CRISPR-Based Therapeutic Gene Editing for Duchenne Muscular Dystrophy: Advances, Challenges and Perspectives
Abstract
1. Introduction
2. Dystrophin
3. CRISPR Systems
4. CRISPR-Driven Therapeutic Strategies
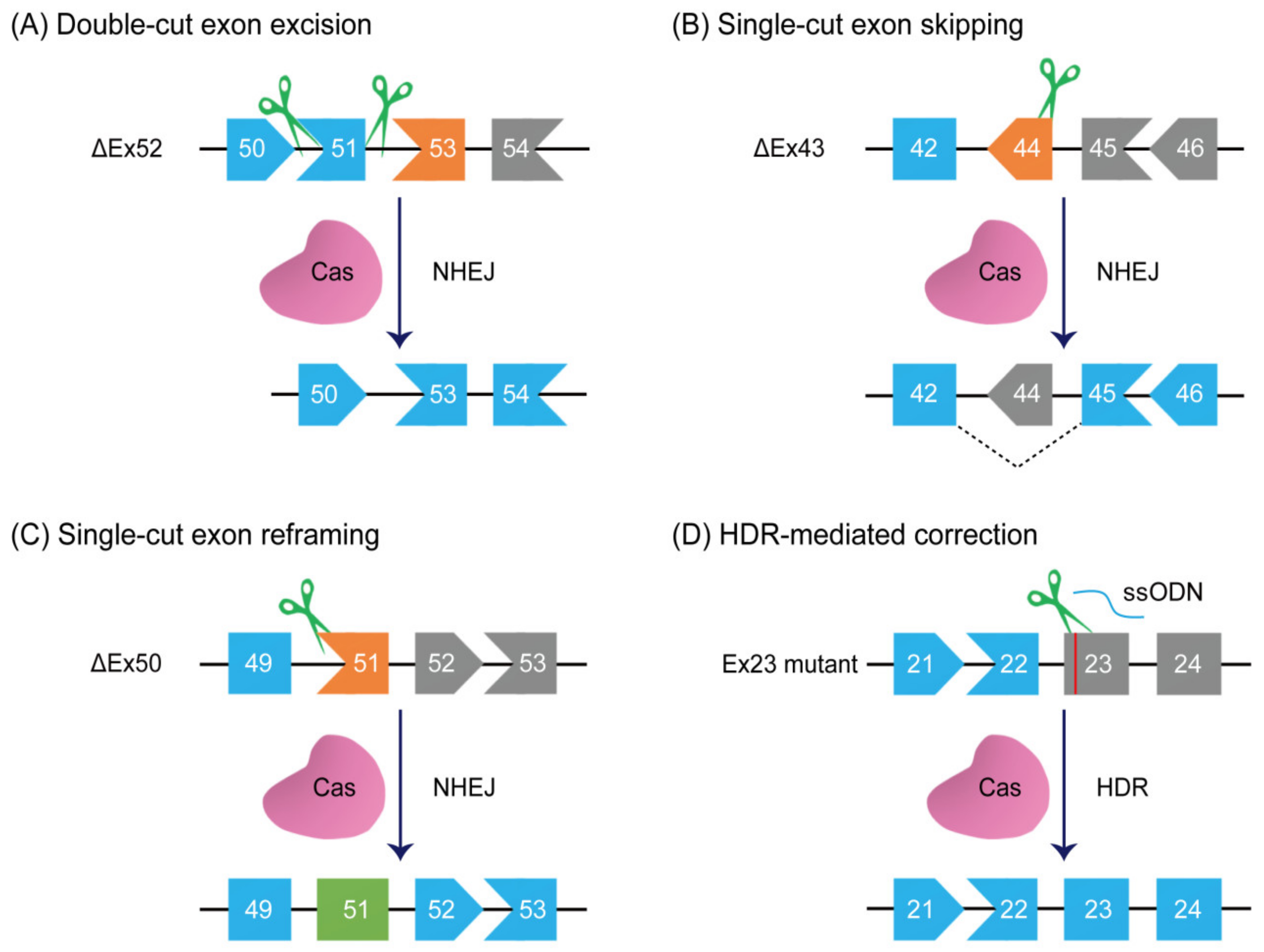
5. Double-Cut Exon Excision
6. Single-Cut Exon Skipping and Reframing
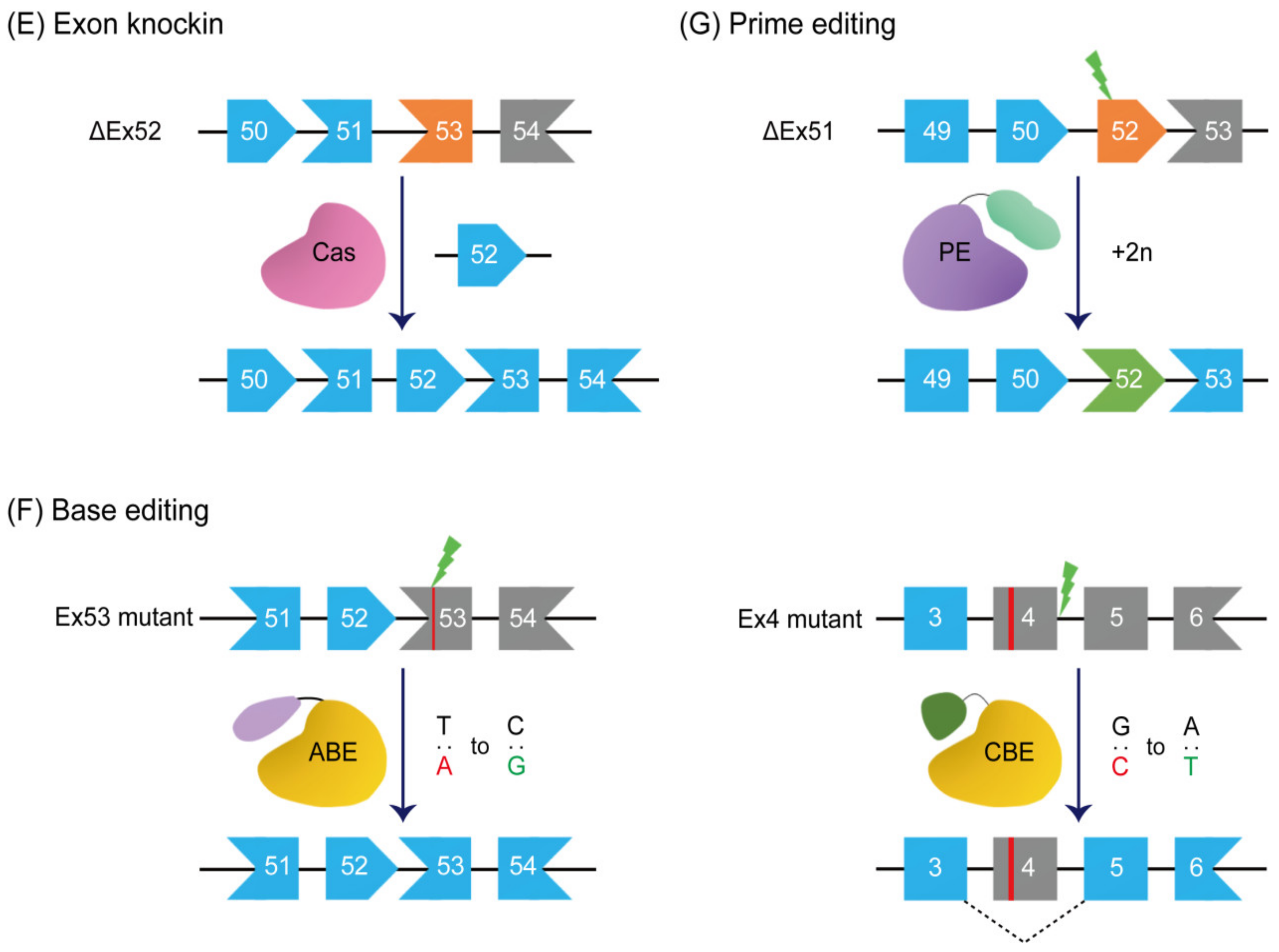
7. Exon Knockin
8. Base Editing
9. Prime Editing
10. DMD Animal Models
11. Future Challenges and Prospects
11.1. Safety
11.2. Immunogenicity
11.3. Off-Target Activity
11.4. Durability
12. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mendell, J.R.; Shilling, C.; Leslie, N.D.; Flanigan, K.M.; al-Dahhak, R.; Gastier-Foster, J.; Kneile, K.; Dunn, D.M.; Duval, B.; Aoyagi, A.; et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann. Neurol. 2012, 71, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Muntoni, F.; Torelli, S.; Ferlini, A. Dystrophin and mutations: One gene, several proteins, multiple phenotypes. Lancet Neurol. 2003, 2, 731–740. [Google Scholar] [CrossRef]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef] [PubMed]
- Constantin, B. Dystrophin complex functions as a scaffold for signalling proteins. Biochim. Biophys. Acta 2014, 1838, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Zhao, J.; Yue, Y.; Duan, D. α2 and α3 helices of dystrophin R16 and R17 frame a microdomain in the α1 helix of dystrophin R17 for neuronal NOS binding. Proc. Natl. Acad. Sci. USA 2013, 110, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E.; Muntoni, F. Muscular dystrophies. Lancet 2013, 381, 845–860. [Google Scholar] [CrossRef]
- Bladen, C.L.; Salgado, D.; Monges, S.; Foncuberta, M.E.; Kekou, K.; Kosma, K.; Dawkins, H.; Lamont, L.; Roy, A.J.; Chamova, T.; et al. The TREAT-NMD DMD Global Database: Analysis of more than 7000 Duchenne muscular dystrophy mutations. Hum. Mutat. 2015, 36, 395–402. [Google Scholar] [CrossRef]
- Echigoya, Y.; Lim, K.R.Q.; Nakamura, A.; Yokota, T. Multiple Exon Skipping in the Duchenne Muscular Dystrophy Hot Spots: Prospects and Challenges. J. Pers. Med. 2018, 8, 41. [Google Scholar] [CrossRef]
- Gloss, D.; Moxley, R.T., 3rd; Ashwal, S.; Oskoui, M. Practice guideline update summary: Corticosteroid treatment of Duchenne muscular dystrophy: Report of the Guideline Development Subcommittee of the American Academy of Neurology. Neurology 2016, 86, 465–472. [Google Scholar] [CrossRef]
- Birnkrant, D.J.; Bushby, K.; Bann, C.M.; Apkon, S.D.; Blackwell, A.; Colvin, M.K.; Cripe, L.; Herron, A.R.; Kennedy, A.; Kinnett, K.; et al. Diagnosis and management of Duchenne muscular dystrophy, part 3: Primary care, emergency management, psychosocial care, and transitions of care across the lifespan. Lancet Neurol. 2018, 17, 445–455. [Google Scholar] [CrossRef]
- Wilton-Clark, H.; Yokota, T. Antisense and Gene Therapy Options for Duchenne Muscular Dystrophy Arising from Mutations in the N-Terminal Hotspot. Genes 2022, 13, 257. [Google Scholar] [CrossRef]
- Zhang, T.; Kong, X. Recent advances of glucocorticoids in the treatment of Duchenne muscular dystrophy (Review). Exp. Ther. Med. 2021, 21, 447. [Google Scholar] [CrossRef]
- Ciafaloni, E.; Moxley, R.T. Treatment options for Duchenne muscular dystrophy. Curr. Treat. Options Neurol. 2008, 10, 86–93. [Google Scholar] [CrossRef]
- Dubinin, M.V.; Talanov, E.Y.; Tenkov, K.S.; Starinets, V.S.; Belosludtseva, N.V.; Belosludtsev, K.N. The Effect of Deflazacort Treatment on the Functioning of Skeletal Muscle Mitochondria in Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2020, 21, 8763. [Google Scholar] [CrossRef]
- Hörster, I.; Weigt-Usinger, K.; Carmann, C.; Chobanyan-Jürgens, K.; Köhler, C.; Schara, U.; Kayacelebi, A.A.; Beckmann, B.; Tsikas, D.; Lücke, T. The L-arginine/NO pathway and homoarginine are altered in Duchenne muscular dystrophy and improved by glucocorticoids. Amino Acids 2015, 47, 1853–1863. [Google Scholar] [CrossRef]
- Podkalicka, P.; Mucha, O.; Dulak, J.; Loboda, A. Targeting angiogenesis in Duchenne muscular dystrophy. Cell. Mol. Life Sci. 2019, 76, 1507–1528. [Google Scholar] [CrossRef]
- Miyatake, S.; Shimizu-Motohashi, Y.; Takeda, S.; Aoki, Y. Anti-inflammatory drugs for Duchenne muscular dystrophy: Focus on skeletal muscle-releasing factors. Drug Des. Dev. Ther. 2016, 10, 2745–2758. [Google Scholar]
- Shimizu-Motohashi, Y.; Miyatake, S.; Komaki, H.; Takeda, S.; Aoki, Y. Recent advances in innovative therapeutic approaches for Duchenne muscular dystrophy: From discovery to clinical trials. Am. J. Transl. Res. 2016, 8, 2471–2489. [Google Scholar]
- Goemans, N.; Buyse, G. Current treatment and management of dystrophinopathies. Curr. Treat. Options Neurol. 2014, 16, 287. [Google Scholar] [CrossRef]
- Ricotti, V.; Ridout, D.A.; Scott, E.; Quinlivan, R.; Robb, S.A.; Manzur, A.Y.; Muntoni, F.; NorthStar Clinical, N. Long-term benefits and adverse effects of intermittent versus daily glucocorticoids in boys with Duchenne muscular dystrophy. J. Neurol. Neurosurg. Psychiatry 2013, 84, 698–705. [Google Scholar] [CrossRef]
- Hammers, D.W.; Hart, C.C.; Patsalos, A.; Matheny, M.K.; Wright, L.A.; Nagy, L.; Sweeney, H.L. Glucocorticoids counteract hypertrophic effects of myostatin inhibition in dystrophic muscle. JCI Insight 2020, 5, e133276. [Google Scholar] [CrossRef]
- Schneider, A.E.; Aartsma-Rus, A. Developments in reading frame restoring therapy approaches for Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 2021, 21, 343–359. [Google Scholar] [CrossRef]
- Charleston, J.S.; Schnell, F.J.; Dworzak, J.; Donoghue, C.; Lewis, S.; Chen, L.; Young, G.D.; Milici, A.J.; Voss, J.; DeAlwis, U.; et al. Eteplirsen treatment for Duchenne muscular dystrophy: Exon skipping and dystrophin production. Neurology 2018, 90, e2146–e2154. [Google Scholar] [CrossRef]
- Frank, D.E.; Schnell, F.J.; Akana, C.; El-Husayni, S.H.; Desjardins, C.A.; Morgan, J.; Charleston, J.S.; Sardone, V.; Domingos, J.; Dickson, G.; et al. Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology 2020, 94, e2270–e2282. [Google Scholar] [CrossRef]
- Clemens, P.R.; Rao, V.K.; Connolly, A.M.; Harper, A.D.; Mah, J.K.; Smith, E.C.; McDonald, C.M.; Zaidman, C.M.; Morgenroth, L.P.; Osaki, H.; et al. Safety, Tolerability, and Efficacy of Viltolarsen in Boys With Duchenne Muscular Dystrophy Amenable to Exon 53 Skipping: A Phase 2 Randomized Clinical Trial. JAMA Neurol. 2020, 77, 982–991. [Google Scholar] [CrossRef]
- Wagner, K.R.; Kuntz, N.L.; Koenig, E.; East, L.; Upadhyay, S.; Han, B.; Shieh, P.B. Safety, tolerability, and pharmacokinetics of casimersen in patients with Duchenne muscular dystrophy amenable to exon 45 skipping: A randomized, double-blind, placebo-controlled, dose-titration trial. Muscle Nerve 2021, 64, 285–292. [Google Scholar] [CrossRef]
- Lehto, T.; Castillo Alvarez, A.; Gauck, S.; Gait, M.J.; Coursindel, T.; Wood, M.J.; Lebleu, B.; Boisguerin, P. Cellular trafficking determines the exon skipping activity of Pip6a-PMO in mdx skeletal and cardiac muscle cells. Nucleic Acids Res. 2014, 42, 3207–3217. [Google Scholar] [CrossRef]
- Lim, K.R.; Maruyama, R.; Yokota, T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Dev. Ther. 2017, 11, 533–545. [Google Scholar] [CrossRef]
- Nelson, C.E.; Robinson-Hamm, J.N.; Gersbach, C.A. Genome engineering: A new approach to gene therapy for neuromuscular disorders. Nat. Rev. Neurol. 2017, 13, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.N. Toward the correction of muscular dystrophy by gene editing. Proc. Natl. Acad. Sci. USA 2021, 118, e2004840117. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A. The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.D.; Richardson, C.D.; Corn, J.E. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019, 21, 1468–1478. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Amoasii, L.; Mireault, A.A.; McAnally, J.R.; Li, H.; Sanchez-Ortiz, E.; Bhattacharyya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016, 351, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Hakim, C.H.; Wasala, N.B.; Nelson, C.E.; Wasala, L.P.; Yue, Y.; Louderman, J.A.; Lessa, T.B.; Dai, A.; Zhang, K.; Jenkins, G.J.; et al. AAV CRISPR editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight 2018, 3, e124297. [Google Scholar] [CrossRef]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Castellanos Rivera, R.M.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef]
- Nelson, C.E.; Wu, Y.; Gemberling, M.P.; Oliver, M.L.; Waller, M.A.; Bohning, J.D.; Robinson-Hamm, J.N.; Bulaklak, K.; Castellanos Rivera, R.M.; Collier, J.H.; et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019, 25, 427–432. [Google Scholar] [CrossRef]
- Young, C.S.; Hicks, M.R.; Ermolova, N.V.; Nakano, H.; Jan, M.; Younesi, S.; Karumbayaram, S.; Kumagai-Cresse, C.; Wang, D.; Zack, J.A.; et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell 2016, 18, 533–540. [Google Scholar] [CrossRef]
- Moretti, A.; Fonteyne, L.; Giesert, F.; Hoppmann, P.; Meier, A.B.; Bozoglu, T.; Baehr, A.; Schneider, C.M.; Sinnecker, D.; Klett, K.; et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat. Med. 2020, 26, 207–214. [Google Scholar] [CrossRef]
- Duchêne, B.L.; Cherif, K.; Iyombe-Engembe, J.P.; Guyon, A.; Rousseau, J.; Ouellet, D.L.; Barbeau, X.; Lague, P.; Tremblay, J.P. CRISPR-Induced Deletion with SaCas9 Restores Dystrophin Expression in Dystrophic Models In Vitro and In Vivo. Mol. Ther. 2018, 26, 2604–2616. [Google Scholar] [CrossRef]
- Chemello, F.; Bassel-Duby, R.; Olson, E.N. Correction of muscular dystrophies by CRISPR gene editing. J. Clin. Investig. 2020, 130, 2766–2776. [Google Scholar] [CrossRef]
- Choi, E.; Koo, T. CRISPR technologies for the treatment of Duchenne muscular dystrophy. Mol. Ther. 2021, 29, 3179–3191. [Google Scholar] [CrossRef]
- Xu, L.; Lau, Y.S.; Gao, Y.; Li, H.; Han, R. Life-Long AAV-Mediated CRISPR Genome Editing in Dystrophic Heart Improves Cardiomyopathy without Causing Serious Lesions in mdx Mice. Mol. Ther. 2019, 27, 1407–1414. [Google Scholar] [CrossRef]
- Karri, D.R.; Zhang, Y.; Chemello, F.; Min, Y.L.; Huang, J.; Kim, J.; Mammen, P.P.A.; Xu, L.; Liu, N.; Bassel-Duby, R.; et al. Long-term maintenance of dystrophin expression and resistance to injury of skeletal muscle in gene edited DMD mice. Mol. Ther. Nucleic Acids 2022, 28, 154–167. [Google Scholar] [CrossRef]
- Bergmann, O.; Zdunek, S.; Felker, A.; Salehpour, M.; Alkass, K.; Bernard, S.; Sjostrom, S.L.; Szewczykowska, M.; Jackowska, T.; Dos Remedios, C.; et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell 2015, 161, 1566–1575. [Google Scholar] [CrossRef]
- Takeda, S.; Clemens, P.R.; Hoffman, E.P. Exon-Skipping in Duchenne Muscular Dystrophy. J. Neuromuscul. Dis. 2021, 8, S343–S358. [Google Scholar] [CrossRef]
- Wasala, N.B.; Chen, S.J.; Duan, D. Duchenne muscular dystrophy animal models for high-throughput drug discovery and precision medicine. Expert Opin. Drug Discov. 2020, 15, 443–456. [Google Scholar] [CrossRef]
- Lionarons, J.M.; Hoogland, G.; Hendriksen, R.G.F.; Faber, C.G.; Hellebrekers, D.M.J.; Van Koeveringe, G.A.; Schipper, S.; Vles, J.S.H. Dystrophin is expressed in smooth muscle and afferent nerve fibers in the rat urinary bladder. Muscle Nerve 2019, 60, 202–210. [Google Scholar] [CrossRef]
- Elangkovan, N.; Dickson, G. Gene Therapy for Duchenne Muscular Dystrophy. J. Neuromuscul. Dis. 2021, 8, S303–S316. [Google Scholar] [CrossRef]
- Murphy, S.; Zweyer, M.; Henry, M.; Meleady, P.; Mundegar, R.R.; Swandulla, D.; Ohlendieck, K. Proteomic analysis of the sarcolemma-enriched fraction from dystrophic mdx-4cv skeletal muscle. J. Proteom. 2019, 191, 212–227. [Google Scholar] [CrossRef]
- Zabłocka, B.; Górecki, D.C.; Zabłocki, K. Disrupted Calcium Homeostasis in Duchenne Muscular Dystrophy: A Common Mechanism behind Diverse Consequences. Int. J. Mol. Sci. 2021, 22, 11040. [Google Scholar] [CrossRef] [PubMed]
- Fong, P.Y.; Turner, P.R.; Denetclaw, W.F.; Steinhardt, R.A. Increased activity of calcium leak channels in myotubes of Duchenne human and mdx mouse origin. Science 1990, 250, 673–676. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.R.; Fong, P.Y.; Denetclaw, W.F.; Steinhardt, R.A. Increased calcium influx in dystrophic muscle. J. Cell Biol. 1991, 115, 1701–1712. [Google Scholar] [CrossRef] [PubMed]
- Rando, T.A.; Disatnik, M.H.; Yu, Y.; Franco, A. Muscle cells from mdx mice have an increased susceptibility to oxidative stress. Neuromuscul. Disord. 1998, 8, 14–21. [Google Scholar] [CrossRef]
- Kuznetsov, A.V.; Winkler, K.; Wiedemann, F.R.; von Bossanyi, P.; Dietzmann, K.; Kunz, W.S. Impaired mitochondrial oxidative phosphorylation in skeletal muscle of the dystrophin-deficient mdx mouse. Mol. Cell Biochem. 1998, 183, 87–96. [Google Scholar] [CrossRef]
- Timpani, C.A.; Hayes, A.; Rybalka, E. Revisiting the dystrophin-ATP connection: How half a century of research still implicates mitochondrial dysfunction in Duchenne Muscular Dystrophy aetiology. Med. Hypotheses 2015, 85, 1021–1033. [Google Scholar] [CrossRef]
- Even, P.C.; Decrouy, A.; Chinet, A. Defective regulation of energy metabolism in mdx-mouse skeletal muscles. Biochem. J. 1994, 304 Pt 2, 649–654. [Google Scholar] [CrossRef]
- De Palma, C.; Morisi, F.; Cheli, S.; Pambianco, S.; Cappello, V.; Vezzoli, M.; Rovere-Querini, P.; Moggio, M.; Ripolone, M.; Francolini, M.; et al. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis. 2012, 3, e418. [Google Scholar] [CrossRef]
- Brenman, J.E.; Chao, D.S.; Xia, H.; Aldape, K.; Bredt, D.S. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell 1995, 82, 743–752. [Google Scholar] [CrossRef]
- Smith, M.A.; Schnellmann, R.G. Calpains, mitochondria, and apoptosis. Cardiovasc. Res. 2012, 96, 32–37. [Google Scholar] [CrossRef]
- Chen, Y.W.; Nagaraju, K.; Bakay, M.; McIntyre, O.; Rawat, R.; Shi, R.; Hoffman, E.P. Early onset of inflammation and later involvement of TGFbeta in Duchenne muscular dystrophy. Neurology 2005, 65, 826–834. [Google Scholar] [CrossRef]
- Szigyarto, C.A.; Spitali, P. Biomarkers of Duchenne muscular dystrophy: Current findings. Degener. Neurol. Neuromuscul. Dis. 2018, 8, 1–13. [Google Scholar]
- Yucel, N.; Chang, A.C.; Day, J.W.; Rosenthal, N.; Blau, H.M. Humanizing the mdx mouse model of DMD: The long and the short of it. NPJ Regen. Med. 2018, 3, 4. [Google Scholar] [CrossRef]
- Kornegay, J.N. The golden retriever model of Duchenne muscular dystrophy. Skelet Muscle 2017, 7, 9. [Google Scholar] [CrossRef]
- Brinkmeyer-Langford, C.; Chu, C.; Balog-Alvarez, C.; Yu, X.; Cai, J.J.; Nabity, M.; Kornegay, J.N. Expression profiling of disease progression in canine model of Duchenne muscular dystrophy. PLoS ONE 2018, 13, e0194485. [Google Scholar] [CrossRef]
- Min, Y.L.; Bassel-Duby, R.; Olson, E.N. CRISPR Correction of Duchenne Muscular Dystrophy. Annu Rev Med 2019, 70, 239–255. [Google Scholar] [CrossRef]
- Monaco, A.P.; Bertelson, C.J.; Liechti-Gallati, S.; Moser, H.; Kunkel, L.M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988, 2, 90–95. [Google Scholar] [CrossRef]
- Andrews, J.G.; Wahl, R.A. Duchenne and Becker muscular dystrophy in adolescents: Current perspectives. Adolesc. Health Med. Ther 2018, 9, 53–63. [Google Scholar] [CrossRef]
- Flanigan, K.M. Duchenne and Becker muscular dystrophies. Neurol. Clin. 2014, 32, 671–688. [Google Scholar] [CrossRef]
- Ferreiro, V.; Giliberto, F.; Muñiz, G.M.; Francipane, L.; Marzese, D.M.; Mampel, A.; Roqué, M.; Frechtel, G.D.; Szijan, I. Asymptomatic Becker muscular dystrophy in a family with a multiexon deletion. Muscle Nerve 2009, 39, 239–243. [Google Scholar] [CrossRef]
- Wang, B.; Li, J.; Fu, F.H.; Xiao, X. Systemic human minidystrophin gene transfer improves functions and life span of dystrophin and dystrophin/utrophin-deficient mice. J. Orthop. Res. 2009, 27, 421–426. [Google Scholar] [CrossRef]
- Le Guiner, C.; Servais, L.; Montus, M.; Larcher, T.; Fraysse, B.; Moullec, S.; Allais, M.; François, V.; Dutilleul, M.; Malerba, A.; et al. Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy. Nat. Commun. 2017, 8, 16105. [Google Scholar] [CrossRef]
- Koppanati, B.M.; Li, J.; Reay, D.P.; Wang, B.; Daood, M.; Zheng, H.; Xiao, X.; Watchko, J.F.; Clemens, P.R. Improvement of the mdx mouse dystrophic phenotype by systemic in utero AAV8 delivery of a minidystrophin gene. Gene Ther. 2010, 17, 1355–1362. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wasala, N.B.; Shin, J.H.; Lai, Y.; Yue, Y.; Montanaro, F.; Duan, D. Cardiac-Specific Expression of ΔH2-R15 Mini-Dystrophin Normalized All Electrocardiogram Abnormalities and the End-Diastolic Volume in a 23-Month-Old Mouse Model of Duchenne Dilated Cardiomyopathy. Hum. Gene Ther. 2018, 29, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.E.; McPhee, S.W.; Li, C.; Gray, S.J.; Samulski, J.J.; Camp, A.S.; Li, J.; Wang, B.; Monahan, P.E.; Rabinowitz, J.E.; et al. Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol. Ther. 2012, 20, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Gao, Y.; Gao, K.; Yang, H. CRISPR/Cas: A potential gene-editing tool in the nervous system. Cell Regen. 2020, 9, 12. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Zuo, Z.; Liu, J. Cas9-catalyzed DNA Cleavage Generates Staggered Ends: Evidence from Molecular Dynamics Simulations. Sci. Rep. 2016, 5, 37584. [Google Scholar] [CrossRef]
- Lemos, B.R.; Kaplan, A.C.; Bae, J.E.; Ferrazzoli, A.E.; Kuo, J.; Anand, R.P.; Waterman, D.P.; Haber, J.E. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc. Natl. Acad. Sci. USA 2018, 115, E2040–E2047. [Google Scholar] [CrossRef]
- Kakarougkas, A.; Jeggo, P.A. DNA DSB repair pathway choice: An orchestrated handover mechanism. Br. J. Radiol. 2014, 87, 20130685. [Google Scholar] [CrossRef]
- Moynahan, M.E.; Jasin, M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010, 11, 196–207. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar] [CrossRef]
- Xie, H.; Tang, L.; He, X.; Liu, X.; Zhou, C.; Liu, J.; Ge, X.; Li, J.; Liu, C.; Zhao, J.; et al. SaCas9 Requires 5′-NNGRRT-3′ PAM for Sufficient Cleavage and Possesses Higher Cleavage Activity than SpCas9 or FnCpf1 in Human Cells. Biotechnol. J. 2018, 13, e1700561. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar]
- Wu, Z.; Yang, H.; Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010, 18, 80–86. [Google Scholar]
- Tran, M.H.; Park, H.; Nobles, C.L.; Karunadharma, P.; Pan, L.; Zhong, G.; Wang, H.; He, W.; Ou, T.; Crynen, G.; et al. A more efficient CRISPR-Cas12a variant derived from Lachnospiraceae bacterium MA2020. Mol. Ther. Nucleic Acids 2021, 24, 40–53. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.H.; Kim, J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef]
- Esvelt, K.M.; Mali, P.; Braff, J.L.; Moosburner, M.; Yaung, S.J.; Church, G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 2013, 10, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Lee, C.M.; Gasiunas, G.; Davis, T.H.; Cradick, T.J.; Siksnys, V.; Bao, G.; Cathomen, T.; Mussolino, C. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol. Ther. 2016, 24, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Zhang, Y.; Propson, N.E.; Howden, S.E.; Chu, L.F.; Sontheimer, E.J.; Thomson, J.A. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 2013, 110, 15644–15649. [Google Scholar] [CrossRef] [PubMed]
- Edraki, A.; Mir, A.; Ibraheim, R.; Gainetdinov, I.; Yoon, Y.; Song, C.Q.; Cao, Y.; Gallant, J.; Xue, W.; Rivera-Pérez, J.A.; et al. A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Mol. Cell 2019, 73, 714–726. [Google Scholar] [CrossRef]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef]
- Yan, W.X.; Hunnewell, P.; Alfonse, L.E.; Carte, J.M.; Keston-Smith, E.; Sothiselvam, S.; Garrity, A.J.; Chong, S.; Makarova, K.S.; Koonin, E.V.; et al. Functionally diverse type V CRISPR-Cas systems. Science 2019, 363, 88–91. [Google Scholar] [CrossRef]
- Strecker, J.; Jones, S.; Koopal, B.; Schmid-Burgk, J.; Zetsche, B.; Gao, L.; Makarova, K.S.; Koonin, E.V.; Zhang, F. Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 2019, 10, 212. [Google Scholar] [CrossRef]
- Liu, J.J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019, 566, 218–223. [Google Scholar] [CrossRef]
- Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F.; et al. Structure and Engineering of Francisella novicida Cas9. Cell 2016, 164, 950–961. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef]
- Kulcsár, P.I.; Tálas, A.; Huszár, K.; Ligeti, Z.; Tóth, E.; Weinhardt, N.; Fodor, E.; Welker, E. Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage. Genome Biol. 2017, 18, 190. [Google Scholar] [CrossRef]
- Casini, A.; Olivieri, M.; Petris, G.; Montagna, C.; Reginato, G.; Maule, G.; Lorenzin, F.; Prandi, D.; Romanel, A.; Demichelis, F.; et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018, 36, 265–271. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef]
- Lee, J.K.; Jeong, E.; Lee, J.; Jung, M.; Shin, E.; Kim, Y.H.; Lee, K.; Jung, I.; Kim, D.; Kim, S.; et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018, 9, 3048. [Google Scholar] [CrossRef]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef]
- Chatterjee, P.; Lee, J.; Nip, L.; Koseki, S.R.T.; Tysinger, E.; Sontheimer, E.J.; Jacobson, J.M.; Jakimo, N. A Cas9 with PAM recognition for adenine dinucleotides. Nat. Commun. 2020, 11, 2474. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Topkar, V.V.; Zheng, Z.; Joung, J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015, 33, 1293–1298. [Google Scholar] [CrossRef]
- Gao, L.; Cox, D.B.T.; Yan, W.X.; Manteiga, J.C.; Schneider, M.W.; Yamano, T.; Nishimasu, H.; Nureki, O.; Crosetto, N.; Zhang, F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017, 35, 789–792. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Sousa, A.A.; Walton, R.T.; Tak, Y.E.; Hsu, J.Y.; Clement, K.; Welch, M.M.; Horng, J.E.; Malagon-Lopez, J.; Scarfò, I.; et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019, 37, 276–282. [Google Scholar] [CrossRef]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Porto, E.M.; Komor, A.C.; Slaymaker, I.M.; Yeo, G.W. Base editing: Advances and therapeutic opportunities. Nat. Rev. Drug Discov. 2020, 19, 839–859. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022, 40, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Wojtal, D.; Kemaladewi, D.U.; Malam, Z.; Abdullah, S.; Wong, T.W.; Hyatt, E.; Baghestani, Z.; Pereira, S.; Stavropoulos, J.; Mouly, V.; et al. Spell Checking Nature: Versatility of CRISPR/Cas9 for Developing Treatments for Inherited Disorders. Am. J. Hum. Genet. 2016, 98, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.W.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.W.; Rangarajan, S.; Shivalila, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013, 23, 1163–1171. [Google Scholar] [CrossRef]
- Long, C.; McAnally, J.R.; Shelton, J.M.; Mireault, A.A.; Bassel-Duby, R.; Olson, E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 2014, 345, 1184–1188. [Google Scholar] [CrossRef]
- Maino, E.; Wojtal, D.; Evagelou, S.L.; Farheen, A.; Wong, T.W.Y.; Lindsay, K.; Scott, O.; Rizvi, S.Z.; Hyatt, E.; Rok, M.; et al. Targeted genome editing in vivo corrects a Dmd duplication restoring wild-type dystrophin expression. EMBO Mol. Med. 2021, 13, e13228. [Google Scholar] [CrossRef]
- Xu, L.; Park, K.H.; Zhao, L.; Xu, J.; El Refaey, M.; Gao, Y.; Zhu, H.; Ma, J.; Han, R. CRISPR-mediated Genome Editing Restores Dystrophin Expression and Function in mdx Mice. Mol. Ther. 2016, 24, 564–569. [Google Scholar] [CrossRef]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef]
- Bengtsson, N.E.; Hall, J.K.; Odom, G.L.; Phelps, M.P.; Andrus, C.R.; Hawkins, R.D.; Hauschka, S.D.; Chamberlain, J.R.; Chamberlain, J.S. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 2017, 8, 14454. [Google Scholar] [CrossRef]
- Bengtsson, N.E.; Tasfaout, H.; Hauschka, S.D.; Chamberlain, J.S. Dystrophin Gene-Editing Stability Is Dependent on Dystrophin Levels in Skeletal but Not Cardiac Muscles. Mol. Ther. 2021, 29, 1070–1085. [Google Scholar] [CrossRef]
- Young, C.S.; Mokhonova, E.; Quinonez, M.; Pyle, A.D.; Spencer, M.J. Creation of a Novel Humanized Dystrophic Mouse Model of Duchenne Muscular Dystrophy and Application of a CRISPR/Cas9 Gene Editing Therapy. J. Neuromuscul. Dis. 2017, 4, 139–145. [Google Scholar] [CrossRef]
- El Refaey, M.; Xu, L.; Gao, Y.; Canan, B.D.; Adesanya, T.M.A.; Warner, S.C.; Akagi, K.; Symer, D.E.; Mohler, P.J.; Ma, J.; et al. In Vivo Genome Editing Restores Dystrophin Expression and Cardiac Function in Dystrophic Mice. Circ. Res. 2017, 121, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.L.; Li, H.; Rodriguez-Caycedo, C.; Mireault, A.A.; Huang, J.; Shelton, J.M.; McAnally, J.R.; Amoasii, L.; Mammen, P.P.A.; Bassel-Duby, R.; et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 2019, 5, eaav4324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nishiyama, T.; Li, H.; Huang, J.; Atmanli, A.; Sanchez-Ortiz, E.; Wang, Z.; Mireault, A.A.; Mammen, P.P.A.; Bassel-Duby, R.; et al. A consolidated AAV system for single-cut CRISPR correction of a common Duchenne muscular dystrophy mutation. Mol. Ther. Methods Clin. Dev. 2021, 22, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.L.; Chemello, F.; Li, H.; Rodriguez-Caycedo, C.; Sanchez-Ortiz, E.; Mireault, A.A.; McAnally, J.R.; Shelton, J.M.; Zhang, Y.; Bassel-Duby, R.; et al. Correction of Three Prominent Mutations in Mouse and Human Models of Duchenne Muscular Dystrophy by Single-Cut Genome Editing. Mol. Ther. 2020, 28, 2044–2055. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, H.; Nishiyama, T.; McAnally, J.R.; Sanchez-Ortiz, E.; Huang, J.; Mammen, P.P.A.; Bassel-Duby, R.; Olson, E.N. A humanized knockin mouse model of Duchenne muscular dystrophy and its correction by CRISPR-Cas9 therapeutic gene editing. Mol. Ther. Nucleic Acids 2022, 29, 525–537. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, H.; Min, Y.L.; Sanchez-Ortiz, E.; Huang, J.; Mireault, A.A.; Shelton, J.M.; Kim, J.; Mammen, P.P.A.; Bassel-Duby, R.; et al. Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci. Adv. 2020, 6, eaay6812. [Google Scholar] [CrossRef]
- Koo, T.; Lu-Nguyen, N.B.; Malerba, A.; Kim, E.; Kim, D.; Cappellari, O.; Cho, H.Y.; Dickson, G.; Popplewell, L.; Kim, J.S. Functional Rescue of Dystrophin Deficiency in Mice Caused by Frameshift Mutations Using Campylobacter jejuni Cas9. Mol. Ther. 2018, 26, 1529–1538. [Google Scholar] [CrossRef]
- Amoasii, L.; Long, C.; Li, H.; Mireault, A.A.; Shelton, J.M.; Sanchez-Ortiz, E.; McAnally, J.R.; Bhattacharyya, S.; Schmidt, F.; Grimm, D.; et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl. Med. 2017, 9, eaan8081. [Google Scholar] [CrossRef]
- Amoasii, L.; Li, H.; Zhang, Y.; Min, Y.L.; Sanchez-Ortiz, E.; Shelton, J.M.; Long, C.; Mireault, A.A.; Bhattacharyya, S.; McAnally, J.R.; et al. In vivo non-invasive monitoring of dystrophin correction in a new Duchenne muscular dystrophy reporter mouse. Nat. Commun. 2019, 10, 4537. [Google Scholar] [CrossRef]
- Amoasii, L.; Hildyard, J.C.W.; Li, H.; Sanchez-Ortiz, E.; Mireault, A.; Caballero, D.; Harron, R.; Stathopoulou, T.R.; Massey, C.; Shelton, J.M.; et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 2018, 362, 86–91. [Google Scholar] [CrossRef]
- Hakim, C.H.; Kumar, S.R.P.; Pérez-López, D.O.; Wasala, N.B.; Zhang, D.; Yue, Y.; Teixeira, J.; Pan, X.; Zhang, K.; Million, E.D.; et al. Cas9-specific immune responses compromise local and systemic AAV CRISPR therapy in multiple dystrophic canine models. Nat. Commun. 2021, 12, 6769. [Google Scholar] [CrossRef]
- Zhang, Y.; Long, C.; Li, H.; McAnally, J.R.; Baskin, K.K.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci. Adv. 2017, 3, e1602814. [Google Scholar] [CrossRef]
- Mata López, S.; Balog-Alvarez, C.; Vitha, S.; Bettis, A.K.; Canessa, E.H.; Kornegay, J.N.; Nghiem, P.P. Challenges associated with homologous directed repair using CRISPR-Cas9 and TALEN to edit the DMD genetic mutation in canine Duchenne muscular dystrophy. PLoS ONE 2020, 15, e0228072. [Google Scholar]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef]
- Pickar-Oliver, A.; Gough, V.; Bohning, J.D.; Liu, S.; Robinson-Hamm, J.N.; Daniels, H.; Majoros, W.H.; Devlin, G.; Asokan, A.; Gersbach, C.A. Full-length dystrophin restoration via targeted exon integration by AAV-CRISPR in a humanized mouse model of Duchenne muscular dystrophy. Mol. Ther. 2021, 29, 3243–3257. [Google Scholar] [CrossRef]
- Chemello, F.; Chai, A.C.; Li, H.; Rodriguez-Caycedo, C.; Sanchez-Ortiz, E.; Atmanli, A.; Mireault, A.A.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci. Adv. 2021, 7, eabg4910. [Google Scholar] [CrossRef]
- Ryu, S.M.; Koo, T.; Kim, K.; Lim, K.; Baek, G.; Kim, S.T.; Kim, H.S.; Kim, D.E.; Lee, H.; Chung, E.; et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018, 36, 536–539. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, C.; Li, H.; Wang, P.; Gao, Y.; Mokadam, N.A.; Ma, J.; Arnold, W.D.; Han, R. Efficient precise in vivo base editing in adult dystrophic mice. Nat. Commun. 2021, 12, 3719. [Google Scholar] [CrossRef]
- Li, J.; Wang, K.; Zhang, Y.; Qi, T.; Yuan, J.; Zhang, L.; Qiu, H.; Wang, J.; Yang, H.T.; Dai, Y.; et al. Therapeutic Exon Skipping Through a CRISPR-Guided Cytidine Deaminase Rescues Dystrophic Cardiomyopathy in Vivo. Circulation 2021, 144, 1760–1776. [Google Scholar] [CrossRef]
- Long, C.; Li, H.; Tiburcy, M.; Rodriguez-Caycedo, C.; Kyrychenko, V.; Zhou, H.; Zhang, Y.; Min, Y.L.; Shelton, J.M.; Mammen, P.P.A.; et al. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv. 2018, 4, eaap9004. [Google Scholar] [CrossRef]
- Lattanzi, A.; Duguez, S.; Moiani, A.; Izmiryan, A.; Barbon, E.; Martin, S.; Mamchaoui, K.; Mouly, V.; Bernardi, F.; Mavilio, F.; et al. Correction of the Exon 2 Duplication in DMD Myoblasts by a Single CRISPR/Cas9 System. Mol. Ther. Nucleic Acids 2017, 7, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Pini, V.; Mariot, V.; Dumonceaux, J.; Counsell, J.; O’Neill, H.C.; Farmer, S.; Conti, F.; Muntoni, F. Transiently expressed CRISPR/Cas9 induces wild-type dystrophin in vitro in DMD patient myoblasts carrying duplications. Sci. Rep. 2022, 12, 3756. [Google Scholar] [CrossRef]
- Wang, D.N.; Wang, Z.Q.; Jin, M.; Lin, M.T.; Wang, N. CRISPR/Cas9-based genome editing for the modification of multiple duplications that cause Duchenne muscular dystrophy. Gene Ther. 2022. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Ousterout, D.G.; Kabadi, A.M.; Thakore, P.I.; Majoros, W.H.; Reddy, T.E. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015, 6, 6244. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Shi, H.; Gou, S.; Wang, X.; Li, L.; Jin, Q.; Wu, H.; Zhang, H.; Li, Y.; Wang, L.; et al. In vivo genome editing in mouse restores dystrophin expression in Duchenne muscular dystrophy patient muscle fibers. Genome Med. 2021, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Maggio, I.; Liu, J.; Janssen, J.M.; Chen, X.; Gonçalves, M.A. Adenoviral vectors encoding CRISPR/Cas9 multiplexes rescue dystrophin synthesis in unselected populations of DMD muscle cells. Sci. Rep. 2016, 6, 37051. [Google Scholar] [CrossRef] [PubMed]
- Maggio, I.; Stefanucci, L.; Janssen, J.M.; Liu, J.; Chen, X.; Mouly, V.; Gonçalves, M.A. Selection-free gene repair after adenoviral vector transduction of designer nucleases: Rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016, 44, 1449–1470. [Google Scholar] [CrossRef] [PubMed]
- Brescia, M.; Janssen, J.M.; Liu, J.; Gonçalves, M. High-Capacity Adenoviral Vectors Permit Robust and Versatile Testing of DMD Gene Repair Tools and Strategies in Human Cells. Cells 2020, 9, 869. [Google Scholar] [CrossRef]
- Kyrychenko, V.; Kyrychenko, S.; Tiburcy, M.; Shelton, J.M.; Long, C.; Schneider, J.W.; Zimmermann, W.H.; Bassel-Duby, R.; Olson, E.N. Functional correction of dystrophin actin binding domain mutations by genome editing. JCI Insight 2017, 2, e95918. [Google Scholar] [CrossRef]
- Matre, P.R.; Mu, X.; Wu, J.; Danila, D.; Hall, M.A.; Kolonin, M.G.; Darabi, R.; Huard, J. CRISPR/Cas9-Based Dystrophin Restoration Reveals a Novel Role for Dystrophin in Bioenergetics and Stress Resistance of Muscle Progenitors. Stem Cells 2019, 37, 1615–1628. [Google Scholar] [CrossRef]
- Li, H.L.; Fujimoto, N.; Sasakawa, N.; Shirai, S.; Ohkame, T.; Sakuma, T.; Tanaka, M.; Amano, N.; Watanabe, A.; Sakurai, H.; et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 2015, 4, 143–154. [Google Scholar] [CrossRef]
- Yuan, J.; Ma, Y.; Huang, T.; Chen, Y.; Peng, Y.; Li, B.; Li, J.; Zhang, Y.; Song, B.; Sun, X.; et al. Genetic Modulation of RNA Splicing with a CRISPR-Guided Cytidine Deaminase. Mol. Cell 2018, 72, 380–394. [Google Scholar] [CrossRef]
- Happi Mbakam, C.; Rousseau, J.; Tremblay, G.; Yameogo, P.; Tremblay, J.P. Prime Editing Permits the Introduction of Specific Mutations in the Gene Responsible for Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2022, 23, 6160. [Google Scholar] [CrossRef]
- Tuffery-Giraud, S.; Béroud, C.; Leturcq, F.; Yaou, R.B.; Hamroun, D.; Michel-Calemard, L.; Moizard, M.P.; Bernard, R.; Cossée, M.; Boisseau, P.; et al. Genotype-phenotype analysis in 2405 patients with a dystrophinopathy using the UMD-DMD database: A model of nationwide knowledgebase. Hum. Mutat. 2009, 30, 934–945. [Google Scholar] [CrossRef]
- Taylor, P.J.; Maroulis, S.; Mullan, G.L.; Pedersen, R.L.; Baumli, A.; Elakis, G.; Piras, S.; Walsh, C.; Prósper-Gutiérrez, B.; De La Puente-Alonso, F.; et al. Measurement of the clinical utility of a combined mutation detection protocol in carriers of Duchenne and Becker muscular dystrophy. J. Med. Genet. 2007, 44, 368–372. [Google Scholar] [CrossRef]
- Van Putten, M.; Hulsker, M.; Nadarajah, V.D.; van Heiningen, S.H.; van Huizen, E.; van Iterson, M.; Admiraal, P.; Messemaker, T.; den Dunnen, J.T.; t Hoen, P.A.; et al. The effects of low levels of dystrophin on mouse muscle function and pathology. PLoS ONE 2012, 7, e31937. [Google Scholar] [CrossRef]
- Van Putten, M.; van der Pijl, E.M.; Hulsker, M.; Verhaart, I.E.; Nadarajah, V.D.; van der Weerd, L.; Aartsma-Rus, A. Low dystrophin levels in heart can delay heart failure in mdx mice. J. Mol. Cell. Cardiol. 2014, 69, 17–23. [Google Scholar] [CrossRef]
- Jin, Y.; Shen, Y.; Su, X.; Weintraub, N.L.; Tang, Y. Effective restoration of dystrophin expression in iPSC (Mdx)-derived muscle progenitor cells using the CRISPR/Cas9 system and homology-directed repair technology. Comput. Struct. Biotechnol. J. 2020, 18, 765–773. [Google Scholar] [CrossRef]
- Zhang, Y.; Long, C.; Bassel-Duby, R.; Olson, E.N. Myoediting: Toward Prevention of Muscular Dystrophy by Therapeutic Genome Editing. Physiol. Rev. 2018, 98, 1205–1240. [Google Scholar] [CrossRef]
- Xie, R.; Wang, X.; Wang, Y.; Ye, M.; Zhao, Y.; Yandell, B.S.; Gong, S. pH-Responsive Polymer Nanoparticles for Efficient Delivery of Cas9 Ribonucleoprotein with or without Donor DNA. Adv. Mater. 2022, 34, e2110618. [Google Scholar] [CrossRef]
- Suzuki, K.; Tsunekawa, Y.; Hernandez-Benitez, R.; Wu, J.; Zhu, J.; Kim, E.J.; Hatanaka, F.; Yamamoto, M.; Araoka, T.; Li, Z.; et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016, 540, 144–149. [Google Scholar] [CrossRef]
- Suzuki, K.; Izpisua Belmonte, J.C. In vivo genome editing via the HITI method as a tool for gene therapy. J. Hum. Genet. 2018, 63, 157–164. [Google Scholar] [CrossRef]
- Newby, G.A.; Liu, D.R. In vivo somatic cell base editing and prime editing. Mol. Ther. 2021, 29, 3107–3124. [Google Scholar] [CrossRef]
- Zhou, C.; Sun, Y.; Yan, R.; Liu, Y.; Zuo, E.; Gu, C.; Han, L.; Wei, Y.; Hu, X.; Zeng, R.; et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 2019, 571, 275–278. [Google Scholar] [CrossRef]
- Grünewald, J.; Zhou, R.; Garcia, S.P.; Iyer, S.; Lareau, C.A.; Aryee, M.J.; Joung, J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 2019, 569, 433–437. [Google Scholar] [CrossRef]
- Jin, S.; Zong, Y.; Gao, Q.; Zhu, Z.; Wang, Y.; Qin, P.; Liang, C.; Wang, D.; Qiu, J.L.; Zhang, F.; et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 2019, 364, 292–295. [Google Scholar] [CrossRef]
- Lee, H.K.; Willi, M.; Miller, S.M.; Kim, S.; Liu, C.; Liu, D.R.; Hennighausen, L. Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat. Commun. 2018, 9, 4804. [Google Scholar] [CrossRef]
- Chen, P.J.; Hussmann, J.A.; Yan, J.; Knipping, F.; Ravisankar, P.; Chen, P.F.; Chen, C.; Nelson, J.W.; Newby, G.A.; Sahin, M.; et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 2021, 184, 5635–5652. [Google Scholar] [CrossRef]
- Chamberlain, J.S.; Metzger, J.; Reyes, M.; Townsend, D.; Faulkner, J.A. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007, 21, 2195–2204. [Google Scholar] [CrossRef]
- Pastoret, C.; Sebille, A. mdx mice show progressive weakness and muscle deterioration with age. J. Neurol. Sci. 1995, 129, 97–105. [Google Scholar] [CrossRef]
- Lefaucheur, J.P.; Pastoret, C.; Sebille, A. Phenotype of dystrophinopathy in old mdx mice. Anat. Rec. 1995, 242, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.R.Q.; Nguyen, Q.; Dzierlega, K.; Huang, Y.; Yokota, T. CRISPR-Generated Animal Models of Duchenne Muscular Dystrophy. Genes 2020, 11, 342. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Ge, W.; Li, N.; Liu, Q.; Chen, F.; Yang, X.; Huang, X.; Ouyang, Z.; Zhang, Q.; Zhao, Y.; et al. Efficient base editing for multiple genes and loci in pigs using base editors. Nat. Commun. 2019, 10, 2852. [Google Scholar] [CrossRef] [PubMed]
- Klymiuk, N.; Blutke, A.; Graf, A.; Krause, S.; Burkhardt, K.; Wuensch, A.; Krebs, S.; Kessler, B.; Zakhartchenko, V.; Kurome, M.; et al. Dystrophin-deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle. Hum. Mol. Genet. 2013, 22, 4368–4382. [Google Scholar] [CrossRef]
- Valentine, B.A.; Cooper, B.J.; de Lahunta, A.; O’Quinn, R.; Blue, J.T. Canine X-linked muscular dystrophy. An animal model of Duchenne muscular dystrophy: Clinical studies. J. Neurol. Sci. 1988, 88, 69–81. [Google Scholar] [CrossRef]
- Sui, T.; Lau, Y.S.; Liu, D.; Liu, T.; Xu, L.; Gao, Y.; Lai, L.; Li, Z.; Han, R. A novel rabbit model of Duchenne muscular dystrophy generated by CRISPR/Cas9. Dis. Models Mech. 2018, 11, dmm032201. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Y.; Kang, Y.; Yang, W.; Niu, Y.; Guo, X.; Tu, Z.; Si, C.; Wang, H.; Xing, R.; et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum. Mol. Genet. 2015, 24, 3764–3774. [Google Scholar] [CrossRef]
- McGreevy, J.W.; Hakim, C.H.; McIntosh, M.A.; Duan, D. Animal models of Duchenne muscular dystrophy: From basic mechanisms to gene therapy. Dis. Models Mech. 2015, 8, 195–213. [Google Scholar] [CrossRef]
- Oh, H.J.; Chung, E.; Kim, J.; Kim, M.J.; Kim, G.A.; Lee, S.H.; Ra, K.; Eom, K.; Park, S.; Chae, J.H.; et al. Generation of a Dystrophin Mutant in Dog by Nuclear Transfer Using CRISPR/Cas9-Mediated Somatic Cells: A Preliminary Study. Int. J. Mol. Sci. 2022, 23, 2898. [Google Scholar] [CrossRef]
- Egorova, T.V.; Zotova, E.D. CRISPR/Cas9-generated mouse model of Duchenne muscular dystrophy recapitulating a newly identified large 430 kb deletion in the human DMD gene. Dis. Models Mech. 2019, 12, dmm037655. [Google Scholar] [CrossRef]
- Deconinck, A.E.; Rafael, J.A.; Skinner, J.A.; Brown, S.C.; Potter, A.C.; Metzinger, L.; Watt, D.J.; Dickson, J.G.; Tinsley, J.M.; Davies, K.E. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell 1997, 90, 717–727. [Google Scholar] [CrossRef]
- Perkins, K.J.; Davies, K.E. The role of utrophin in the potential therapy of Duchenne muscular dystrophy. Neuromuscul. Disord. 2002, 12, S78–S89. [Google Scholar] [CrossRef]
- Smith, B.F.; Yue, Y.; Woods, P.R.; Kornegay, J.N.; Shin, J.H.; Williams, R.R.; Duan, D. An intronic LINE-1 element insertion in the dystrophin gene aborts dystrophin expression and results in Duchenne-like muscular dystrophy in the corgi breed. Lab. Investig. 2011, 91, 216–231. [Google Scholar] [CrossRef]
- Cozzi, F.; Cerletti, M.; Luvoni, G.C.; Lombardo, R.; Brambilla, P.G.; Faverzani, S.; Blasevich, F.; Cornelio, F.; Pozza, O.; Mora, M. Development of muscle pathology in canine X-linked muscular dystrophy. II. Quantitative characterization of histopathological progression during postnatal skeletal muscle development. Acta Neuropathol. 2001, 101, 469–478. [Google Scholar] [CrossRef]
- Wells, D.J. Tracking progress: An update on animal models for Duchenne muscular dystrophy. Dis. Models Mech. 2018, 11, dmm035774. [Google Scholar] [CrossRef]
- Samulski, R.J.; Muzyczka, N. AAV-Mediated Gene Therapy for Research and Therapeutic Purposes. Annu. Rev. Virol. 2014, 1, 427–451. [Google Scholar] [CrossRef]
- Crudele, J.M.; Chamberlain, J.S. AAV-based gene therapies for the muscular dystrophies. Hum. Mol. Genet. 2019, 28, R102–R107. [Google Scholar] [CrossRef]
- Frontera, W.R.; Ochala, J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015, 96, 183–195. [Google Scholar] [CrossRef]
- Kornegay, J.N.; Li, J.; Bogan, J.R.; Bogan, D.J.; Chen, C.; Zheng, H.; Wang, B.; Qiao, C.; Howard, J.F., Jr.; Xiao, X. Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs. Mol. Ther. 2010, 18, 1501–1508. [Google Scholar] [CrossRef]
- Hinderer, C.; Katz, N.; Buza, E.L.; Dyer, C.; Goode, T.; Bell, P.; Richman, L.K.; Wilson, J.M. Severe Toxicity in Nonhuman Primates and Piglets Following High-Dose Intravenous Administration of an Adeno-Associated Virus Vector Expressing Human SMN. Hum. Gene Ther. 2018, 29, 285–298. [Google Scholar] [CrossRef]
- Buscara, L.; Gross, D.A.; Daniele, N. Of rAAV and Men: From Genetic Neuromuscular Disorder Efficacy and Toxicity Preclinical Studies to Clinical Trials and Back. J. Pers. Med. 2020, 10, 258. [Google Scholar] [CrossRef]
- Ren, C.; Kumar, S.; Shaw, D.R.; Ponnazhagan, S. Genomic stability of self-complementary adeno-associated virus 2 during early stages of transduction in mouse muscle in vivo. Hum. Gene Ther. 2005, 16, 1047–1057. [Google Scholar] [CrossRef]
- McCarty, D.M. Self-complementary AAV vectors; advances and applications. Mol. Ther. 2008, 16, 1648–1656. [Google Scholar] [CrossRef]
- Boutin, S.; Monteilhet, V.; Veron, P.; Leborgne, C.; Benveniste, O.; Montus, M.F.; Masurier, C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: Implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010, 21, 704–712. [Google Scholar] [CrossRef]
- Louis Jeune, V.; Joergensen, J.A.; Hajjar, R.J.; Weber, T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum. Gene Ther. Methods 2013, 24, 59–67. [Google Scholar] [CrossRef]
- Moreno, A.M.; Palmer, N.; Alemán, F.; Chen, G.; Pla, A.; Jiang, N.; Leong Chew, W.; Law, M.; Mali, P. Immune-orthogonal orthologues of AAV capsids and of Cas9 circumvent the immune response to the administration of gene therapy. Nat. Biomed. Eng. 2019, 3, 806–816. [Google Scholar] [CrossRef]
- Chew, W.L.; Tabebordbar, M.; Cheng, J.K.; Mali, P.; Wu, E.Y.; Ng, A.H.; Zhu, K.; Wagers, A.J.; Church, G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 2016, 13, 868–874. [Google Scholar]
- Charlesworth, C.T.; Deshpande, P.S.; Dever, D.P.; Camarena, J.; Lemgart, V.T.; Cromer, M.K.; Vakulskas, C.A.; Collingwood, M.A.; Zhang, L.; Bode, N.M.; et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019, 25, 249–254. [Google Scholar] [CrossRef]
- Wagner, D.L.; Amini, L.; Wendering, D.J.; Burkhardt, L.M.; Akyüz, L.; Reinke, P.; Volk, H.D.; Schmueck-Henneresse, M. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat. Med. 2019, 25, 242–248. [Google Scholar] [CrossRef]
- Wienert, B.; Shin, J.; Zelin, E.; Pestal, K.; Corn, J.E. In vitro-transcribed guide RNAs trigger an innate immune response via the RIG-I pathway. PLoS Biol. 2018, 16, e2005840. [Google Scholar] [CrossRef]
- Kim, S.; Koo, T.; Jee, H.G.; Cho, H.Y.; Lee, G.; Lim, D.G.; Shin, H.S.; Kim, J.S. CRISPR RNAs trigger innate immune responses in human cells. Genome Res. 2018, 28, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Ferdosi, S.R.; Ewaisha, R.; Moghadam, F.; Krishna, S.; Park, J.G.; Ebrahimkhani, M.R.; Kiani, S.; Anderson, K.S. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat. Commun. 2019, 10, 1842. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Cradick, T.J.; Brown, M.T.; Deshmukh, H.; Ranjan, P.; Sarode, N.; Wile, B.M.; Vertino, P.M.; Stewart, F.J.; Bao, G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014, 42, 7473–7485. [Google Scholar] [CrossRef] [PubMed]
- Murugan, K.; Suresh, S.K.; Seetharam, A.S.; Severin, A.J.; Sashital, D.G. Systematic in vitro specificity profiling reveals nicking defects in natural and engineered CRISPR-Cas9 variants. Nucleic Acids Res. 2021, 49, 4037–4053. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Tsai, S.Q.; Prew, M.S.; Nguyen, N.T.; Welch, M.M.; Lopez, J.M.; McCaw, Z.R.; Aryee, M.J.; Joung, J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016, 34, 869–874. [Google Scholar] [CrossRef]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef]
- Kocak, D.D.; Josephs, E.A.; Bhandarkar, V.; Adkar, S.S.; Kwon, J.B.; Gersbach, C.A. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019, 37, 657–666. [Google Scholar] [CrossRef]
- Lau, C.H.; Suh, Y. In vivo genome editing in animals using AAV-CRISPR system: Applications to translational research of human disease. F1000Res. 2017, 6, 2153. [Google Scholar] [CrossRef]
- Duddy, W.; Duguez, S.; Johnston, H.; Cohen, T.V.; Phadke, A.; Gordish-Dressman, H.; Nagaraju, K.; Gnocchi, V.; Low, S.; Partridge, T. Muscular dystrophy in the mdx mouse is a severe myopathy compounded by hypotrophy, hypertrophy and hyperplasia. Skelet Muscle 2015, 5, 16. [Google Scholar] [CrossRef]
- Wong, T.W.Y.; Ahmed, A.; Yang, G.; Maino, E.; Steiman, S.; Hyatt, E.; Chan, P.; Lindsay, K.; Wong, N.; Golebiowski, D.; et al. A novel mouse model of Duchenne muscular dystrophy carrying a multi-exonic Dmd deletion exhibits progressive muscular dystrophy and early-onset cardiomyopathy. Dis. Models Mech. 2020, 13, dmm045369. [Google Scholar] [CrossRef]
- Ancel, S.; Stuelsatz, P.; Feige, J.N. Muscle Stem Cell Quiescence: Controlling Stemness by Staying Asleep. Trends Cell Biol. 2021, 31, 556–568. [Google Scholar] [CrossRef]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef]
- Nance, M.E.; Shi, R.; Hakim, C.H.; Wasala, N.B.; Yue, Y.; Pan, X.; Zhang, T.; Robinson, C.A.; Duan, S.X.; Yao, G.; et al. AAV9 Edits Muscle Stem Cells in Normal and Dystrophic Adult Mice. Mol. Ther. 2019, 27, 1568–1585. [Google Scholar] [CrossRef]
- Kwon, J.B.; Ettyreddy, A.R.; Vankara, A.; Bohning, J.D.; Devlin, G.; Hauschka, S.D.; Asokan, A.; Gersbach, C.A. In Vivo Gene Editing of Muscle Stem Cells with Adeno-Associated Viral Vectors in a Mouse Model of Duchenne Muscular Dystrophy. Mol. Ther. Methods Clin. Dev. 2020, 19, 320–329. [Google Scholar] [CrossRef]
- Goldstein, J.M.; Tabebordbar, M.; Zhu, K.; Wang, L.D.; Messemer, K.A.; Peacker, B.; Ashrafi Kakhki, S.; Gonzalez-Celeiro, M.; Shwartz, Y.; Cheng, J.K.W.; et al. In Situ Modification of Tissue Stem and Progenitor Cell Genomes. Cell Rep. 2019, 27, 1254–1264. [Google Scholar] [CrossRef]
| Strategy | Mutation | Nuclease | Target Region | DMD Model | Delivery | Infusion | Reference |
|---|---|---|---|---|---|---|---|
| Double-cut exon exicision | Ex23 mut | SpCas9 | i22, i23 | mdx mice | AAV9 | IM, IV, IP, RO | [34,35] |
| Ex23 mut | SaCas9 | i22, i23 | mdx mice | AAV8 | IM, IV, IP | [36,37] | |
| ΔEx52 | SpCas9 | i50, i51 | DMDΔ52 mice | AAV9 | IM, IV | [39] | |
| ΔEx52 | SaCas9 | Ex47, Ex58 | hDMDΔ52/mdx mice | AAV9 | IV | [40] | |
| Ex23 mut | SaCas9 | i20, i23 | mdx mice | AAVrh74 | IV | [43] | |
| Dup Ex18-30 | SaCas9 | i21 | Dup 18-30 mice | AAV9 | IV | [125] | |
| Ex23 mut | SpCas9 | i20, i23 | mdx mice | AAV | IM | [126] | |
| Ex23 mut | SaCas9 | i22, i23 | mdx mice | AAV9 | IM, IV, IP | [127] | |
| Ex53 mut | SaCas9, SpCas9 | i51, i53 | mdx4Cv mice | AAV6 | IM, RO | [128,129] | |
| ΔEx45 | SpCas9 | i44, i55 | hDMDΔ45/mdxD2 mice | Plasmid | IM | [130] | |
| Ex23 mut | SaCas9 | i20, i23 | mdx/Utr+/− mice | AAV | IV | [131] | |
| Single-cut exon skipping and reframing | ΔEx44 | SpCas9 | Ex45 | DMDΔ44 mice | AAV9 | IM, IP | [132] |
| ΔEx50 | SaCas9-KKH | Ex51 | DMDΔ50 mice | AAV9 | IP | [133] | |
| ΔEx43 | SpCas9 | Ex44 | DMDΔ43 mice | AAV9 | IM | [134] | |
| ΔEx45 | SpCas9 | Ex44 | DMDΔ45 mice | AAV9 | IM | [134] | |
| ΔEx52 | SpCas9 | Ex53 | DMDΔ52 mice | AAV9 | IM | [134] | |
| ΔEx50 | SpCas9-VRQR | Ex51 | DMDΔ50;h51KI mice | AAV9 | IP | [135] | |
| ΔEx44 | SpCas9 | Ex45 | DMDΔ44 mice | AAV9 | IP | [44,136] | |
| Ex23 mut | CjCas9 | Ex23 | DMD/Ex23 mut mice | AAV9 | IM | [137] | |
| ΔEx50 | SpCas9 | Ex51 | DMDΔ50 mice | AAV9 | IM, IP | [138] | |
| ΔEx50 | SpCas9 | Ex51 | ΔEx50-Dmd-Luc mice | AAV9 | IM, IP | [139] | |
| ΔEx50 | SpCas9 | Ex51 | DMDΔ50 dogs | AAV9 | IM, IV | [140] | |
| Pseudo Ex13 | SpCas9 | i13 | WCMD dogs | AAV8 | IM, IV | [141] | |
| Pseudo Ex19 | SpCas9 | i19 | LRMD dogs | AAV8 | IM, IV | [141] | |
| HDR-based correction | Ex23 mut | SpCas9 | Ex23 | mdx mice | Injection | Zygote | [124] |
| Ex53 mut | SpCas9 | Ex53 | mdx4Cv mice | AAV6 | IM | [128] | |
| Ex23 mut | LbCas12a | Ex23 | mdx mice | Injection | Zygote | [142] | |
| i6 mut | SpCas9 | i6 | GRMD dogs | Plasmid | IM | [143] | |
| Ex23 mut | SpCas9 | Ex23 | mdx mice | Nanoparticle | IM | [144] | |
| Exon knockin | Ex51 mut | SaCas9 | Ex52 | hDMDΔ52/mdx mice | AAV9 | IM, IV | [145] |
| Base editing | Ex51 mut | ABEmax-nSpCas9 | Ex50 | DMDΔ51 mice | AAV9 | IM | [146] |
| Ex20 mut | ABE-nSpCas9 | Ex20 | DMD/Ex20* mice | AAV9 | IM | [147] | |
| Ex53 mut | ABE-nSpCas9-iNG | Ex53 | mdx4Cv mice | AAV9 | IV | [148] | |
| Ex4 mut | CBE-nSaCas9 | Ex4 | DmdE4* mice | AAV9 | IP | [149] |
| Strategy | Mutation | Nuclease | Target Region | DMD Model | Delivery | Reference |
|---|---|---|---|---|---|---|
| Double-cut exon exicision | ΔEx46-51 | SpCas9 | i44, i55 | human iPSCs | electroporation | [38] |
| ΔEx52 | SpCas9 | i50, i51 | human iPSCs | AAV6 | [39] | |
| ΔEx49-50, ΔEx50-52, ΔEx51-53, ΔEx51-56 | SaCas9 | Ex47, Ex58 | human myoblasts | lentivirus | [40] | |
| Dup Ex18-30 | SpCas9 | i27 | human myoblasts | lentivirus | [122] | |
| Dup Ex55-59 | SpCas9 | i54 | human iPSCs | nucleofection | [150] | |
| Dup Ex2 | SpCas9 | i2 | human myoblasts | lentivirus | [151] | |
| Dup Ex3-16 | SpCas9 | i9 | human myoblasts | lentivirus | [152] | |
| Dup Ex18-25 | SpCas9 | i25 | human myoblasts | lentivirus | [153] | |
| ΔEx48-50 | SpCas9 | i50, i51 | human myoblasts | electroporation | [154] | |
| ΔEx48-50 | SpCas9 | i44, i55 | human myoblasts | electroporation | [154] | |
| ΔEx51 | SpCas9, AsCas12a | i44, i55; i45, i54 | human myoblasts | electroporation | [155] | |
| ΔEx45-52 | SpCas9 | i52, i53 | human myoblasts | adenovirus | [156,157] | |
| ΔEx45-52, ΔEx48-50 | SpCas9, eSpCas9(1.1) | i43, i54 | human myoblasts | adenovirus | [156,157,158] | |
| ΔEx8-9 | SpCas9 | i2, i7; i5, i7; i6, i11 | human iPSCs | nucleofection | [159] | |
| ΔEx3-7 | SpCas9 | i7, i9 | human iPSCs | nucleofection | [159] | |
| Ex23 mut | SpCas9 | i22, i23 | mouse myoblasts | lipotransfection | [160] | |
| Single-cut exon skipping and reframing | ΔEx44 | SpCas9 | Ex45 | human iPSCs | nucleofection | [132] |
| ΔEx48-50 | SaCas9-KKH | Ex51 | human iPSCs | nucleofection | [133] | |
| ΔEx43, ΔEx45 | SpCas9 | Ex44 | human iPSCs | nucleofection | [134] | |
| ΔEx52 | SpCas9 | Ex51 | human iPSCs | nucleofection | [134] | |
| ΔEx48-50 | SpCas9-VRQR | Ex51 | human iPSCs | nucleofection | [135] | |
| ΔEx48-50 | LbCas12a, AsCas12a | Ex51 | human iPSCs | nucleofection | [142] | |
| ΔEx48-50 | SpCas9 | Ex51 | human iPSCs | nucleofection | [150] | |
| Pseudo Ex47 | SpCas9 | i47 | human iPSCs | nucleofection | [150] | |
| ΔEx48-50 | eSpCas9(1.1) | Ex51 | human myoblasts | adenovirus | [158] | |
| HDR-based knockin | i6 mut | SpCas9 | i6 | canine myoblasts | nucleofection | [143] |
| ΔEx44 | SpCas9 | Ex44 | human iPSCs | electroporation | [161] | |
| Base editing | ΔEx51 | ABEmax-nSpCas9 | Ex50 | human iPSCs | nucleofection | [146] |
| ΔEx51 | CBE-nSaCas9 | Ex50 | human iPSCs | lipotransfection | [162] | |
| Prime editing | ΔEx51 | PE2-dSpCas9 | Ex52 | human iPSCs | nucleofection | [146] |
| Ex6 mut | PE2-nSpCas9 | Ex6 | human myoblasts | electroporation | [163] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.; Wei, T.; Yang, H.; Li, G.; Li, H. CRISPR-Based Therapeutic Gene Editing for Duchenne Muscular Dystrophy: Advances, Challenges and Perspectives. Cells 2022, 11, 2964. https://doi.org/10.3390/cells11192964
Chen G, Wei T, Yang H, Li G, Li H. CRISPR-Based Therapeutic Gene Editing for Duchenne Muscular Dystrophy: Advances, Challenges and Perspectives. Cells. 2022; 11(19):2964. https://doi.org/10.3390/cells11192964
Chicago/Turabian StyleChen, Guofang, Tingyi Wei, Hui Yang, Guoling Li, and Haisen Li. 2022. "CRISPR-Based Therapeutic Gene Editing for Duchenne Muscular Dystrophy: Advances, Challenges and Perspectives" Cells 11, no. 19: 2964. https://doi.org/10.3390/cells11192964
APA StyleChen, G., Wei, T., Yang, H., Li, G., & Li, H. (2022). CRISPR-Based Therapeutic Gene Editing for Duchenne Muscular Dystrophy: Advances, Challenges and Perspectives. Cells, 11(19), 2964. https://doi.org/10.3390/cells11192964






