Biochemical Pathways of Cellular Mechanosensing/Mechanotransduction and Their Role in Neurodegenerative Diseases Pathogenesis
Abstract
1. Introduction
2. Mechanobiology—A Brief Overview
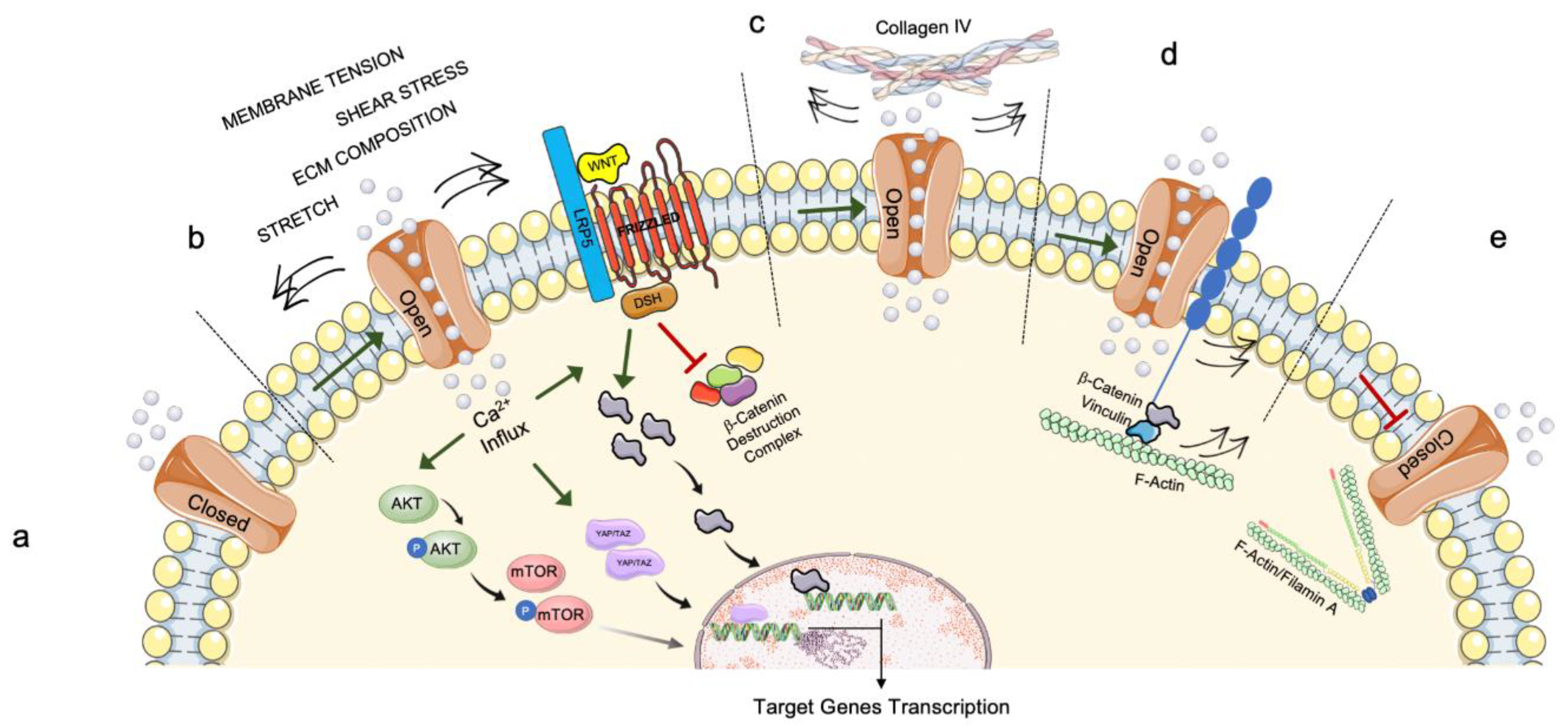
2.1. Mechanosensing
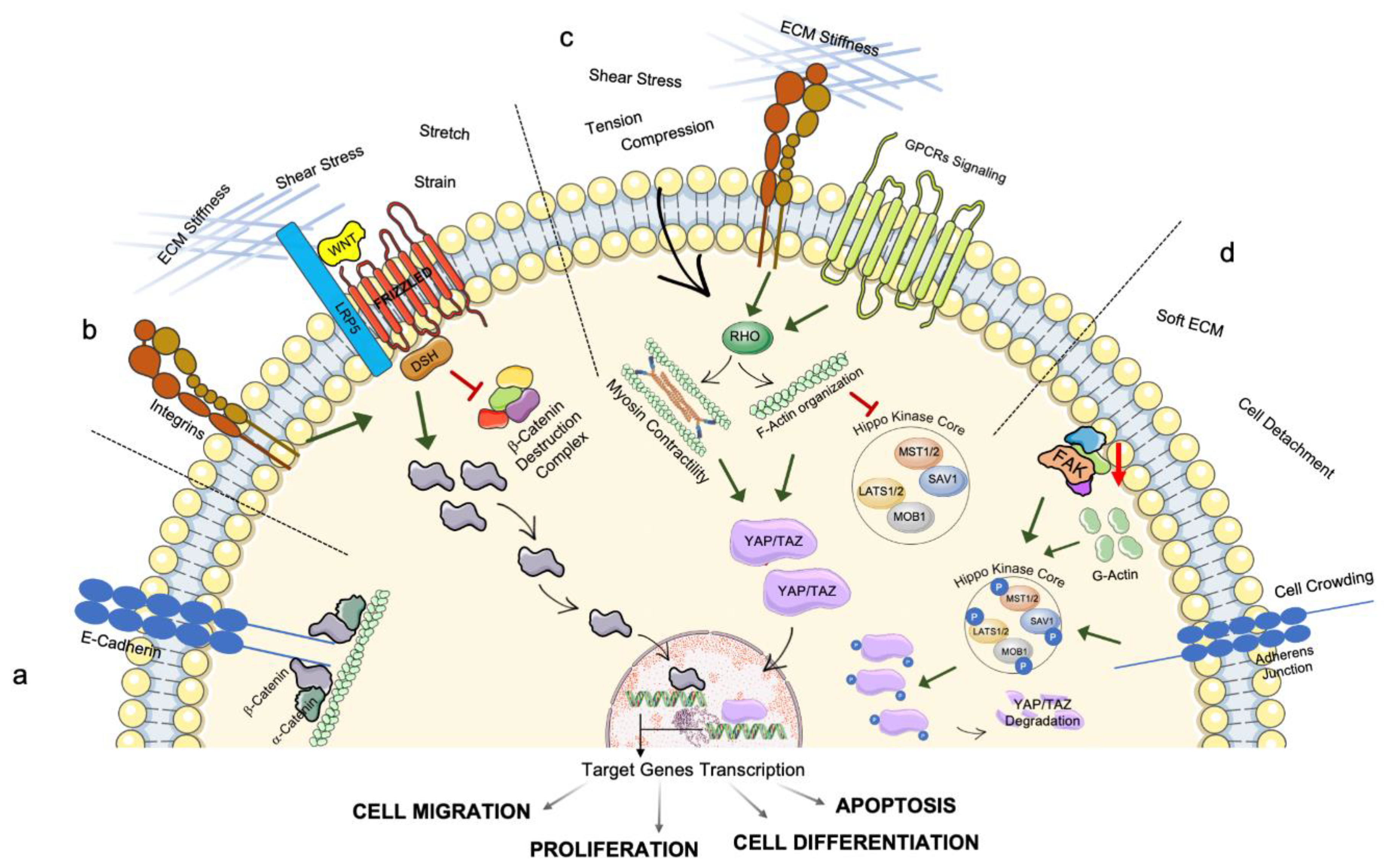
2.2. Mechanotransduction
3. Intracellular Transmission of Mechanosensing and Mechanotransduction Pathways: The Involvement of Organelles’ Homeostasis and Dysfunction
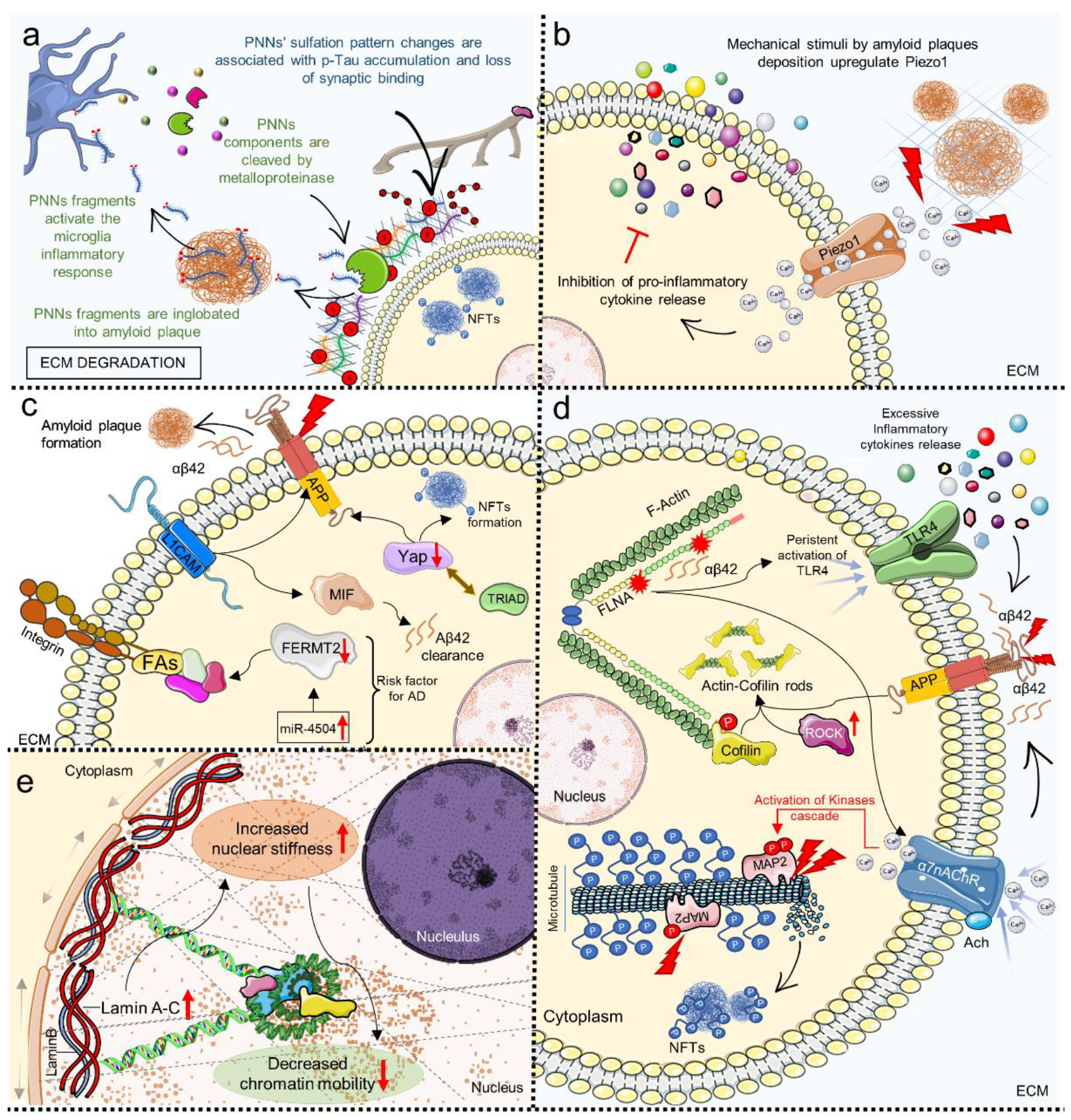
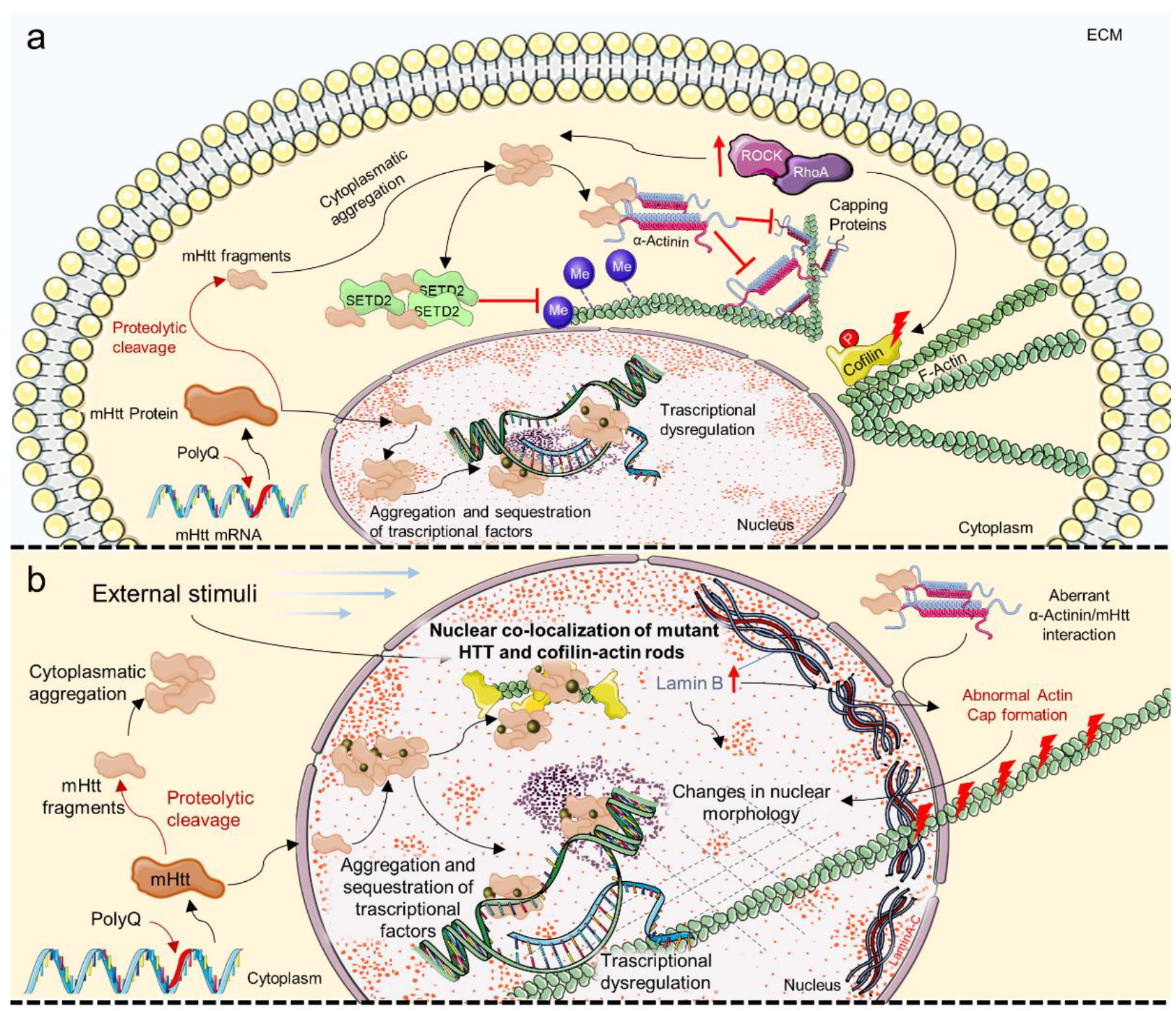
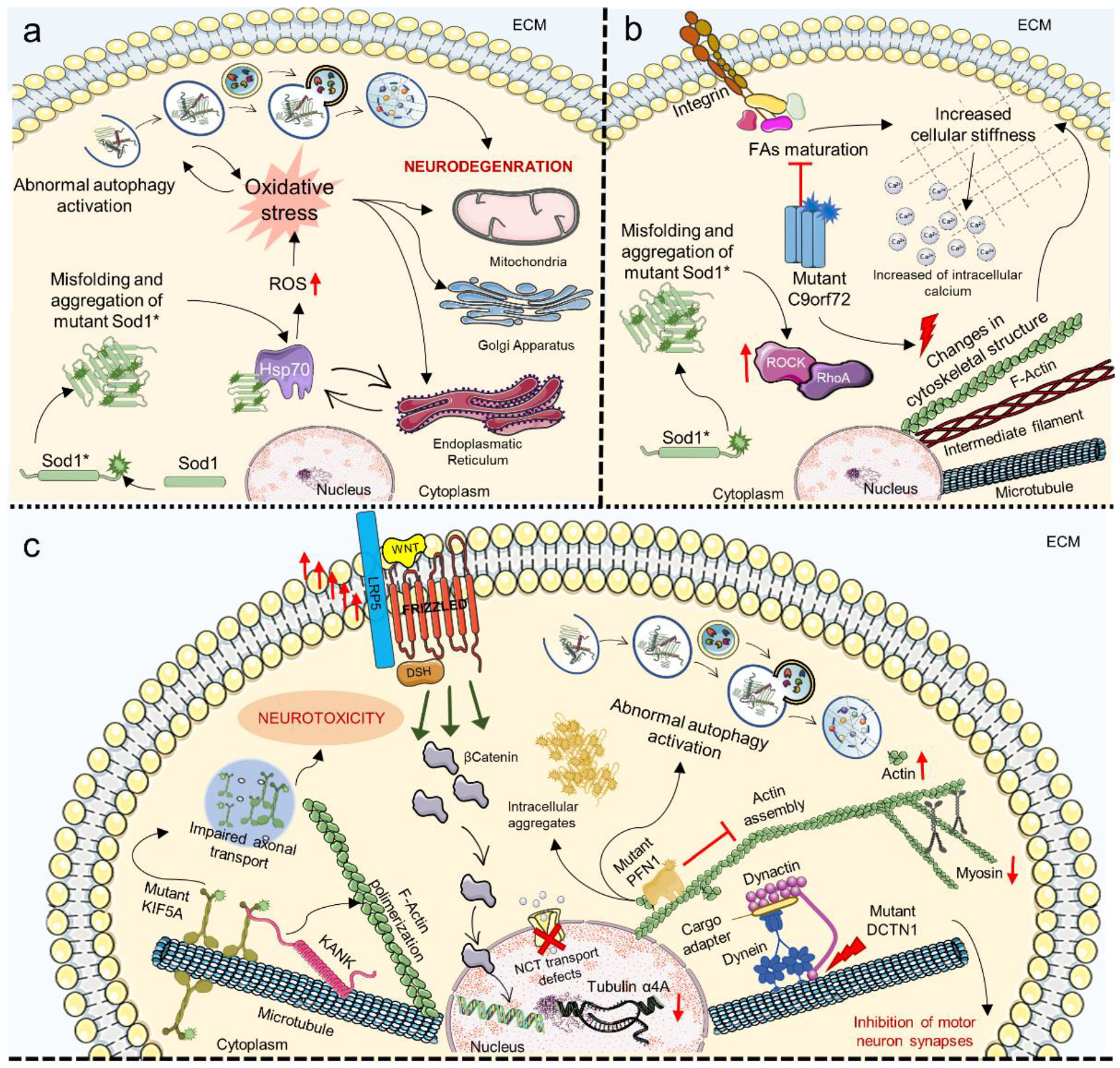
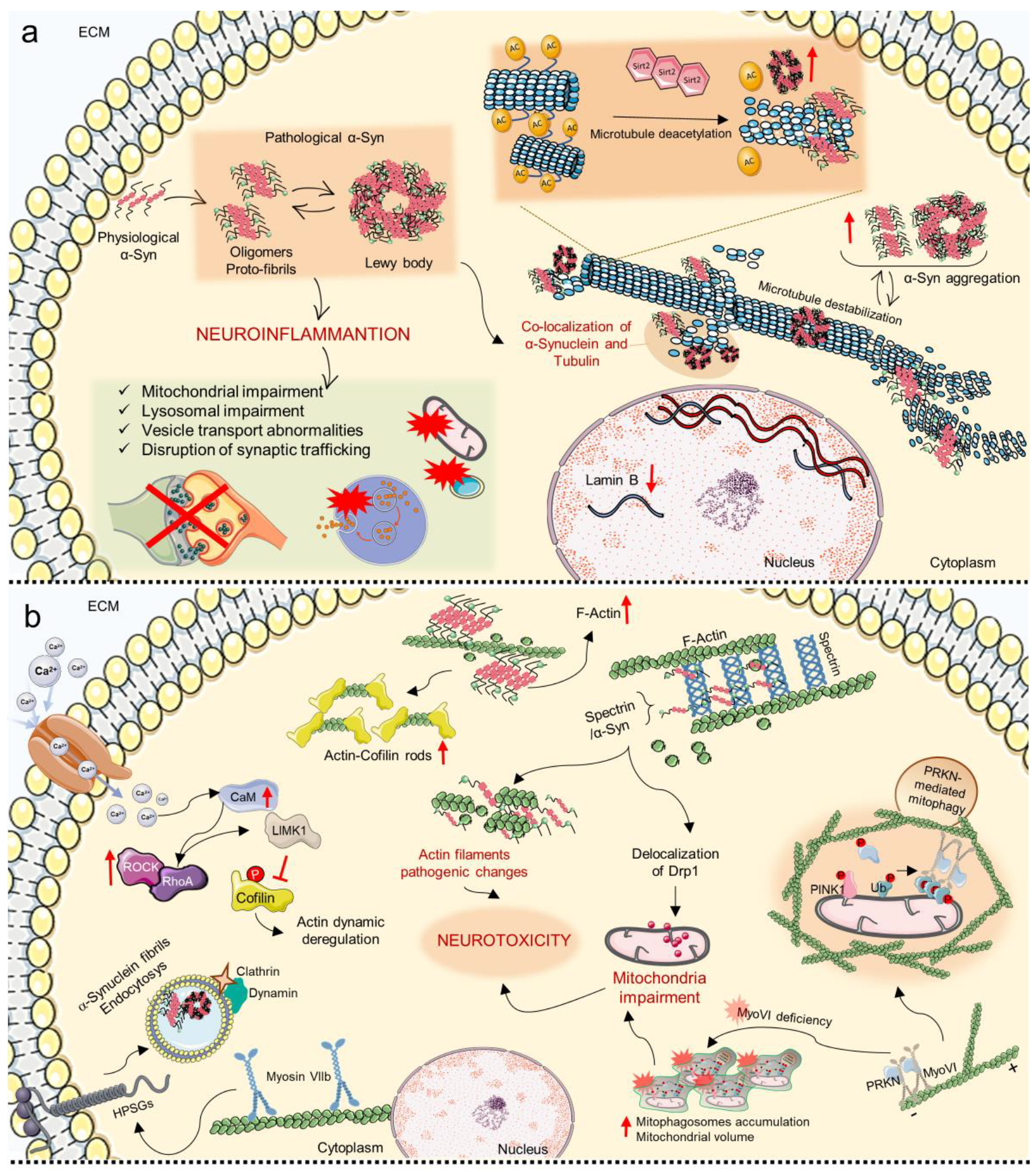
4. Mechanosensing and Mechanotransduction in Neurodegenerative Diseases
5. Mechanosensing and Mechanotransduction Pathways in Alzheimer’s Disease
6. Mechanosensing and Mechanotransduction Pathways in Huntington’s Disease
7. Mechanosensing and Mechanotransduction Pathways in Amyotrophic Lateral Sclerosis
8. Mechanosensing and Mechanotransduction Pathways in Parkinson’s Disease
9. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Niethammer, P. Components and Mechanisms of Nuclear Mechanotransduction. Annu. Rev. Cell Dev. Biol. 2021, 37, 233–256. [Google Scholar] [CrossRef]
- Pennacchio, F.A.; Nastały, P.; Poli, A.; Maiuri, P. Tailoring Cellular Function: The Contribution of the Nucleus in Mechanotransduction. Front. Bioeng. Biotechnol. 2021, 8, 596746. [Google Scholar] [CrossRef] [PubMed]
- Motz, C.T.; Kabat, V.; Saxena, T.; Bellamkonda, R.V.; Zhu, C. Neuromechanobiology: An Expanding Field Driven by the Force of Greater Focus. Adv. Healthc. Mater. 2021, 10, 2100102. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jung, W.-H.; Pittman, M.; Chen, J.; Chen, Y. The Effects of Stiffness, Viscosity, and Geometry of Microenvironment in Homeostasis, Aging and Diseases. J. Biomech. Eng. 2020, 142, 100804. [Google Scholar] [CrossRef] [PubMed]
- Tortorella, I.; Argentati, C.; Emiliani, C.; Martino, S.; Morena, F. The role of physical cues in the development of stem cell-derived organoids. Eur. Biophys. J. 2021, 51, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Argentati, C.; Tortorella, I.; Bazzucchi, M.; Morena, F.; Martino, S. Harnessing the Potential of Stem Cells for Disease Modeling: Progress and Promises. J. Pers. Med. 2020, 10, 8. [Google Scholar] [CrossRef]
- Abdeljawad, M.B.; Carette, X.; Argentati, C.; Martino, S.; Gonon, M.-F.; Odent, J.; Morena, F.; Mincheva, R.; Raquez, J.-M. Interfacial Compatibilization into PLA/Mg Composites for Improved In Vitro Bioactivity and Stem Cell Adhesion. Molecules 2021, 26, 5944. [Google Scholar] [CrossRef]
- Argentati, C.; Morena, F.; Fontana, C.; Tortorella, I.; Emiliani, C.; Latterini, L.; Zampini, G.; Martino, S. Functionalized Silica Star-Shaped Nanoparticles and Human Mesenchymal Stem Cells: An In Vitro Model. Nanomaterials 2021, 11, 779. [Google Scholar] [CrossRef]
- Morena, F.; Argentati, C.; Soccio, M.; Bicchi, I.; Luzi, F.; Torre, L.; Munari, A.; Emiliani, C.; Gigli, M.; Lotti, N.; et al. Unpatterned Bioactive Poly(Butylene 1,4-Cyclohexanedicarboxylate)-Based Film Fast Induced Neuronal-Like Differentiation of Human Bone Marrow-Mesenchymal Stem Cells. Int. J. Mol. Sci. 2020, 21, 9274. [Google Scholar] [CrossRef]
- Luzi, F.; Tortorella, I.; Di Michele, A.; Dominici, F.; Argentati, C.; Morena, F.; Torre, L.; Puglia, D.; Martino, S. Novel Nanocomposite PLA Films with Lignin/Zinc Oxide Hybrids: Design, Characterization, Interaction with Mesenchymal Stem Cells. Nanomaterials 2020, 10, 2176. [Google Scholar] [CrossRef]
- Argentati, C.; Morena, F.; Montanucci, P.; Rallini, M.; Basta, G.; Calabrese, N.; Calafiore, R.; Cordellini, M.; Emiliani, C.; Armentano, I.; et al. Surface hydrophilicity of poly(l-Lactide) acid polymer film changes the human adult adipose stem cell architecture. Polymers 2018, 10, 140. [Google Scholar] [CrossRef] [PubMed]
- Morena, F.; Armentano, I.; Montanucci, P.; Argentati, C.; Fortunati, E.; Montesano, S.; Bicchi, I.; Pescara, T.; Pennoni, I.; Mattioli, S.; et al. Design of a nanocomposite substrate inducing adult stem cell assembly and progression toward an epiblast-like or primitive endoderm-like phenotype via mechanotransduction. Biomaterials 2017, 144, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Aragona, M.; Sifrim, A.; Malfait, M.; Song, Y.; Van Herck, J.; Dekoninck, S.; Gargouri, S.; Lapouge, G.; Swedlund, B.; Dubois, C.; et al. Mechanisms of stretch-mediated skin expansion at single-cell resolution. Nature 2020, 584, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Gütl, D.; Zheden, V.; Heisenberg, C.-P. Lateral Inhibition in Cell Specification Mediated by Mechanical Signals Modulating TAZ Activity. Cell 2019, 176, 1379–1392.e14. [Google Scholar] [CrossRef]
- Pocaterra, A.; Santinon, G.; Romani, P.; Brian, I.; Dimitracopoulos, A.; Ghisleni, A.; Carnicer-Lombarte, A.; Forcato, M.; Braghetta, P.; Montagner, M.; et al. F-actin dynamics regulates mammalian organ growth and cell fate maintenance. J. Hepatol. 2019, 71, 130–142. [Google Scholar] [CrossRef]
- Mohammed, D.; Versaevel, M.; Bruyère, C.; Alaimo, L.; Luciano, M.; Vercruysse, E.; Procès, A.; Gabriele, S. Innovative Tools for Mechanobiology: Unraveling Outside-In and Inside-Out Mechanotransduction. Front. Bioeng. Biotechnol. 2019, 7, 162. [Google Scholar] [CrossRef]
- Scheuren, A.; Wehrle, E.; Flohr, F.; Müller, R. Bone mechanobiology in mice: Toward single-cell in vivo mechanomics. Biomech. Model. Mechanobiol. 2017, 16, 2017–2034. [Google Scholar] [CrossRef]
- Verbruggen, S. Mechanobiology in Health and Disease; Elsevier: Amsterdam, The Netherlands, 2018; ISBN 9780128129524. [Google Scholar]
- Yu, W.; Sharma, S.; Rao, E.; Rowat, A.C.; Gimzewski, J.K.; Han, D.; Rao, J. Cancer cell mechanobiology: A new frontier for cancer research. J. Natl. Cancer Cent. 2021, 2, 10–17. [Google Scholar] [CrossRef]
- Liang, C.; Huang, M.; Li, T.; Li, L.; Sussman, H.; Dai, Y.; Siemann, D.W.; Xie, M.; Tang, X. Towards an integrative understanding of cancer mechanobiology: Calcium, YAP, and microRNA under biophysical forces. Soft Matter 2022, 18, 1112–1148. [Google Scholar] [CrossRef]
- Choudhury, A.R.; Gupta, S.; Chaturvedi, P.K.; Kumar, N.; Pandey, D. Mechanobiology of Cancer Stem Cells and Their Niche. Cancer Microenviron. 2019, 12, 17–27. [Google Scholar] [CrossRef]
- Armistead, F.; De Pablo, J.G.; Bloomfield-Gadêlha, H.; Peyman, S.A.; Evans, S.D. Physical Biomarkers of Disease Progression: On-Chip Monitoring of Changes in Mechanobiology of Colorectal Cancer Cells. Sci. Rep. 2020, 10, 3254. [Google Scholar] [CrossRef]
- Broders-Bondon, F.; Ho-Bouldoires, T.H.N.; Sanchez, M.E.F.; Farge, E. Mechanotransduction in tumor progression: The dark side of the force. J. Cell Biol. 2018, 217, 1571–1587. [Google Scholar] [CrossRef] [PubMed]
- Tschumperlin, D.J.; Ligresti, G.; Hilscher, M.B.; Shah, V.H. Mechanosensing and fibrosis. J. Clin. Investig. 2018, 128, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Tschumperlin, D.J.; Lagares, D. Mechano-therapeutics: Targeting Mechanical Signaling in Fibrosis and Tumor Stroma. Pharmacol. Ther. 2020, 212, 107575. [Google Scholar] [CrossRef]
- Harn, H.I.; Ogawa, R.; Hsu, C.; Hughes, M.W.; Tang, M.; Chuong, C. The tension biology of wound healing. Exp. Dermatol. 2019, 28, 464–471. [Google Scholar] [CrossRef]
- Humphrey, J.D.; Schwartz, M.A. Vascular Mechanobiology: Homeostasis, Adaptation, and Disease. Annu. Rev. Biomed. Eng. 2021, 23, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Jorba, I.; Mostert, D.; Hermans, L.H.; van der Pol, A.; Kurniawan, N.A.; Bouten, C.V. In Vitro Methods to Model Cardiac Mechanobiology in Health and Disease. Tissue Eng. Part C Methods 2021, 27, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Ta, H.T.; Nguyen, N. Mechanobiology in cardiology: Micro- and nanotechnologies to probe mechanosignaling. VIEW 2021, 2, 20200080. [Google Scholar] [CrossRef]
- Garoffolo, G.; Pesce, M. Mechanotransduction in the Cardiovascular System: From Developmental Origins to Homeostasis and Pathology. Cells 2019, 8, 1607. [Google Scholar] [CrossRef]
- Jabre, S.; Hleihel, W.; Coirault, C. Nuclear Mechanotransduction in Skeletal Muscle. Cells 2021, 10, 318. [Google Scholar] [CrossRef]
- Guo, X.E.; Hung, C.T.; Sandell, L.J.; Silva, M.J. Musculoskeletal mechanobiology: A new era for MechanoMedicine. J. Orthop. Res. 2018, 36, 531–532. [Google Scholar] [CrossRef]
- van Ingen, M.J.A.; Kirby, T.J. LINCing Nuclear Mechanobiology With Skeletal Muscle Mass and Function. Front. Cell Dev. Biol. 2021, 9, 1924. [Google Scholar] [CrossRef]
- Khuu, S.; Fernandez, J.W.; Handsfield, G.G. A Coupled Mechanobiological Model of Muscle Regeneration In Cerebral Palsy. Front. Bioeng. Biotechnol. 2021, 9, 689714. [Google Scholar] [CrossRef] [PubMed]
- Clippinger, S.R.; Cloonan, P.E.; Greenberg, L.; Ernst, M.; Stump, W.T.; Greenberg, M.J. Disrupted mechanobiology links the molecular and cellular phenotypes in familial dilated cardiomyopathy. Proc. Natl. Acad. Sci. USA 2019, 116, 17831–17840. [Google Scholar] [CrossRef]
- Barnes, J.M.; Przybyla, L.; Weaver, V.M. Tissue mechanics regulate brain development, homeostasis and disease. J. Cell Sci. 2017, 130, 71–82. [Google Scholar] [CrossRef]
- Marinval, N.; Chew, S.Y. Mechanotransduction assays for neural regeneration strategies: A focus on glial cells. APL Bioeng. 2021, 5, 021505. [Google Scholar] [CrossRef]
- Procès, A.; Luciano, M.; Kalukula, Y.; Ris, L.; Gabriele, S. Multiscale Mechanobiology in Brain Physiology and Diseases. Front. Cell Dev. Biol. 2022, 10, 64. [Google Scholar] [CrossRef]
- Argentati, C.; Morena, F.; Tortorella, I.; Bazzucchi, M.; Porcellati, S.; Emiliani, C.; Martino, S. Insight into Mechanobiology: How stem cells feel mechanical forces and orchestrate biobical functions. Int. J. Mol. Sci. 2019, 20, 5337. [Google Scholar] [CrossRef] [PubMed]
- Maurer, M.; Lammerding, J. The Driving Force: Nuclear Mechanotransduction in Cellular Function, Fate, and Disease. Annu. Rev. Biomed. Eng. 2019, 21, 443–468. [Google Scholar] [CrossRef] [PubMed]
- Ingber, D.E. Tensegrity: The Architectural Basis of Cellular. Annu. Rev. Physiol. 1997, 59, 575–599. [Google Scholar] [CrossRef] [PubMed]
- Ingber, D.E.; Wang, N.; Stamenovic, D. Tensegrity, cellular biophysics, and the mechanics of living systems. Rep. Prog. Phys. 2014, 77, 046603. [Google Scholar] [CrossRef] [PubMed]
- Ingber, D.E. From mechanobiology to developmentally inspired engineering. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170323. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, S.; Manneville, J.-B. Intracellular mechanics: Connecting rheology and mechanotransduction. Curr. Opin. Cell Biol. 2019, 56, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Mierke, C.T. Viscoelasticity, Like Forces, Plays a Role in Mechanotransduction. Front. Cell Dev. Biol. 2022, 10, 789841. [Google Scholar] [CrossRef] [PubMed]
- Corominas-Murtra, B.; Petridou, N.I. Viscoelastic Networks: Forming Cells and Tissues. Front. Phys. 2021, 9, 666916. [Google Scholar] [CrossRef]
- Uray, I.P.; Uray, K. Mechanotransduction at the Plasma Membrane-Cytoskeleton Interface. Int. J. Mol. Sci. 2021, 22, 11566. [Google Scholar] [CrossRef] [PubMed]
- Lavrenyuk, K.; Conway, D.; Dahl, K.N. Imaging methods in mechanosensing: A historical perspective and visions for the future. Mol. Biol. Cell 2021, 32, 842–854. [Google Scholar] [CrossRef]
- Chugh, M.; Munjal, A.; Megason, S.G. Hydrostatic pressure as a driver of cell and tissue morphogenesis. Semin. Cell Dev. Biol. 2022; (in press). [CrossRef]
- Jin, P.; Jan, L.Y.; Jan, Y.-N. Mechanosensitive Ion Channels: Structural Features Relevant to Mechanotransduction Mechanisms. Annu. Rev. Neurosci. 2020, 43, 207–229. [Google Scholar] [CrossRef]
- Swaminathan, V.; Gloerich, M. Decoding mechanical cues by molecular mechanotransduction. Curr. Opin. Cell Biol. 2021, 72, 72–80. [Google Scholar] [CrossRef]
- Qin, L.; He, T.; Chen, S.; Yang, D.; Yi, W.; Cao, H.; Xiao, G. Roles of mechanosensitive channel Piezo1/2 proteins in skeleton and other tissues. Bone Res. 2021, 91, 44. [Google Scholar] [CrossRef]
- Pathak, M.M.; Nourse, J.L.; Tran, T.; Hwe, J.; Arulmoli, J.; Dai Trang, T.L.; Bernardis, E.; Flanagan, L.A.; Tombola, F. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 16148–16153. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.; Cui, Y.; Wang, H.; Jiang, J.; Zhang, T.; Sun, S.; Zhou, Z.; Zhong, Y.; Xiao, B. Astrocytic Piezo1-mediated mechanotransduction determines adult neurogenesis and cognitive functions. Neuron 2022, 110, 2984–2999.e8. [Google Scholar] [CrossRef] [PubMed]
- Alper, S. Genetic Diseases of PIEZO1 and PIEZO2 Dysfunction. Curr. Top. Membr. 2017, 79, 97–134. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, J.; Zhao, X.; Li, J.; Chen, Y. Piezo channels in the urinary system. Exp. Mol. Med. 2022, 54, 697–710. [Google Scholar] [CrossRef]
- Velasco-Estevez, M.; Gadalla, K.; Liñan-Barba, N.; Cobb, S.; Dev, K.K.; Sheridan, G.K. Inhibition of Piezo1 attenuates demyelination in the central nervous system. Glia 2020, 68, 356–375. [Google Scholar] [CrossRef]
- Lai, A.; Cox, C.D.; Sekar, N.C.; Thurgood, P.; Jaworowski, A.; Peter, K.; Baratchi, S. Mechanosensing by Piezo1 and its implications for physiology and various pathologies. Biol. Rev. 2022, 97, 604–614. [Google Scholar] [CrossRef]
- Yang, K.; He, X.; Wu, Z.; Yin, Y.; Pan, H.; Zhao, X.; Sun, T. The emerging roles of piezo1 channels in animal models of multiple sclerosis. Front. Immunol. 2022, 13, 5322. [Google Scholar] [CrossRef]
- Zhu, W.; Guo, S.; Homilius, M.; Nsubuga, C.; Wright, S.H.; Quan, D.; Kc, A.; Eddy, S.S.; Victorio, R.A.; Beerens, M.; et al. PIEZO1 mediates a mechanothrombotic pathway in diabetes. Sci. Transl. Med. 2022, 14, eabk1707. [Google Scholar] [CrossRef]
- Sforna, L.; Michelucci, A.; Morena, F.; Argentati, C.; Franciolini, F.; Vassalli, M.; Martino, S.; Catacuzzeno, L. Piezo1 controls cell volume and migration by modulating swelling-activated chloride current through Ca2+ influx. J. Cell. Physiol. 2022, 237, 1857–1870. [Google Scholar] [CrossRef] [PubMed]
- Gaub, B.M.; Müller, D.J. Mechanical Stimulation of Piezo1 Receptors Depends on Extracellular Matrix Proteins and Directionality of Force. Nano Lett. 2017, 17, 2064–2072. [Google Scholar] [CrossRef]
- Retailleau, K.; Duprat, F.; Arhatte, M.; Ranade, S.S.; Peyronnet, R.; Martins, J.R.; Jodar, M.; Moro, C.; Offermanns, S.; Feng, Y.; et al. Piezo1 in Smooth Muscle Cells Is Involved in Hypertension-Dependent Arterial Remodeling. Cell Rep. 2015, 13, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Ridone, P.; Vassalli, M.; Martinac, B. Piezo1 mechanosensitive channels: What are they and why are they important. Biophys. Rev. 2019, 11, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.-C.; Chen, C.-C. Force From Filaments: The Role of the Cytoskeleton and Extracellular Matrix in the Gating of Mechanosensitive Channels. Front. Cell Dev. Biol. 2022, 10, 998. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, J.; Yang, X.; Zhou, G.; Wang, L.; Xiao, B. Tethering Piezo channels to the actin cytoskeleton for mechanogating via the cadherin-β-catenin mechanotransduction complex. Cell Rep. 2022, 38, 110342. [Google Scholar] [CrossRef] [PubMed]
- Wiedmann, F.; Rinné, S.; Donner, B.; Decher, N.; Katus, H.A.; Schmidt, C. Mechanosensitive TREK-1 two-pore-domain potassium (K2P) channels in the cardiovascular system. Prog. Biophys. Mol. Biol. 2020, 159, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Yap, A.S.; Duszyc, K.; Viasnoff, V. Mechanosensing and Mechanotransduction at Cell–Cell Junctions. Cold Spring Harb. Perspect. Biol. 2017, 10, a028761. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wang, K.-C.; Meng, Z. Mechanoregulation of YAP and TAZ in Cellular Homeostasis and Disease Progression. Front. Cell Dev. Biol. 2021, 9, 673599. [Google Scholar] [CrossRef]
- Martino, F.; Perestrelo, A.R.; Vinarský, V.; Pagliari, S.; Forte, G. Cellular mechanotransduction: From tension to function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef]
- Astudillo, P. Extracellular matrix stiffness and Wnt/β-catenin signaling in physiology and disease. Biochem. Soc. Trans. 2020, 48, 1187–1198. [Google Scholar] [CrossRef]
- Amit, C.; Padmanabhan, P.; Narayanan, J. Deciphering the mechanoresponsive role of β-catenin in keratoconus epithelium. Sci. Rep. 2020, 10, 21382. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yu, C.; Li, F.; Zuo, Y.; Wang, Y.; Yao, L.; Wu, C.; Wang, C.; Ye, L. Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduct. Target. Ther. 2021, 6, 307. [Google Scholar] [CrossRef]
- del Río Hernández, A.; Warboys, C.M. Mechanoactivation of Wnt/β-catenin pathways in health and disease. Emerg. Top. Life Sci. 2018, 2, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Hammes, S.R. Paxillin actions in the nucleus. Steroids 2018, 133, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Noh, K.; Bach, D.-H.; Choi, H.-J.; Kim, M.S.; Wu, S.Y.; Pradeep, S.; Ivan, C.; Cho, M.-S.; Bayraktar, E.; Rodriguez-Aguayo, C.; et al. The hidden role of paxillin: Localization to nucleus promotes tumor angiogenesis. Oncogene 2020, 40, 384–395. [Google Scholar] [CrossRef]
- Silva, A.J.D.; Hästbacka, H.S.E.; Puustinen, M.C.; Pessa, J.C.; Goult, B.T.; Jacquemet, G.; Henriksson, E.; Sistonen, L. A subpopulation of Talin 1 resides in the nucleus and regulates gene expression. bioRxiv 2022. bioRxiv:2022.03.15.484419. [Google Scholar]
- LeBlanc, L.; Ramirez, N.; Kim, J. Context-dependent roles of YAP/TAZ in stem cell fates and cancer. Cell. Mol. Life Sci. 2021, 78, 4201–4219. [Google Scholar] [CrossRef]
- He, L.; Pratt, H.; Gao, M.; Wei, F.; Weng, Z.; Struhl, K. YAP and TAZ are transcriptional co-activators of AP-1 proteins and STAT3 during breast cellular transformation. eLife 2021, 10, e67312. [Google Scholar] [CrossRef]
- Dupont, S. Regulation of YAP/TAZ Activity by Mechanical Cues: An Experimental Overview. Methods Mol. Biol. 2018, 1893, 183–202. [Google Scholar] [CrossRef]
- Burridge, K.; Monaghan-Benson, E.; Graham, D.M. Mechanotransduction: From the cell surface to the nucleus via RhoA. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180229. [Google Scholar] [CrossRef] [PubMed]
- Crosas-Molist, E.; Samain, R.; Kohlhammer, L.; Orgaz, J.L.; George, S.L.; Maiques, O.; Barcelo, J.; Sanz-Moreno, V. Rho GTPase signaling in cancer progression and dissemination. Physiol. Rev. 2022, 102, 455–510. [Google Scholar] [CrossRef] [PubMed]
- Boyle, S.T.; Kular, J.; Nobis, M.; Ruszkiewicz, A.; Timpson, P.; Samuel, M.S. Acute compressive stress activates RHO/ROCK-mediated cellular processes. Small GTPases 2020, 11, 354–370. [Google Scholar] [CrossRef] [PubMed]
- Totaro, A.; Zhuang, Q.; Panciera, T.; Battilana, G.; Azzolin, L.; Brumana, G.; Gandin, A.; Brusatin, G.; Cordenonsi, M.; Piccolo, S. Cell phenotypic plasticity requires autophagic flux driven by YAP/TAZ mechanotransduction. Proc. Natl. Acad. Sci. USA 2019, 116, 17848–17857. [Google Scholar] [CrossRef] [PubMed]
- Boyko, S.; Surewicz, W.K. Tau liquid–liquid phase separation in neurodegenerative diseases. Trends Cell Biol. 2022, 32, 611–623. [Google Scholar] [CrossRef]
- Wiersma, V.; Rigort, R.; Polymenidou, M. Tau: A phase in the crowd. EMBO J. 2022, 41, e111425. [Google Scholar] [CrossRef] [PubMed]
- Kanekura, K.; Kuroda, M. How can we interpret the relationship between liquid-liquid phase separation and amyotrophic lateral sclerosis? Lab. Investig. 2022, 102, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Carey, J.L.; Guo, L. Liquid-Liquid Phase Separation of TDP-43 and FUS in Physiology and Pathology of Neurodegenerative Diseases. Front. Mol. Biosci. 2022, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Zbinden, A.; Pérez-Berlanga, M.; De Rossi, P.; Polymenidou, M. Phase Separation and Neurodegenerative Diseases: A Disturbance in the Force. Dev. Cell 2020, 55, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Morrow, C.S.; Porter, T.J.; Xu, N.; Arndt, Z.P.; Ako-Asare, K.; Heo, H.; Thompson, E.A.; Moore, D.L. Vimentin Coordinates Protein Turnover at the Aggresome during Neural Stem Cell Quiescence Exit. Cell Stem Cell 2020, 26, 558–568.e9. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Kornmann, B. Mechanical forces on cellular organelles. J. Cell Sci. 2018, 131, jcs218479. [Google Scholar] [CrossRef]
- Phuyal, S.; Baschieri, F. Endomembranes: Unsung Heroes of Mechanobiology? Front. Bioeng. Biotechnol. 2020, 8, 1243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, J.; Quan, Z.; Li, H.; Qing, H. Mitochondria and Other Organelles in Neural Development and Their Potential as Therapeutic Targets in Neurodegenerative Diseases. Front. Neurosci. 2022, 16, 418. [Google Scholar] [CrossRef]
- Lu, M.; Ward, E.; van Tartwijk, F.W.; Kaminski, C.F. Advances in the study of organelle interactions and their role in neurodegenerative diseases enabled by super-resolution microscopy. Neurobiol. Dis. 2021, 159, 105475. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.S.; Yoon, C.W.; Wang, Q.; Moon, S.; Koo, K.M.; Jung, H.; Chen, R.; Jiang, L.; Lu, G.; Fernandez, A.; et al. Focused Ultrasound Stimulates ER Localized Mechanosensitive PANNEXIN-1 to Mediate Intracellular Calcium Release in Invasive Cancer Cells. Front. Cell Dev. Biol. 2020, 8, 504. [Google Scholar] [CrossRef] [PubMed]
- Nava, M.; Miroshnikova, Y.A.; Biggs, L.; Whitefield, D.B.; Metge, F.; Boucas, J.; Vihinen, H.; Jokitalo, E.; Li, X.; Arcos, J.M.G.; et al. Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage. Cell 2020, 181, 800–817.e22. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Erady, C.; Balasubramanian, N. Cell-matrix adhesion controls Golgi organization and function through Arf1 activation in anchorage-dependent cells. J. Cell Sci. 2018, 131, jcs215855. [Google Scholar] [CrossRef]
- Rafiq, N.B.M.; Nishimura, Y.; Plotnikov, S.V.; Thiagarajan, V.; Zhang, Z.; Shi, S.; Natarajan, M.; Viasnoff, V.; Kanchanawong, P.; Jones, G.E.; et al. A mechano-signalling network linking microtubules, myosin IIA filaments and integrin-based adhesions. Nat. Mater. 2019, 18, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Romani, P.; Brian, I.; Santinon, G.; Pocaterra, A.; Audano, M.; Pedretti, S.; Mathieu, S.; Forcato, M.; Bicciato, S.; Manneville, J.-B.; et al. Extracellular matrix mechanical cues regulate lipid metabolism through Lipin-1 and SREBP. Nat. Cell Biol. 2019, 21, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Romani, P.; Valcarcel-Jimenez, L.; Frezza, C.; Dupont, S. Crosstalk between mechanotransduction and metabolism. Nat. Rev. Mol. Cell Biol. 2020, 22, 22–38. [Google Scholar] [CrossRef]
- Elia, I.; Broekaert, D.; Christen, S.; Boon, R.; Radaelli, E.; Orth, M.; Verfaillie, C.; Grünewald, T.G.P.; Fendt, S.-M. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 2017, 8, 15267. [Google Scholar] [CrossRef] [PubMed]
- Hawk, M.A.; Gorsuch, C.L.; Fagan, P.; Lee, C.; Kim, S.E.; Hamann, J.C.; Mason, J.A.; Weigel, K.J.; Tsegaye, M.A.; Shen, L.; et al. RIPK1-mediated induction of mitophagy compromises the viability of extracellular-matrix-detached cells. Nat. Cell Biol. 2018, 20, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Helle, S.C.J.; Feng, Q.; Aebersold, M.J.; Hirt, L.; Grüter, R.R.; Vahid, A.; Sirianni, A.; Mostowy, S.; Snedeker, J.G.; Šarić, A.; et al. Mechanical force induces mitochondrial fission. eLife 2017, 6, e30292. [Google Scholar] [CrossRef] [PubMed]
- Mahecic, D.; Carlini, L.; Kleele, T.; Colom, A.; Goujon, A.; Matile, S.; Roux, A.; Manley, S. Mitochondrial membrane tension governs fission. Cell Rep. 2021, 35, 108947. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Qiu, H. New Insights Into Energy Substrate Utilization and Metabolic Remodeling in Cardiac Physiological Adaption. Front. Physiol. 2022, 13, 831829. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; Karwi, Q.G.; Tian, R.; Wende, A.R.; Abel, E.D. Cardiac Energy Metabolism in Heart Failure. Circ. Res. 2021, 128, 1487–1513. [Google Scholar] [CrossRef] [PubMed]
- Tharp, K.M.; Higuchi-Sanabria, R.; Timblin, G.A.; Ford, B.; Garzon-Coral, C.; Schneider, C.; Muncie, J.M.; Stashko, C.; Daniele, J.R.; Moore, A.S.; et al. Adhesion-mediated mechanosignaling forces mitohormesis. Cell Metab. 2021, 33, 1322–1341.e13. [Google Scholar] [CrossRef] [PubMed]
- Bevere, M.; Morabito, C.; Mariggiò, M.A.; Guarnieri, S. The Oxidative Balance Orchestrates the Main Keystones of the Functional Activity of Cardiomyocytes. Oxid. Med. Cell. Longev. 2022, 2022, 7714542. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Qi, Y.; Ye, Y.; Yue, P.; Zhang, D.; Li, Y. Mechanotranduction Pathways in the Regulation of Mitochondrial Homeostasis in Cardiomyocytes. Front. Cell Dev. Biol. 2021, 8, 625089. [Google Scholar] [CrossRef]
- Pavel, M.; Renna, M.; Park, S.J.; Menzies, F.M.; Ricketts, T.; Füllgrabe, J.; Ashkenazi, A.; Frake, R.A.; Lombarte, A.C.; Bento, C.F.; et al. Contact inhibition controls cell survival and proliferation via YAP/TAZ-autophagy axis. Nat. Commun. 2018, 9, 2961. [Google Scholar] [CrossRef] [PubMed]
- Stürner, E.; Behl, C. The Role of the Multifunctional BAG3 Protein in Cellular Protein Quality Control and in Disease. Front. Mol. Neurosci. 2017, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Kirk, J.A.; Cheung, J.Y.; Feldman, A.M. Therapeutic targeting of BAG3: Considering its complexity in cancer and heart disease. J. Clin. Investig. 2021, 131(16), e149415. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Deaton, C.A.; Johnson, G.V. Commentary: BAG3 as a Mediator of Endosome Function and Tau Clearance. Neuroscience, 2022; (in press). [CrossRef]
- Basu, S.; Singh, M.; Verma, M.; Rachana, R. Multifarious Role of BAG3 in Neurodegenerative Disorders. In Quality Control of Cellular Protein in Neurodegenerative Disorders; Uddin, M., Ashraf, G., Eds.; IGI Global: Hershey, Pennsylvania, 2020; pp. 261–281. [Google Scholar] [CrossRef]
- Gamerdinger, M.; Kaya, A.M.; Wolfrum, U.; Clement, A.M.; Behl, C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011, 12, 149–156. [Google Scholar] [CrossRef]
- Agarwal, R.; Paulo, J.A.; Toepfer, C.N.; Ewoldt, J.K.; Sundaram, S.; Chopra, A.; Zhang, Q.; Gorham, J.; DePalma, S.R.; Chen, C.S.; et al. Filamin C Cardiomyopathy Variants Cause Protein and Lysosome Accumulation. Circ. Res. 2021, 129, 751–766. [Google Scholar] [CrossRef] [PubMed]
- Thottacherry, J.J.; Kosmalska, A.J.; Kumar, A.; Vishen, A.S.; Elosegui-Artola, A.; Pradhan, S.; Sharma, S.; Singh, P.P.; Guadamillas, M.C.; Chaudhary, N.; et al. Mechanochemical feedback control of dynamin independent endocytosis modulates membrane tension in adherent cells. Nat. Commun. 2018, 9, 4217. [Google Scholar] [CrossRef]
- Molnár, M.; Sőth, Á.; Simon-Vecsei, Z. Pathways of integrins in the endo-lysosomal system. Biol. Futur. 2022, 73, 171–185. [Google Scholar] [CrossRef]
- Shang, X.; Böker, K.O.; Taheri, S.; Lehmann, W.; Schilling, A.F. Extracellular Vesicles Allow Epigenetic Mechanotransduction between Chondrocytes and Osteoblasts. Int. J. Mol. Sci. 2021, 22, 13282. [Google Scholar] [CrossRef] [PubMed]
- Scerra, G.; De Pasquale, V.; Pavone, L.M.; Caporaso, M.G.; Mayer, A.; Renna, M.; D’Agostino, M. Early onset effects of single substrate accumulation recapitulate major features of LSD in patient-derived lysosomes. iScience 2021, 24, 102707. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Nola, S.; Bovio, S.; Bun, P.; Coppey-Moisan, M.; Lafont, F.; Galli, T. Biomechanical Control of Lysosomal Secretion Via the VAMP7 Hub: A Tug-of-War between VARP and LRRK1. iScience 2018, 4, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Engelke, K.; Ebert, R.; Müller-Deubert, S.; Rudert, M.; Ziouti, F.; Jundt, F.; Felsenberg, D.; Jakob, F. Interactions between Muscle and Bone—Where Physics Meets Biology. Biomolecules 2020, 10, 432. [Google Scholar] [CrossRef]
- Kamienieva, I.; Duszyński, J.; Szczepanowska, J. Multitasking guardian of mitochondrial quality: Parkin function and Parkinson’s disease. Transl. Neurodegener. 2021, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Bartolák-Suki, E.; Imsirovic, J.; Nishibori, Y.; Krishnan, R.; Suki, B. Regulation of Mitochondrial Structure and Dynamics by the Cytoskeleton and Mechanical Factors. Int. J. Mol. Sci. 2017, 18, 1812. [Google Scholar] [CrossRef]
- Greenwell, A.A.; Gopal, K.; Ussher, J.R. Myocardial Energy Metabolism in Non-ischemic Cardiomyopathy. Front. Physiol. 2020, 11, 570421. [Google Scholar] [CrossRef] [PubMed]
- Kashihara, T.; Sadoshima, J. Role of YAP/TAZ in Energy Metabolism in the Heart. J. Cardiovasc. Pharmacol. 2019, 74, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Malvezzi, C.C.; Cabassi, A.; Miragoli, M. Mitochondrial mechanosensor in cardiovascular diseases. Vasc. Biol. 2020, 2, R85–R92. [Google Scholar] [CrossRef] [PubMed]
- Saftig, P.; Puertollano, R. How Lysosomes Sense, Integrate, and Cope with Stress. Trends Biochem. Sci. 2021, 46, 97–112. [Google Scholar] [CrossRef]
- Zatyka, M.; Sarkar, S.; Barrett, T. Autophagy in Rare (NonLysosomal) Neurodegenerative Diseases. J. Mol. Biol. 2020, 432, 2735–2753. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Pedro, J.M.B.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 2019, 18, 923–948. [Google Scholar] [CrossRef] [PubMed]
- Wallings, R.L.; Humble, S.W.; Ward, M.E.; Wade-Martins, R. Lysosomal Dysfunction at the Centre of Parkinson’s Disease and Frontotemporal Dementia/Amyotrophic Lateral Sclerosis. Trends Neurosci. 2019, 42, 899–912. [Google Scholar] [CrossRef]
- Root, J.; Merino, P.; Nuckols, A.; Johnson, M.; Kukar, T. Lysosome dysfunction as a cause of neurodegenerative diseases: Lessons from frontotemporal dementia and amyotrophic lateral sclerosis. Neurobiol. Dis. 2021, 154, 105360. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, C.; Sun, J.; Shen, H.-M.; Wang, J.; Yang, C. Impairment of the autophagy–lysosomal pathway in Alzheimer’s diseases: Pathogenic mechanisms and therapeutic potential. Acta Pharm. Sin. B 2022, 12, 1019–1040. [Google Scholar] [CrossRef]
- Darios, F.; Stevanin, G. Impairment of Lysosome Function and Autophagy in Rare Neurodegenerative Diseases. J. Mol. Biol. 2020, 432, 2714–2734. [Google Scholar] [CrossRef] [PubMed]
- Tiribuzi, R.; Orlacchio, A.; Crispoltoni, L.; Maiotti, M.; Zampolini, M.; De Angeliz, M.; Mecocci, P.; Cecchetti, R.; Bernardi, G.; Datti, A.; et al. Lysosomal β-Galactosidase and β-Hexosaminidase Activities Correlate with Clinical Stages of Dementia Associated with Alzheimer’s Disease and Type 2 Diabetes Mellitus. J. Alzheimer Dis. 2011, 24, 785–797. [Google Scholar] [CrossRef]
- Martino, S.; Emiliani, C.; Tancini, B.; Severini, G.M.; Chigorno, V.; Bordignon, C.; Sonnino, S.; Orlacchio, A. Absence of Metabolic Cross-correction in Tay-Sachs Cells: Implications for gene therapy. J. Biol. Chem. 2002, 277, 20177–20184. [Google Scholar] [CrossRef] [PubMed]
- Santambrogio, S.; Ricca, A.; Maderna, C.; Ieraci, A.; Aureli, M.; Sonnino, S.; Kulik, W.; Aimar, P.; Bonfanti, L.; Martino, S.; et al. The galactocerebrosidase enzyme contributes to maintain a functional neurogenic niche during early post-natal CNS development. Hum. Mol. Genet. 2012, 21, 4732–4750. [Google Scholar] [CrossRef] [PubMed]
- Frati, G.; Luciani, M.; Meneghini, V.; De Cicco, S.; Ståhlman, M.; Blomqvist, M.; Grossi, S.; Filocamo, M.; Morena, F.; Menegon, A.; et al. Human iPSC-based models highlight defective glial and neuronal differentiation from neural progenitor cells in metachromatic leukodystrophy. Cell Death Dis. 2018, 9, 698. [Google Scholar] [CrossRef] [PubMed]
- Meneghini, V.; Lattanzi, A.; Tiradani, L.; Bravo, G.; Morena, F.; Sanvito, F.; Calabria, A.; Bringas, J.; Fisher-Perkins, J.M.; Dufour, J.P.; et al. Pervasive supply of therapeutic lysosomal enzymes in the CNS of normal and Krabbe-affected non-human primates by intracerebral lentiviral gene therapy. EMBO Mol. Med. 2016, 8, 489–510. [Google Scholar] [CrossRef] [PubMed]
- Martino, S.; di Girolamo, I.; Cavazzin, C.; Tiribuzi, R.; Galli, R.; Rivaroli, A.; Valsecchi, M.; Sandhoff, K.; Sonnino, S.; Vescovi, A.; et al. Neural precursor cell cultures from GM2 gangliosidosis animal models recapitulate the biochemical and molecular hallmarks of the brain pathology. J. Neurochem. 2009, 109, 135–147. [Google Scholar] [CrossRef]
- Fumagalli, F.; Calbi, V.; Sora, M.G.N.; Sessa, M.; Baldoli, C.; Rancoita, P.M.V.; Ciotti, F.; Sarzana, M.; Fraschini, M.; Zambon, A.A.; et al. Lentiviral haematopoietic stem-cell gene therapy for early-onset metachromatic leukodystrophy: Long-term results from a non-randomised, open-label, phase 1/2 trial and expanded access. Lancet 2022, 399, 372–383. [Google Scholar] [CrossRef]
- Parenti, G.; Medina, D.L.; Ballabio, A. The rapidly evolving view of lysosomal storage diseases. EMBO Mol. Med. 2021, 13, e12836. [Google Scholar] [CrossRef]
- Lakpa, K.L.; Khan, N.; Afghah, Z.; Chen, X.; Geiger, J.D. Lysosomal Stress Response (LSR): Physiological Importance and Pathological Relevance. J. Neuroimmune Pharmacol. 2021, 16, 219–237. [Google Scholar] [CrossRef]
- Malik, B.R.; Maddison, D.C.; Smith, G.A.; Peters, O.M. Autophagic and endo-lysosomal dysfunction in neurodegenerative disease. Mol. Brain 2019, 12, 100. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.G.; Liu, A.P. Mechanical Regulation of Endocytosis: New Insights and Recent Advances. Adv. Biosyst. 2020, 4, 1900278. [Google Scholar] [CrossRef] [PubMed]
- Franze, K.; Janmey, P.A.; Guck, J. Mechanics in Neuronal Development and Repair. Annu. Rev. Biomed. Eng. 2013, 15, 227–251. [Google Scholar] [CrossRef]
- Oliveri, H.; Goriely, A. Mathematical models of neuronal growth. Biomech. Model. Mechanobiol. 2022, 21, 89–118. [Google Scholar] [CrossRef] [PubMed]
- Franze, K. Integrating Chemistry and Mechanics: The Forces Driving Axon Growth. Annu. Rev. Cell Dev. Biol. 2020, 36, 61–83. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, C.; Zhao, H. Mechanical properties of brain tissue based on microstructure. J. Mech. Behav. Biomed. Mater. 2021, 126, 104924. [Google Scholar] [CrossRef]
- Chighizola, M.; Dini, T.; Lenardi, C.; Milani, P.; Podestà, A.; Schulte, C. Mechanotransduction in neuronal cell development and functioning. Biophys. Rev. 2019, 11, 701–720. [Google Scholar] [CrossRef]
- Long, K.R.; Huttner, W.B. How the extracellular matrix shapes neural development. Open Biol. 2019, 9, 180216. [Google Scholar] [CrossRef]
- Segel, M.; Neumann, B.; Hill, M.F.E.; Weber, I.P.; Viscomi, C.; Zhao, C.; Young, A.; Agley, C.C.; Thompson, A.J.; Gonzalez, G.A.; et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature 2019, 573, 130–134. [Google Scholar] [CrossRef]
- Babu, P.K.V.; Radmacher, M. Mechanics of Brain Tissues Studied by Atomic Force Microscopy: A Perspective. Front. Neurosci. 2019, 13, 600. [Google Scholar] [CrossRef]
- Roth, J.G.; Huang, M.S.; Li, T.L.; Feig, V.R.; Jiang, Y.; Cui, B.; Greely, H.T.; Bao, Z.; Paşca, S.P.; Heilshorn, S.C. Advancing models of neural development with biomaterials. Nat. Rev. Neurosci. 2021, 22, 593–615. [Google Scholar] [CrossRef] [PubMed]
- Ławkowska, K.; Pokrywczyńska, M.; Koper, K.; Kluth, L.A.; Drewa, T.; Adamowicz, J. Application of Graphene in Tissue Engineering of the Nervous System. Int. J. Mol. Sci. 2021, 23, 33. [Google Scholar] [CrossRef] [PubMed]
- Kothapalli, C.; Mahajan, G.; Farrell, K. Substrate stiffness induced mechanotransduction regulates temporal evolution of human fetal neural progenitor cell phenotype, differentiation, and biomechanics. Biomater. Sci. 2020, 8, 5452–5464. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, A.; Ahmadi, H.; Taheri, B.; Hosseinzadeh, S.; Fatahi, Y.; Soleimani, M.; Atyabi, F.; Dinarvand, R. Appropriate Scaffold Selection for CNS Tissue Engineering. Avicenna J. Med. Biotechnol. 2020, 12, 203–220. [Google Scholar] [PubMed]
- Linka, K.; Reiter, N.; Würges, J.; Schicht, M.; Bräuer, L.; Cyron, C.J.; Paulsen, F.; Budday, S. Unraveling the Local Relation Between Tissue Composition and Human Brain Mechanics Through Machine Learning. Front. Bioeng. Biotechnol. 2021, 9, 704738. [Google Scholar] [CrossRef]
- Menichetti, A.; Bartsoen, L.; Depreitere, B.; Sloten, J.V.; Famaey, N. A Machine Learning Approach to Investigate the Uncertainty of Tissue-Level Injury Metrics for Cerebral Contusion. Front. Bioeng. Biotechnol. 2021, 9, 714128. [Google Scholar] [CrossRef]
- Schroder, A.; Lawrence, T.; Voets, N.; Garcia-Gonzalez, D.; Jones, M.; Peña, J.-M.; Jerusalem, A. A Machine Learning Enhanced Mechanistic Simulation Framework for Functional Deficit Prediction in TBI. Front. Bioeng. Biotechnol. 2021, 9, 50. [Google Scholar] [CrossRef]
- Sack, I.; Beierbach, B.; Wuerfel, J.; Klatt, D.; Hamhaber, U.; Papazoglou, S.; Martus, P.; Braun, J. The impact of aging and gender on brain viscoelasticity. NeuroImage 2009, 46, 652–657. [Google Scholar] [CrossRef]
- Arani, A.; Murphy, M.C.; Glaser, K.J.; Manduca, A.; Lake, D.S.; Kruse, S.A.; Jack, C.R.; Ehman, R.L.; Huston, J. Measuring the effects of aging and sex on regional brain stiffness with MR elastography in healthy older adults. NeuroImage 2015, 111, 59–64. [Google Scholar] [CrossRef]
- Takamura, T.; Motosugi, U.; Sasaki, Y.; Rt, T.K.; Sato, K.; Glaser, K.J.; Ehman, R.L.; Onishi, H. Influence of Age on Global and Regional Brain Stiffness in Young and Middle-Aged Adults. J. Magn. Reson. Imaging 2019, 51, 727–733. [Google Scholar] [CrossRef]
- Yin, Z.; Romano, A.J.; Manduca, A.; Ehman, R.L.; Huston, J. Stiffness and Beyond: What MR Elastography Can Tell Us About Brain Structure and Function Under Physiologic and Pathologic Conditions. Top. Magn. Reson. Imaging 2018, 27, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Hiscox, L.V.; Schwarb, H.; McGarry, M.D.; Johnson, C.L. Aging brain mechanics: Progress and promise of magnetic resonance elastography. NeuroImage 2021, 232, 117889. [Google Scholar] [CrossRef] [PubMed]
- Delgorio, P.L.; Hiscox, L.V.; Daugherty, A.M.; Sanjana, F.; Pohlig, R.T.; Ellison, J.M.; Martens, C.R.; Schwarb, H.; McGarry, M.D.J.; Johnson, C.L. Effect of Aging on the Viscoelastic Properties of Hippocampal Subfields Assessed with High-Resolution MR Elastography. Cereb. Cortex 2021, 31, 2799–2811. [Google Scholar] [CrossRef] [PubMed]
- Ngo, M.T.; Harley, B.A.C. Progress in mimicking brain microenvironments to understand and treat neurological disorders. APL Bioeng. 2021, 5, 020902. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, K.; Carman-Esparza, C.M.; Munson, J.M. Methods to measure, model and manipulate fluid flow in brain. J. Neurosci. Methods 2019, 333, 108541. [Google Scholar] [CrossRef] [PubMed]
- Cobbaut, M.; Karagil, S.; Bruno, L.; Loza, M.D.C.D.D.L.; Mackenzie, F.; Stolinski, M.; Elbediwy, A. Dysfunctional Mechanotransduction through the YAP/TAZ/Hippo Pathway as a Feature of Chronic Disease. Cells 2020, 9, 151. [Google Scholar] [CrossRef]
- Muñoz-Lasso, D.C.; Romá-Mateo, C.; Pallardó, F.V.; Gonzalez-Cabo, P. Much More Than a Scaffold: Cytoskeletal Proteins in Neurological Disorders. Cells 2020, 9, 358. [Google Scholar] [CrossRef]
- Gharaba, S.; Paz, O.; Feld, L.; Abashidze, A.; Weinrab, M.; Wolfenson, H.; Weil, M. Perturbed actin cap and nuclear morphology in primary fibroblasts of Huntington’s disease patients as a new phenotypic marker for personalized drug evaluation. bioRxiv 2022. bioRxiv:2022.04.03.486386. [Google Scholar]
- Castellanos-Montiel, M.J.; Chaineau, M.; Durcan, T.M. The Neglected Genes of ALS: Cytoskeletal Dynamics Impact Synaptic Degeneration in ALS. Front. Cell. Neurosci. 2020, 14, 380. [Google Scholar] [CrossRef]
- Rush, T.; Martinez-Hernandez, J.; Dollmeyer, M.; Frandemiche, M.L.; Borel, E.; Boisseau, S.; Jacquier-Sarlin, M.; Buisson, A. Synaptotoxicity in Alzheimer’s Disease Involved a Dysregulation of Actin Cytoskeleton Dynamics through Cofilin 1 Phosphorylation. J. Neurosci. 2018, 38, 10349–10361. [Google Scholar] [CrossRef]
- Wang, Q.; Yuan, W.; Yang, X.; Wang, Y.; Li, Y.; Qiao, H. Role of Cofilin in Alzheimer’s Disease. Front. Cell Dev. Biol. 2020, 8, 1357. [Google Scholar] [CrossRef]
- Kang, D.E.; Woo, J.A. Cofilin, a Master Node Regulating Cytoskeletal Pathogenesis in Alzheimer’s Disease. J. Alzheimers. Dis. 2019, 72, S131–S144. [Google Scholar] [CrossRef] [PubMed]
- Lapeña-Luzón, T.; Rodríguez, L.R.; Beltran-Beltran, V.; Benetó, N.; Pallardó, F.V.; Gonzalez-Cabo, P. Cofilin and Neurodegeneration: New Functions for an Old but Gold Protein. Brain Sci. 2021, 11, 954. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.I.; Blaabjerg, M.; Freude, K.; Meyer, M. RhoA Signaling in Neurodegenerative Diseases. Cells 2022, 11, 1520. [Google Scholar] [CrossRef] [PubMed]
- Oliveira da Silva, M.I.; Liz, M.A. Linking Alpha-Synuclein to the Actin Cytoskeleton: Consequences to Neuronal Function. Front. Cell Dev. Biol. 2020, 8, 787. [Google Scholar] [CrossRef]
- Richardson, D.; McEntagart, M.M.; Isaacs, J.D. DCTN1-related Parkinson-plus disorder (Perry syndrome). Pract. Neurol. 2020, 20, 317–319. [Google Scholar] [CrossRef]
- Burns, L.H.; Wang, H.-Y. Altered filamin A enables amyloid beta-induced tau hyperphosphorylation and neuroinflammation in Alzheimer’s disease. Neuroimmunol. Neuroinflamm. 2017, 4, 263–271. [Google Scholar] [CrossRef]
- Seervai, R.N.H.; Jangid, R.K.; Karki, M.; Tripathi, D.N.; Jung, S.Y.; Kearns, S.E.; Verhey, K.J.; Cianfrocco, M.A.; Millis, B.A.; Tyska, M.J.; et al. The Huntingtin-interacting protein SETD2/HYPB is an actin lysine methyltransferase. Sci. Adv. 2020, 6, eabb7854. [Google Scholar] [CrossRef]
- Dourlen, P.; Kilinc, D.; Malmanche, N.; Chapuis, J.; Lambert, J.C. The new genetic landscape of Alzheimer’s disease: From amyloid cascade to genetically driven synaptic failure hypothesis? Acta Neuropathol. 2019, 138, 221–236. [Google Scholar] [CrossRef]
- Eysert, F.; Coulon, A.; Boscher, E.; Vreulx, A.-C.; Flaig, A.; Mendes, T.; Hughes, S.; Grenier-Boley, B.; Hanoulle, X.; Demiautte, F.; et al. Alzheimer’s genetic risk factor FERMT2 (Kindlin-2) controls axonal growth and synaptic plasticity in an APP-dependent manner. Mol. Psychiatry 2020, 26, 5592–5607. [Google Scholar] [CrossRef]
- Nicolas, A.; Kenna, K.P.; Renton, A.E.; Ticozzi, N.; Faghri, F.; Chia, R.; Dominov, J.A.; Kenna, B.J.; Nalls, M.A.; Keagle, P.; et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 2018, 97, 1268–1283.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cooper-Knock, J.; Weimer, A.K.; Shi, M.; Moll, T.; Marshall, J.N.; Harvey, C.; Nezhad, H.G.; Franklin, J.; Souza, C.D.S.; et al. Genome-wide identification of the genetic basis of amyotrophic lateral sclerosis. Neuron 2022, 110, 992–1008.e11. [Google Scholar] [CrossRef] [PubMed]
- Gil, L.; Niño, S.A.; Capdeville, G.; Jiménez-Capdeville, M.E. Aging and Alzheimer’s disease connection: Nuclear Tau and lamin A. Neurosci. Lett. 2021, 749, 135741. [Google Scholar] [CrossRef] [PubMed]
- Alcalá-Vida, R.; Garcia-Forn, M.; Castany-Pladevall, C.; Creus-Muncunill, J.; Ito, Y.; Blanco, E.; Golbano, A.; Crespí-Vázquez, K.; Parry, A.; Slater, G.; et al. Neuron type-specific increase in lamin B1 contributes to nuclear dysfunction in Huntington’s disease. EMBO Mol. Med. 2020, 13, e12105. [Google Scholar] [CrossRef] [PubMed]
- Matias, I.; Diniz, L.P.; Damico, I.V.; Araujo, A.P.B.; Neves, L.D.S.; Vargas, G.; Leite, R.E.P.; Suemoto, C.K.; Nitrini, R.; Jacob-Filho, W.; et al. Loss of lamin-B1 and defective nuclear morphology are hallmarks of astrocyte senescence in vitro and in the aging human hippocampus. Aging Cell 2021, 21, e13521. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Vargas, J.; Castro-Álvarez, J.; Zapata-Berruecos, J.; Abdul-Rahim, K.; Arteaga-Noriega, A. Neurodegeneration and convergent factors contributing to the deterioration of the cytoskeleton in Alzheimer’s disease, cerebral ischemia and multiple sclerosis (Review). Biomed. Rep. 2022, 16, 27. [Google Scholar] [CrossRef]
- DeGiosio, R.A.; Grubisha, M.J.; MacDonald, M.L.; McKinney, B.C.; Camacho, C.J.; Sweet, R.A. More than a marker: Potential pathogenic functions of MAP2. Front. Mol. Neurosci. 2022, 15, 974890. [Google Scholar] [CrossRef]
- Cabrera, J.R.; Lucas, J.J. MAP2 Splicing is Altered in Huntington’s Disease. Brain Pathol. 2016, 27, 181–189. [Google Scholar] [CrossRef]
- Oeckl, P.; Weydt, P.; Thal, D.R.; Weishaupt, J.H.; Ludolph, A.C.; Otto, M. Proteomics in cerebrospinal fluid and spinal cord suggests UCHL1, MAP2 and GPNMB as biomarkers and underpins importance of transcriptional pathways in amyotrophic lateral sclerosis. Acta Neuropathol. 2019, 139, 119–134. [Google Scholar] [CrossRef]
- Varga, B.; Martin-Fernandez, M.; Hilaire, C.; Sanchez-Vicente, A.; Areias, J.; Salsac, C.; Cuisinier, F.J.G.; Raoul, C.; Scamps, F.; Gergely, C. Myotube elasticity of an amyotrophic lateral sclerosis mouse model. Sci. Rep. 2018, 8, 5917. [Google Scholar] [CrossRef]
- Jun, M.-H.; Jang, J.-W.; Jeon, P.; Lee, S.-K.; Lee, S.-H.; Choi, H.-E.; Lee, Y.-K.; Choi, H.; Park, S.-W.; Kim, J.; et al. Nonmuscle myosin IIB regulates Parkin-mediated mitophagy associated with amyotrophic lateral sclerosis-linked TDP. Cell Death Dis. 2020, 11, 952. [Google Scholar] [CrossRef]
- Hu, J.; Lin, S.L.; Schachner, M. A fragment of cell adhesion molecule L1 reduces amyloid-β plaques in a mouse model of Alzheimer’s disease. Cell Death Dis. 2022, 13, 48. [Google Scholar] [CrossRef]
- Chou, C.-C.; Zhang, Y.; Umoh, M.E.; Vaughan, S.W.; Lorenzini, I.; Liu, F.; Sayegh, M.; Donlin-Asp, P.; Chen, Y.H.; Duong, D.; et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 2018, 21, 228–239. [Google Scholar] [CrossRef]
- Fallini, C.; Khalil, B.; Smith, C.L.; Rossoll, W. Traffic jam at the nuclear pore: All roads lead to nucleocytoplasmic transport defects in ALS/FTD. Neurobiol. Dis. 2020, 140, 104835. [Google Scholar] [CrossRef] [PubMed]
- Velasco, M.; Mampay, M.; Boutin, H.; Chaney, A.; Warn, P.; Sharp, A.; Burgess, E.; Moeendarbary, E.; Dev, K.K.; Sheridan, G.K. Infection Augments Expression of Mechanosensing Piezo1 Channels in Amyloid Plaque-Reactive Astrocytes. Front. Aging Neurosci. 2018, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Velasco, M.; Rolle, S.O.; Mampay, M.; Dev, K.K.; Sheridan, G.K. Piezo1 regulates calcium oscillations and cytokine release from astrocytes. Glia 2019, 68, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Ordonez, D.G.; Lee, M.K.; Feany, M.B. α-synuclein Induces Mitochondrial Dysfunction through Spectrin and the Actin Cytoskeleton. Neuron 2017, 97, 108–124.e6. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, G.; Calogero, A.M.; Rolando, C. Microtubule acetylation: A reading key to neural physiology and degeneration. Neurosci. Lett. 2021, 755, 135900. [Google Scholar] [CrossRef]
- Billon, C.; Adham, S.; Poblete, N.H.; Legrand, A.; Frank, M.; Chiche, L.; Zuily, S.; Benistan, K.; Savale, L.; Zaafrane-Khachnaoui, K.; et al. Cardiovascular and connective tissue disorder features in FLNA-related PVNH patients: Progress towards a refined delineation of this syndrome. Orphanet J. Rare Dis. 2021, 16, 504. [Google Scholar] [CrossRef]
- Sasaki, E.; Byrne, A.T.; Phelan, E.; Cox, D.W.; Reardon, W. A review of filamin A mutations and associated interstitial lung disease. Eur. J. Pediatr. 2018, 178, 121–129. [Google Scholar] [CrossRef]
- Castaño-Jaramillo, L.M.; Lugo-Reyes, S.O.; Cruz Muñoz, M.E.; Scheffler-Mendoza, S.C.; Duran McKinster, C.; Yamazaki-Nakashimada, M.A.; Espinosa-Padilla, S.E.; Saez-de-Ocariz Gutierrez, M.d.M. Diagnostic and therapeutic caveats in Griscelli syndrome. Scand. J. Immunol. 2021, 93, e13034. [Google Scholar] [CrossRef] [PubMed]
- Griscelli Syndrome Type 1 (Concept Id: C1859194)-MedGen-NCBI. Available online: https://www.ncbi.nlm.nih.gov/medgen/347092 (accessed on 21 September 2022).
- Vontell, R.; Supramaniam, V.G.; Davidson, A.; Thornton, C.; Marnerides, A.; Holder-Espinasse, M.; Lillis, S.; Yau, S.; Jansson, M.; Hagberg, H.E.; et al. Post-mortem Characterisation of a Case With an ACTG1 Variant, Agenesis of the Corpus Callosum and Neuronal Heterotopia. Front. Physiol. 2019, 10, 623. [Google Scholar] [CrossRef]
- Parker, F.; Baboolal, T.G.; Peckham, M. Actin Mutations and Their Role in Disease. Int. J. Mol. Sci. 2020, 21, 3371. [Google Scholar] [CrossRef] [PubMed]
- Argentati, C.; Tortorella, I.; Bazzucchi, M.; Emiliani, C.; Morena, F.; Martino, S. The Other Side of Alzheimer’s Disease: Influence of Metabolic Disorder Features for Novel Diagnostic Biomarkers. J. Pers. Med. 2020, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Gerischer, L.M.; Fehlner, A.; Köbe, T.; Prehn, K.; Antonenko, D.; Grittner, U.; Braun, J.; Sack, I.; Flöel, A. Combining viscoelasticity, diffusivity and volume of the hippocampus for the diagnosis of Alzheimer’s disease based on magnetic resonance imaging. NeuroImage. Clin. 2017, 18, 485–493. [Google Scholar] [CrossRef]
- Hiscox, L.V.; Johnson, C.L.; McGarry, M.D.J.; Marshall, H.; Ritchie, C.W.; Van Beek, E.J.R.; Roberts, N.; Starr, J.M. Mechanical property alterations across the cerebral cortex due to Alzheimer’s disease. Brain Commun. 2019, 2, fcz049. [Google Scholar] [CrossRef]
- Fawcett, J.W.; Oohashi, T.; Pizzorusso, T. The roles of perineuronal nets and the perinodal extracellular matrix in neuronal function. Nat. Rev. Neurosci. 2019, 20, 451–465. [Google Scholar] [CrossRef]
- Logsdon, A.F.; Francis, K.L.; Richardson, N.E.; Hu, S.J.; Faber, C.L.; Phan, B.A.; Nguyen, V.; Setthavongsack, N.; Banks, W.A.; Woltjer, R.L.; et al. Decoding perineuronal net glycan sulfation patterns in the Alzheimer’s disease brain. Alzheimer Dement. 2021, 18, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Reichelt, A.C. Is loss of perineuronal nets a critical pathological event in Alzheimer’s disease? EBioMedicine 2020, 59, 102946. [Google Scholar] [CrossRef]
- Lu, Z.; Li, H.; Hou, C.; Peng, Y.; Long, J.; Liu, J. Endogenously generated amyloid-β increases stiffness in human neuroblastoma cells. Eur. Biophys. J. 2016, 46, 415–424. [Google Scholar] [CrossRef]
- Bruno, L.; Karagil, S.; Mahmood, A.; Elbediwy, A.; Stolinski, M.; Mackenzie, F.E. Mechanosensing and the Hippo Pathway in Microglia: A Potential Link to Alzheimer’s Disease Pathogenesis? Cells 2021, 10, 3144. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhu, H.; Yang, Q.; Shen, H.; Chai, G.; Zhang, B.; Chen, S.; Chen, Q.; Cai, Z.; Li, X.; et al. Microglial Piezo1 senses Aβ fibrils stiffness to restrict Alzheimer’s disease. bioRxiv 2022. bioRxiv:2022.03.23.485446. [Google Scholar] [CrossRef]
- Hall, C.M.; Moeendarbary, E.; Sheridan, G.K. Mechanobiology of the brain in ageing and Alzheimer’s disease. Eur. J. Neurosci. 2020, 53, 3851–3878. [Google Scholar] [CrossRef]
- Xu, J.; Patassini, S.; Rustogi, N.; Riba-Garcia, I.; Hale, B.D.; Phillips, A.M.; Waldvogel, H.; Haines, R.; Bradbury, P.; Stevens, A.; et al. Regional protein expression in human Alzheimer’s brain correlates with disease severity. Commun. Biol. 2019, 2, 43. [Google Scholar] [CrossRef]
- Tanaka, H.; Homma, H.; Fujita, K.; Kondo, K.; Yamada, S.; Jin, X.; Waragai, M.; Ohtomo, G.; Iwata, A.; Tagawa, K.; et al. YAP-dependent necrosis occurs in early stages of Alzheimer’s disease and regulates mouse model pathology. Nat. Commun. 2020, 11, 507. [Google Scholar] [CrossRef]
- Leshchyns’Ka, I.; Sytnyk, V. Synaptic Cell Adhesion Molecules in Alzheimer’s Disease. Neural Plast. 2016, 2016, 6427537. [Google Scholar] [CrossRef]
- Nielsen, H.; Wennström, M. Cell adhesion molecules in Alzheimer’s disease. Degener. Neurol. Neuromuscul. Dis. 2012, 2, 65–77. [Google Scholar] [CrossRef]
- Ilic, K.; Mlinac-Jerković, K.; Jovanov-Milosevic, N.; Simic, G.; Habek, N.; Bogdanovic, N.; Kalanj-Bognar, S. Hippocampal expression of cell-adhesion glycoprotein neuroplastin is altered in Alzheimer’s disease. J. Cell. Mol. Med. 2018, 23, 1602–1607. [Google Scholar] [CrossRef]
- Kozlova, I.; Sah, S.; Keable, R.; Leshchyns’Ka, I.; Janitz, M.; Sytnyk, V. Cell Adhesion Molecules and Protein Synthesis Regulation in Neurons. Front. Mol. Neurosci. 2020, 13, 592126. [Google Scholar] [CrossRef]
- Simufilam (PTI-125). Available online: https://alzheimersnewstoday.com/pti-125/ (accessed on 10 April 2022).
- Simufilam|ALZFORUM. Available online: https://www.alzforum.org/therapeutics/simufilam (accessed on 10 April 2022).
- Wang, H.-Y.; Lee, K.-C.; Pei, Z.; Khan, A.; Bakshi, K.; Burns, L.H. PTI-125 binds and reverses an altered conformation of filamin A to reduce Alzheimer’s disease pathogenesis. Neurobiol. Aging 2017, 55, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Simufilam 50 mg or 100 mg for Mild-to-Moderate Alzheimer’s Disease-Full Text View-ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT05026177 (accessed on 10 April 2022).
- Simufilam 100 mg for Mild-to-Moderate Alzheimer’s Disease-Full Text View-ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT04994483?term=Simufilam&draw=2&rank=2 (accessed on 10 April 2022).
- Bamburg, J.R.; Minamide, L.S.; Wiggan, O.; Tahtamouni, L.H.; Kuhn, T.B. Cofilin and Actin Dynamics: Multiple Modes of Regulation and Their Impacts in Neuronal Development and Degeneration. Cells 2021, 10, 2726. [Google Scholar] [CrossRef]
- Namme, J.N.; Bepari, A.K.; Takebayashi, H. Cofilin Signaling in the CNS Physiology and Neurodegeneration. Int. J. Mol. Sci. 2021, 22, 10727. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gutierrez, R.; Morales, R. The prion-like phenomenon in Alzheimer’s disease: Evidence of pathology transmission in humans. PLoS Pathog. 2020, 16, e1009004. [Google Scholar] [CrossRef]
- Condello, C.; DeGrado, W.F.; Prusiner, S.B. Prion biology: Implications for Alzheimer’s disease therapeutics. Lancet Neurol. 2020, 19, 802–803. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Zhang, L.; Yu, W.; Wang, Y.; Chang, W. Cellular Prion Protein as a Receptor of Toxic Amyloid-β42 Oligomers Is Important for Alzheimer’s Disease. Front. Cell. Neurosci. 2019, 13, 339. [Google Scholar] [CrossRef] [PubMed]
- Shafiq, M.; Zafar, S.; Younas, N.; Noor, A.; Puig, B.; Altmeppen, H.C.; Schmitz, M.; Matschke, J.; Ferrer, I.; Glatzel, M.; et al. Prion protein oligomers cause neuronal cytoskeletal damage in rapidly progressive Alzheimer’s disease. Mol. Neurodegener. 2021, 16, 11. [Google Scholar] [CrossRef]
- Frost, B. Alzheimer’s disease: An acquired neurodegenerative laminopathy. Nucleus 2016, 7, 275–283. [Google Scholar] [CrossRef]
- Vahabikashi, A.; Adam, S.A.; Medalia, O.; Goldman, R.D. Nuclear lamins: Structure and function in mechanobiology. APL Bioeng. 2022, 6, 011503. [Google Scholar] [CrossRef]
- Shin, Y.; Chang, Y.-C.; Lee, D.S.; Berry, J.; Sanders, D.W.; Ronceray, P.; Wingreen, N.S.; Haataja, M.; Brangwynne, C.P. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 2018, 175, 1481–1491.e13. [Google Scholar] [CrossRef]
- Iatrou, A.; Clark, E.M.; Wang, Y. Nuclear dynamics and stress responses in Alzheimer’s disease. Mol. Neurodegener. 2021, 16, 65. [Google Scholar] [CrossRef]
- Gil, L.; Niño, S.A.; Chi-Ahumada, E.; Rodríguez-Leyva, I.; Guerrero, C.; Rebolledo, A.B.; Arias, J.A.; Jiménez-Capdeville, M.E. Perinuclear Lamin A and Nucleoplasmic Lamin B2 Characterize Two Types of Hippocampal Neurons through Alzheimer’s Disease Progression. Int. J. Mol. Sci. 2020, 21, 1841. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-Q.; Ngian, Z.-K.; Koh, T.-W.; Ong, C.-T. Altered stability of nuclear lamin-B marks the onset of aging in male Drosophila. PLoS ONE 2022, 17, e0265223. [Google Scholar] [CrossRef] [PubMed]
- Murray, H.C.; Johnson, K.; Sedlock, A.; Highet, B.; Dieriks, B.V.; Anekal, P.V.; Faull, R.L.M.; Curtis, M.A.; Koretsky, A.; Maric, D. Lamina-specific immunohistochemical signatures in the olfactory bulb of healthy, Alzheimer’s and Parkinson’s disease patients. Commun. Biol. 2022, 5, 88. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wolozin, B. Oligomeric tau disrupts nuclear envelope via binding to lamin proteins and lamin B receptor. Alzheimer Dement. 2021, 17, e054521. [Google Scholar]
- Ippati, S.; Deng, Y.; van der Hoven, J.; Heu, C.; van Hummel, A.; Chua, S.W.; Paric, E.; Chan, G.; Feiten, A.; Fath, T.; et al. Rapid initiation of cell cycle reentry processes protects neurons from amyloid-β toxicity. Proc. Natl. Acad. Sci. USA 2021, 118, e2011876118. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, S.; Rozich, E.; Buttitta, L. Cell Cycle Re-entry in the Nervous System: From Polyploidy to Neurodegeneration. Front. Cell Dev. Biol. 2021, 9, 1511. [Google Scholar] [CrossRef] [PubMed]
- Calzoni, E.; Argentati, C.; Cesaretti, A.; Montegiove, N.; Tortorella, I.; Bazzucchi, M.; Morena, F.; Martino, S.; Emiliani, C. RNA Modifications in Neurodegenerations. In Epitranscriptomics; Springer: Cham, Switzerland, 2021; Volume 12. [Google Scholar]
- Komatsu, H. Innovative Therapeutic Approaches for Huntington’s Disease: From Nucleic Acids to GPCR-Targeting Small Molecules. Front. Cell. Neurosci. 2021, 15, 495. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Flower, M.; Ross, C.A.; Wild, E.J. Huntington disease: New insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 2020, 16, 529–546. [Google Scholar] [CrossRef]
- Taran, A.S.; Shuvalova, L.D.; Lagarkova, M.A.; Alieva, I.B. Huntington’s Disease—An Outlook on the Interplay of the HTT Protein, Microtubules and Actin Cytoskeletal Components. Cells 2020, 9, 1514. [Google Scholar] [CrossRef]
- Maninova, M.; Caslavsky, J.; Vomastek, T. The assembly and function of perinuclear actin cap in migrating cells. Protoplasma 2017, 254, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Hardiman, O.; Al-Chalabi, A.; Chio, A.; Corr, E.M.; Logroscino, G.; Robberecht, W.; Shaw, P.J.; Simmons, Z.; Van Den Berg, L.H. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Prim. 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed]
- Masrori, P.; Van Damme, P. Amyotrophic lateral sclerosis: A clinical review. Eur. J. Neurol. 2020, 27, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Shen, D.; Gao, Y.; Zhou, Q.; Ni, Y.; Meng, H.; Shi, H.; Le, W.; Chen, S.; Chen, S. A perspective on therapies for amyotrophic lateral sclerosis: Can disease progression be curbed? Transl. Neurodegener. 2021, 10, 29. [Google Scholar] [CrossRef]
- McAlary, L.; Chew, Y.L.; Lum, J.S.; Geraghty, N.J.; Yerbury, J.J.; Cashman, N.R. Amyotrophic Lateral Sclerosis: Proteins, Proteostasis, Prions, and Promises. Front. Cell. Neurosci. 2020, 14, 339. [Google Scholar] [CrossRef]
- Bicchi, I.; Morena, F.; Argentati, C.; Nodari, L.R.; Emiliani, C.; Gelati, M.; Vescovi, A.L.; Martino, S. Storage of Mutant Human SOD1 in Non-Neural Cells from the Type-1 Amyotrophic Lateral Sclerosis ratG93A Model Correlated with the Lysosomes’ Dysfunction. Biomedicines 2021, 9, 1080. [Google Scholar] [CrossRef]
- Perciballi, E.; Bovio, F.; Rosati, J.; Arrigoni, F.; D’Anzi, A.; Lattante, S.; Gelati, M.; De Marchi, F.; Lombardi, I.; Ruotolo, G.; et al. Characterization of the p.L145F and p.S135N Mutations in SOD1: Impact on the Metabolism of Fibroblasts Derived from Amyotrophic Lateral Sclerosis Patients. Antioxidants 2022, 11, 815. [Google Scholar] [CrossRef]
- Chen, J.; Bassot, A.; Giuliani, F.; Simmen, T. Amyotrophic Lateral Sclerosis (ALS): Stressed by Dysfunctional Mitochondria-Endoplasmic Reticulum Contacts (MERCs). Cells 2021, 10, 1789. [Google Scholar] [CrossRef]
- Brasil, A.D.A.; de Carvalho, M.D.C.; Gerhardt, E.; Queiroz, D.D.; Pereira, M.D.; Outeiro, T.F.; Eleutherio, E.C.A. Characterization of the activity, aggregation, and toxicity of heterodimers of WT and ALS-associated mutant Sod. Proc. Natl. Acad. Sci. USA 2019, 116, 25991–26000. [Google Scholar] [CrossRef]
- Huang, C.; Yan, S.; Zhang, Z. Maintaining the balance of TDP-43, mitochondria, and autophagy: A promising therapeutic strategy for neurodegenerative diseases. Transl. Neurodegener. 2020, 9, 40. [Google Scholar] [CrossRef]
- Shiota, T.; Nagata, R.; Kikuchi, S.; Nanaura, H.; Matsubayashi, M.; Nakanishi, M.; Kobashigawa, S.; Isozumi, N.; Kiriyama, T.; Nagayama, K.; et al. C9orf72-Derived Proline:Arginine Poly-Dipeptides Modulate Cytoskeleton and Mechanical Stress Response. Front. Cell Dev. Biol. 2022, 10, 630. [Google Scholar] [CrossRef] [PubMed]
- Miceli, M.; Exertier, C.; Cavaglià, M.; Gugole, E.; Boccardo, M.; Casaluci, R.R.; Ceccarelli, N.; De Maio, A.; Vallone, B.; Deriu, M.A. ALS2-Related Motor Neuron Diseases: From Symptoms to Molecules. Biology 2022, 11, 77. [Google Scholar] [CrossRef]
- Bercier, V.; Hubbard, J.M.; Fidelin, K.; Duroure, K.; Auer, T.O.; Revenu, C.; Wyart, C.; Del Bene, F. Dynactin1 depletion leads to neuromuscular synapse instability and functional abnormalities. Mol. Neurodegener. 2019, 14, 27. [Google Scholar] [CrossRef]
- Murk, K.; Ornaghi, M.; Schiweck, J. Profilin Isoforms in Health and Disease–All the Same but Different. Front. Cell Dev. Biol. 2021, 9, 681122. [Google Scholar] [CrossRef] [PubMed]
- Poesen, K.; Van Damme, P. Diagnostic and Prognostic Performance of Neurofilaments in ALS. Front. Neurol. 2019, 9, 1167. [Google Scholar] [CrossRef] [PubMed]
- Helferich, A.M.; Brockmann, S.J.; Reinders, J.; Deshpande, D.; Holzmann, K.; Brenner, D.; Andersen, P.M.; Petri, S.; Thal, D.R.; Michaelis, J.; et al. Dysregulation of a novel miR-1825/TBCB/TUBA4A pathway in sporadic and familial ALS. Cell. Mol. Life Sci. 2018, 75, 4301–4319. [Google Scholar] [CrossRef]
- Nguyen, D.K.; Thombre, R.; Wang, J. Autophagy as a common pathway in amyotrophic lateral sclerosis. Neurosci. Lett. 2018, 697, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Huai, J.; Zhang, Z. Structural Properties and Interaction Partners of Familial ALS-Associated SOD1 Mutants. Front. Neurol. 2019, 10, 527. [Google Scholar] [CrossRef] [PubMed]
- Giampetruzzi, A.; Danielson, E.W.; Gumina, V.; Jeon, M.; Boopathy, S.; Brown, R.H.; Ratti, A.; Landers, J.E.; Fallini, C. Modulation of actin polymerization affects nucleocytoplasmic transport in multiple forms of amyotrophic lateral sclerosis. Nat. Commun. 2019, 10, 3827. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Lusk, C.P. Emerging Connections between Nuclear Pore Complex Homeostasis and ALS. Int. J. Mol. Sci. 2022, 23, 1329. [Google Scholar] [CrossRef]
- Gasset-Rosa, F.; Lu, S.; Yu, H.; Chen, C.; Melamed, Z.; Guo, L.; Shorter, J.; Da Cruz, S.; Cleveland, D.W. Cytoplasmic TDP-43 De-mixing Independent of Stress Granules Drives Inhibition of Nuclear Import, Loss of Nuclear TDP-43, and Cell Death. Neuron 2019, 102, 339–357.e7. [Google Scholar] [CrossRef]
- Ding, B.; Sepehrimanesh, M. Nucleocytoplasmic Transport: Regulatory Mechanisms and the Implications in Neurodegeneration. Int. J. Mol. Sci. 2021, 22, 4165. [Google Scholar] [CrossRef] [PubMed]
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef]
- Ou, Z.; Pan, J.; Tang, S.; Duan, D.; Yu, D.; Nong, H.; Wang, Z. Global Trends in the Incidence, Prevalence, and Years Lived With Disability of Parkinson’s Disease in 204 Countries/Territories From 1990 to 2019. Front. Public Health 2021, 9, 1994. [Google Scholar] [CrossRef]
- Day, J.; Mullin, S. The Genetics of Parkinson’s Disease and Implications for Clinical Practice. Genes 2021, 12, 1006. [Google Scholar] [CrossRef]
- Seebauer, L.; Schneider, Y.; Drobny, A.; Plötz, S.; Koudelka, T.; Tholey, A.; Prots, I.; Winner, B.; Zunke, F.; Winkler, J.; et al. Interaction of Alpha Synuclein and Microtubule Organization Is Linked to Impaired Neuritic Integrity in Parkinson’s Patient-Derived Neuronal Cells. Int. J. Mol. Sci. 2022, 23, 1812. [Google Scholar] [CrossRef] [PubMed]
- Amadeo, A.; Pizzi, S.; Comincini, A.; Modena, D.; Calogero, A.M.; Madaschi, L.; Faustini, G.; Rolando, C.; Bellucci, A.; Pezzoli, G.; et al. The Association between α-Synuclein and α-Tubulin in Brain Synapses. Int. J. Mol. Sci. 2021, 22, 9153. [Google Scholar] [CrossRef]
- Calogero, A.M.; Mazzetti, S.; Pezzoli, G.; Cappelletti, G. Neuronal microtubules and proteins linked to Parkinson’s disease: A relevant interaction? Biol. Chem. 2019, 400, 1099–1112. [Google Scholar] [CrossRef] [PubMed]
- Casagrande, F.V.; Amadeo, A.; Cartelli, D.; Calogero, A.M.; Modena, D.; Costa, I.; Cantele, F.; Onelli, E.; Moscatelli, A.; Ascagni, M.; et al. The imbalance between dynamic and stable microtubules underlies neurodegeneration induced by 2,5-hexanedione. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1866, 165581. [Google Scholar] [CrossRef]
- Kruppa, A.; Kishi-Itakura, C.; Masters, T.; Rorbach, J.; Grice, G.L.; Kendrick-Jones, J.; Nathan, J.; Minczuk, M.; Buss, F. Myosin VI-Dependent Actin Cages Encapsulate Parkin-Positive Damaged Mitochondria. Dev. Cell 2018, 44, 484–499.e6. [Google Scholar] [CrossRef]
- Kruppa, A.J.; Buss, F. Motor proteins at the mitochondria–cytoskeleton interface. J. Cell Sci. 2021, 134, jcs226084. [Google Scholar] [CrossRef]
- Jetto, C.T.; Nambiar, A.; Manjithaya, R. Mitophagy and Neurodegeneration: Between the Knowns and the Unknowns. Front. Cell Dev. Biol. 2022, 10, 226. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, Y.; Lee, J.; Jarnik, M.; Wu, X.; Bonifacino, J.S.; Shen, J.; Ye, Y. A myosin-7B–dependent endocytosis pathway mediates cellular entry of α-synuclein fibrils and polycation-bearing cargos. Proc. Natl. Acad. Sci. USA 2020, 117, 10865–10875. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cué, C.; Rueda, N. Cellular Senescence in Neurodegenerative Diseases. Front. Cell. Neurosci. 2020, 14, 16. [Google Scholar] [CrossRef]
- Paonessa, F.; Evans, L.D.; Solanki, R.; Larrieu, D.; Wray, S.; Hardy, J.; Jackson, S.P.; Livesey, F.J. Microtubules Deform the Nuclear Membrane and Disrupt Nucleocytoplasmic Transport in Tau-Mediated Frontotemporal Dementia. Cell Rep. 2019, 26, 582–593.e5. [Google Scholar] [CrossRef] [PubMed]
- Chinta, S.J.; Woods, G.; Demaria, M.; Rane, A.; Zou, Y.; McQuade, A.; Rajagopalan, S.; Limbad, C.; Madden, D.T.; Campisi, J.; et al. Cellular Senescence Is Induced by the Environmental Neurotoxin Paraquat and Contributes to Neuropathology Linked to Parkinson’s Disease. Cell Rep. 2018, 22, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.; Subramaniam, M.D.; Venkatesan, D.; Cho, S.G.; Ryding, M.; Meyer, M.; Vellingiri, B. Role of RhoA-ROCK signaling in Parkinson’s disease. Eur. J. Pharmacol. 2021, 894, 173815. [Google Scholar] [CrossRef]
- Bogetofte, H.; Jensen, P.; Okarmus, J.; Schmidt, S.I.; Agger, M.; Ryding, M.; Nørregaard, P.; Fenger, C.; Zeng, X.; Graakjær, J.; et al. Perturbations in RhoA signalling cause altered migration and impaired neuritogenesis in human iPSC-derived neural cells with PARK2 mutation. Neurobiol. Dis. 2019, 132, 104581. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, B.; Sharma, S.S. Transient Receptor Potential Channels as an Emerging Target for the Treatment of Parkinson’s Disease: An Insight Into Role of Pharmacological Interventions. Front. Cell Dev. Biol. 2020, 8, 1387. [Google Scholar] [CrossRef]
- Bella, E.D.; Bersano, E.; Antonini, G.; Borghero, G.; Capasso, M.; Caponnetto, C.; Chiò, A.; Corbo, M.; Filosto, M.; Giannini, F.; et al. The unfolded protein response in amyotrophic later sclerosis: Results of a phase 2 trial. Brain 2021, 144, 2635–2647. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Wiedau, M. Therapeutic Approaches Targeting Protein Aggregation in Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2020, 13, 98. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, A.H.S.; Lopes, F.C.; John, E.B.O.; Carlini, C.R.; Ligabue-Braun, R. Modulation of Disordered Proteins with a Focus on Neurodegenerative Diseases and Other Pathologies. Int. J. Mol. Sci. 2019, 20, 1322. [Google Scholar] [CrossRef] [PubMed]
- Armiento, V.; Spanopoulou, A.; Kapurniotu, A. Peptide-Based Molecular Strategies To Interfere with Protein Misfolding, Aggregation, and Cell Degeneration. Angew. Chem. Int. Ed. Engl. 2020, 59, 3372–3384. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Singh, B.K.; Rai, S.N.; Singh, P.; Varshney, R.; Chaturvedi, V.K.; Vamanu, E. Promising drug targets and associated therapeutic interventions in Parkinson’s disease. Neural Regen. Res. 2021, 16, 1730–1739. [Google Scholar] [CrossRef] [PubMed]
- Eisele, Y.S.; Monteiro, C.; Fearns, C.; Encalada, S.E.; Wiseman, R.L.; Powers, E.T.; Kelly, J.W. Targeting protein aggregation for the treatment of degenerative diseases. Nat. Rev. Drug Discov. 2015, 14, 759–780. [Google Scholar] [CrossRef] [PubMed]
| Organelle | Mechanosensing and Mechanotransduction Pathways Involved in Organelles’ Homeostasis and Dysfunction | Ref. |
|---|---|---|
| Endoplasmic Reticulum (ER)  | • Expression of the mechanosensitive channel PANX1 and response to ultrasound stimuli by releasing the signaling molecule Ca2+ | [95] |
| • ER localization of Piezo1 in response to ER membrane tension mediates Ca2+ release | [96] | |
Golgi Apparatus | • Loss of cell adhesion causes Golgi fragmentation and loss of functioning in an integrin-mediated way through the modulation of the Arf1 activity • Increased glycosylation and trafficking of plasma membrane proteins | [97] |
| • RhoA pathway activation is correlated to trans-Golgi vesicles fission via a signaling pathway involving microtubules depolymerization, Myosin IIa and GEF-H1 in integrin-mediated adhesion | [98] | |
| • Lipid metabolic activity modification in response to external mechanical stimuli | [99] | |
Mitochondria | • Suspension culture of epithelial cells induces a reduced usage of glucose for the TCA cycle, which leads to lower ATP production restored by overexpression of ERBB2, an oncogene | [100] |
| • Spheroids from mammary epithelial cells enhance proline metabolism in order to maintain ATP synthesis and intensify the antioxidant activity of mitochondria | [101] | |
| • Cell detachment in cancer cells stimulates mitophagy, a particular type of autophagy targeted to mitochondria, that was proposed to be regulated by the serine/threonine kinase 1 RIPK1, which increases ROS generation and drives non-apoptotic cell death | [100,101,102] | |
| • Mechanical perturbation of cells with intracellular pathogens or extracellular stimulation with AFM compression engages the fission complex via the mitochondrial fission factor mitochondrial membrane | [103] | |
| • Decreasing mitochondrial tension by microtubules depolymerization and Myosin II inhibition reduces the probability of mitochondrial fission | [104] | |
| • FAs Kindlins mitochondrial accumulation in response to ECM stiffening • F-Actin polymerization around mitochondria-ER contact point induces mitochondrial constriction and fission | [100] | |
| • Prolonged mechanical stress causes an increase in glycolysis and glucose oxidation in CMs leading to impairment of mitochondria functioning and compromised ETC | [105,106] | |
| • CMs integrins respond to excessive mechanical load with the involvement of the MAPK and RhoA pathway, which results in ETC dysfunction and insufficient ATP synthesis | [107,108,109] | |
| • YAP activation with Melatonin favors mitochondrial fusion | [109] | |
| Lysosomes, Autophagy Endo-lysosomal system  | • YAP/TAZ activation simulates autophagy by inducing the expression of a RAB7 inhibiting protein, Armus, necessary for the activation of the autophagy flux | [84] |
| • Contact inhibition of cells cultured at high density on soft matrices showed to induce autophagy impairment through inhibition of YAP/TAZ activity axis with consequent loss of stress fibers and MyosinII that maintain the kinases LATS1/2 active | [110] | |
| • Intracellular stress caused by aggregates is implicated in lysosomal functioning and autophagy defects through BAG3 expression | [111,112,113,114,115] | |
| • Mutations leading to misfolded FLNC induce its intracellular accumulation, leading to autophagy activation and increasing lysosomes’ expression in human cardiomyocytes | [116] | |
| • Soft ECM impairs autophagosomes formation | [100] | |
| • Membrane tension regulates the CLIC/GEEC (CG) endocytic pathway through membrane-bound Vinculin that mediates its activation (with high membrane tension) or inhibition (with low tension) | [117] | |
| • Defects in internalization, recycling and lysosomal degradation through the endo-lysosomal compartment of integrins are correlated to pathologic conditions such as cancer and inflammation | [118] | |
| • Extracellular vesicles deriving from arthritic chondrocytes transport miR-221 and act as mediators of mechanical signaling and inhibiting in vitro bone development | [119] | |
| • Accumulation of substrates impose a perturbation in the homeostatic rheology of the cell that causes inhibition of lysosomal trafficking; following microtubules disassembly, enlarged lysosomes with prolonged ER-contact sites are retained in the cytoplasm of fibroblasts from MPS-1 and -3B patients | [120] | |
| • Lysosomal trafficking is regulated by substrate stiffness via different molecular adaptors; LRRK1 induces retrograde transport and perinuclear accumulation in soft matrices while VARP mediates exocytosis in rigid substrates | [121] | |
| • VAMP7 vesicles are important for the regulation of the plasma membrane composition in terms of glyco- and sphingolipids, correlating the sensing of environmental mechanical characteristics and the cellular biochemical response that leads to changes in adhesion and integrin dynamic | [121] | |
| • LAMP1 positive extracellular vesicles release is regulated by increased calcium influx induced by excessive mechanical stress | [122] |
| Genes/Proteins | Function in Homeostasis | Type of Alteration | Neurodegenerative Pathology | Ref. |
|---|---|---|---|---|
| α-Actinin | Scaffolding protein involved in Actin crosslinking | Protein delocalization | HD | [172] |
| Alsin Rho Guanine Nucleotide Exchange Factor | Guanine-nucleotide exchange factor, regulates GTPase activity | Gene mutations (loss of function) | ALS | [173] |
| Cofilin | When dephosphorylated mediates F-Actin disassembly | Increase/decrease in protein activity | AD | [174,175,176,177] |
| Increase protein activity | HD | [178] | ||
| Decrease in protein activity | PD | [179] | ||
| DynActin Subunit 1 | Mediates vesicles retrograde transport by interacting with Dynein | Gene mutations | ALS | [173] |
| Perry Syndrome | [180] | |||
| Filamin A | Scaffold protein required for F-Actin cross-linking | Altered conformation | AD | [181] |
| Histone-lysine N-methyltransferase SETD2 | Actin methylation | Protein activity inhibition | HD | [182] |
| Kindlin-2 | Required for FAs assembly and involved in ECM adhesion, Actin stabilization, and integrin-mediated signaling | Gene downregulation | AD | [183,184] |
| Kinesin 5A | Motor protein involved in spindle formation | Loss-of-function mutations | ALS | [185] |
| KN motif and ankyrin repeat domain-containing protein | Actin polymerization regulation | Gene mutations | ALS | [186] |
| Lamin A | Structural protein of the nuclear envelope | Protein upregulation | AD | [187] |
| Lamin B | Structural protein of the nuclear envelope | Protein upregulation | HD | [188] |
| Protein downregulation | PD | [189] | ||
| Microtubule-associated protein 2 | Essential for microtubule (MTs) assembly through crosslinking with intermediate filaments | Protein hyperphosphorylation | AD | [190,191] |
| Splicing Alteration | HD | [191,192] | ||
| Increased protein levels in CSF | ALS | [193] | ||
| Myosin heavy chain | Motor protein fundamental for cellular contractility | Decreased protein expression | ALS | [194] |
| Myosin IIb | Motor protein involved in Actin organization | Co-localization with TDP-25 | ALS | [195] |
| Neural cell adhesion molecule L1 | Axonal growth, neuronal migration and differentiation | Protein downregulation | AD | [196] |
| Neurofilament light and heavy chains | Neural intermediate filaments | Gene mutations | ALS | [173] |
| Nucleoporins | Mediate nucleocytoplasmatic transport | Protein sequestration | ALS | [197,198] |
| Piezo 1 | Mechanosensitive ion channel, mediates Ca2+ cellular influx | Protein upregulation | AD | [199,200] |
| Profilin 1 | Promotes Actin polymerization | Gene mutations | ALS | [173] |
| Spectrin | Structural protein of the cell membrane | Protein binding by α-Synuclein | PD | [201] |
| Tubulin Alpha 4A | Microtubules component | Gene mutations | ALS | [173] |
| Tubulin | Microtubules constituent | Protein acetylation | PD | [202] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tortorella, I.; Argentati, C.; Emiliani, C.; Morena, F.; Martino, S. Biochemical Pathways of Cellular Mechanosensing/Mechanotransduction and Their Role in Neurodegenerative Diseases Pathogenesis. Cells 2022, 11, 3093. https://doi.org/10.3390/cells11193093
Tortorella I, Argentati C, Emiliani C, Morena F, Martino S. Biochemical Pathways of Cellular Mechanosensing/Mechanotransduction and Their Role in Neurodegenerative Diseases Pathogenesis. Cells. 2022; 11(19):3093. https://doi.org/10.3390/cells11193093
Chicago/Turabian StyleTortorella, Ilaria, Chiara Argentati, Carla Emiliani, Francesco Morena, and Sabata Martino. 2022. "Biochemical Pathways of Cellular Mechanosensing/Mechanotransduction and Their Role in Neurodegenerative Diseases Pathogenesis" Cells 11, no. 19: 3093. https://doi.org/10.3390/cells11193093
APA StyleTortorella, I., Argentati, C., Emiliani, C., Morena, F., & Martino, S. (2022). Biochemical Pathways of Cellular Mechanosensing/Mechanotransduction and Their Role in Neurodegenerative Diseases Pathogenesis. Cells, 11(19), 3093. https://doi.org/10.3390/cells11193093









