobias, P.S.; Soldau, K.; Ulevitch, R.J. Isolation of a lipopolysaccharide-binding acute phase reactant from rabbit serum. J. Exp. Med. 1986, 164, 777–793. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.D.; Tobias, P.S.; Ulevitch, R.J.; Ramos, R.A. Lipopolysaccharide (LPS) binding protein opsonizes LPS-bearing particles for recognition by a novel receptor on macrophages. J. Exp. Med. 1989, 170, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Schumann, R.R.; Leong, S.R.; Flaggs, G.W.; Gray, P.W.; Wright, S.D.; Mathison, J.C.; Tobias, P.S.; Ulevitch, R.J. Structure and Function of Lipopolysaccharide Binding Protein. Science 1990, 249, 1429–1431. [Google Scholar] [CrossRef]
- Wright, S.D.; Ramos, R.A.; Tobias, P.S.; Ulevitch, R.J.; Mathison, J.C. CD14, a Receptor for Complexes of Lipopolysaccharide (LPS) and LPS Binding Protein. Science 1990, 249, 1431–1433. [Google Scholar] [CrossRef]
- Tobias, P.S.; Soldau, K.; Ulevitch, R.J. Identification of a Lipid A Binding Site in the Acute Phase Reactant Lipopolysaccharide Binding Protein. J. Biol. Chem. 1989, 264, 10867–10871. [Google Scholar] [CrossRef]
- Haziot, A.; Ferrero, E.; Köntgen, F.; Hijiya, N.; Yamamoto, S.; Silver, J.; Stewart, C.L.; Goyert, S.M. Resistance to Endotoxin Shock and Reduced Dissemination of Gram-Negative Bacteria in CD14-Deficient Mice. Immunity 1996, 4, 407–414. [Google Scholar] [CrossRef]
- Medzhitov, R.; Preston-Hurlburt, P.; Janeway, C.A. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997, 388, 394–397. [Google Scholar] [CrossRef]
- Nomura, N.; Miyajima, N.; Sazuka, T.; Tanaka, A.; Kawarabayasi, Y.; Sato, S.; Nagase, T.; Seki, N.; Ishikawa, K.-i.; Tabata, S. Prediction of the Coding Sequences of Unidentified Human Genes. I. The Coding Sequences of 40 New Genes (KIAA0001-KIAA0040) Deduced by Analysis of Randomly Sampled cDNA Clones from Human Immature Myeloid Cell Line KG-1. DNA Res. 1994, 1, 27–35. [Google Scholar] [CrossRef]
- Rock, F.L.; Hardiman, G.; Timans, J.C.; Kastelein, R.A.; Bazan, J.F. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA 1998, 95, 588–593. [Google Scholar] [CrossRef] [PubMed]
- SULTZER, B.M. Genetic Control of Leucocyte Responses to Endotoxin. Nature 1968, 219, 1253–1254. [Google Scholar] [CrossRef] [PubMed]
- Rosenstreich, D.L.; Michael Glade, L.; Mergenhagen, S.E. Action of Endotoxin on Lymphoid Cells. J. Infect. Dis. 1977, 136, S239–S245. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, A.; Meo, T. Genetic basis for unresponsiveness to lipopolysaccharide in C57BL/10Cr mice. Immunogenetics 1978, 7, 17–24. [Google Scholar] [CrossRef]
- Peppel, K.; Crawford, D.; Beutler, B. A tumor necrosis factor (TNF) receptor-IgG heavy chain chimeric protein as a bivalent antagonist of TNF activity. J. Exp. Med. 1991, 174, 1483–1489. [Google Scholar] [CrossRef]
- Takeuchi, O.; Hoshino, K.; Kawai, T.; Sanjo, H.; Takada, H.; Ogawa, T.; Takeda, K.; Akira, S. Differential Roles of TLR2 and TLR4 in Recognition of Gram-Negative and Gram-Positive Bacterial Cell Wall Components. Immunity 1999, 11, 443–451. [Google Scholar] [CrossRef]
- Takeuchi, O.; Kawai, T.; Mühlradt, P.F.; Morr, M.; Radolf, J.D.; Zychlinsky, A.; Takeda, K.; Akira, S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 2001, 13, 933–940. [Google Scholar] [CrossRef]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K.; et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [CrossRef]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-Specific Recognition of Single-Stranded RNA via Toll-like Receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ranoa, D.R.E.; Jiang, S.; Mutha, S.K.; Li, X.; Baudry, J.; Tapping, R.I. Human TLRs 10 and 1 Share Common Mechanisms of Innate Immune Sensing but Not Signaling. J. Immunol. 2010, 184, 5094–5103. [Google Scholar] [CrossRef] [PubMed]
- Yarovinsky, F.; Zhang, D.; Andersen, J.F.; Bannenberg, G.L.; Serhan, C.N.; Hayden, M.S.; Hieny, S.; Sutterwala, F.S.; Flavell, R.A.; Ghosh, S.; et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science 2005, 308, 1626–1629. [Google Scholar] [CrossRef]
- Hatai, H.; Lepelley, A.; Zeng, W.; Hayden, M.S.; Ghosh, S. Toll-Like Receptor 11 (TLR11) Interacts with Flagellin and Profilin through Disparate Mechanisms. PLoS ONE 2016, 11, e0148987. [Google Scholar] [CrossRef]
- Oldenburg, M.; Krüger, A.; Ferstl, R.; Kaufmann, A.; Nees, G.; Sigmund, A.; Bathke, B.; Lauterbach, H.; Suter, M.; Dreher, S.; et al. TLR13 Recognizes Bacterial 23 S rRNA Devoid of Erythromycin Resistance–Forming Modification. Science 2012, 337, 1111–1115. [Google Scholar] [CrossRef]
- Jin, M.S.; Kim, S.E.; Heo, J.Y.; Lee, M.E.; Kim, H.M.; Paik, S.-G.; Lee, H.; Lee, J.-O. Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar] [CrossRef]
- Andrade, W.A.; do Carmo Souza, M.; Ramos-Martinez, E.; Nagpal, K.; Dutra, M.S.; Melo, M.B.; Bartholomeu, D.C.; Ghosh, S.; Golenbock, D.T.; Gazzinelli, R.T. Combined Action of Nucleic Acid-Sensing Toll-like Receptors and TLR11/TLR12 Heterodimers Imparts Resistance to Toxoplasma gondii in Mice. Cell Host Microbe 2013, 13, 42–53. [Google Scholar] [CrossRef]
- Rosenstiel, P.; Till, A.; Schreiber, S. NOD-like receptors and human diseases. Microbes Infect. 2007, 9, 648–657. [Google Scholar] [CrossRef]
- Inohara, N.; Koseki, T.; del Peso, L.; Hu, Y.; Yee, C.; Chen, S.; Carrio, R.; Merino, J.; Liu, D.; Ni, J.; et al. Nod1, an Apaf-1-like Activator of Caspase-9 and Nuclear Factor-κB. J. Biol. Chem. 1999, 274, 14560–14567. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Kutikhin, A.; Yuzhalin, A. C-type lectin receptors and RIG-I-like receptors: New points on the oncogenomics map. Cancer Manag. Res. 2012, 4, 39–53. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Neth, O.; Jack, D.L.; Dodds, A.W.; Holzel, H.; Klein, N.J.; Turner, M.W. Mannose-Binding Lectin Binds to a Range of Clinically Relevant Microorganisms and Promotes Complement Deposition. Infect. Immun. 2000, 68, 688–693. [Google Scholar] [CrossRef]
- Wesche, H.; Henzel, W.J.; Shillinglaw, W.; Li, S.; Cao, Z. MyD88: An Adapter That Recruits IRAK to the IL-1 Receptor Complex. Immunity 1997, 7, 837–847. [Google Scholar] [CrossRef]
- O’Neill, L.A.J.; Bowie, A.G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007, 7, 353–364. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sato, S.; Mori, K.; Hoshino, K.; Takeuchi, O.; Takeda, K.; Akira, S. Cutting Edge: A Novel Toll/IL-1 Receptor Domain-Containing Adapter That Preferentially Activates the IFN-β Promoter in the Toll-Like Receptor Signaling. J. Immunol. 2002, 169, 6668–6672. [Google Scholar] [CrossRef]
- Oshiumi, H.; Matsumoto, M.; Funami, K.; Akazawa, T.; Seya, T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3–mediated interferon-β induction. Nat. Immunol. 2003, 4, 161–167. [Google Scholar] [CrossRef]
- Hoebe, K.; Du, X.; Georgel, P.; Janssen, E.; Tabeta, K.; Kim, S.O.; Goode, J.; Lin, P.; Mann, N.; Mudd, S.; et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 2003, 424, 743–748. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sato, S.; Hemmi, H.; Hoshino, K.; Kaisho, T.; Sanjo, H.; Takeuchi, O.; Sugiyama, M.; Okabe, M.; Takeda, K.; et al. Role of Adaptor TRIF in the MyD88-Independent Toll-Like Receptor Signaling Pathway. Science 2003, 301, 640–643. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Rowe, D.C.; Barnes, B.J.; Caffrey, D.R.; Visintin, A.; Latz, E.; Monks, B.; Pitha, P.M.; Golenbock, D.T. LPS-TLR4 Signaling to IRF-3/7 and NF-κB Involves the Toll Adapters TRAM and TRIF. J. Exp. Med. 2003, 198, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Sato, S.; Hemmi, H.; Uematsu, S.; Hoshino, K.; Kaisho, T.; Takeuchi, O.; Takeda, K.; Akira, S. TRAM is specifically involved in the Toll-like receptor 4–mediated MyD88-independent signaling pathway. Nat. Immunol. 2003, 4, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Palsson-McDermott, E.M.; Bowie, A.G.; Jefferies, C.A.; Mansell, A.S.; Brady, G.; Brint, E.; Dunne, A.; Gray, P.; Harte, M.T.; et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature 2001, 413, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.J.; Golenbock, D.; Bowie, A.G. The history of Toll-like receptors—Redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Essuman, K.; Summers, D.W.; Sasaki, Y.; Mao, X.; DiAntonio, A.; Milbrandt, J. The SARM1 Toll/Interleukin-1 Receptor Domain Possesses Intrinsic NAD + Cleavage Activity that Promotes Pathological Axonal Degeneration. Neuron 2017, 93, 1334–1343.e5. [Google Scholar] [CrossRef]
- Essuman, K.; Milbrandt, J.; Dangl, J.L.; Nishimura, M.T. Shared TIR enzymatic functions regulate cell death and immunity across the tree of life. Science 2022, 377, 6614. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiss, H.J.; O’Neill, L.A.J. Of Flies and Men—The Discovery of TLRs. Cells 2022, 11, 3127. https://doi.org/10.3390/cells11193127
Weiss HJ, O’Neill LAJ. Of Flies and Men—The Discovery of TLRs. Cells. 2022; 11(19):3127. https://doi.org/10.3390/cells11193127
Chicago/Turabian StyleWeiss, Hauke Johannes, and Luke Anthony John O’Neill. 2022. "Of Flies and Men—The Discovery of TLRs" Cells 11, no. 19: 3127. https://doi.org/10.3390/cells11193127
APA StyleWeiss, H. J., & O’Neill, L. A. J. (2022). Of Flies and Men—The Discovery of TLRs. Cells, 11(19), 3127. https://doi.org/10.3390/cells11193127