Hydrophobic Bile Salts Induce Pro-Fibrogenic Proliferation of Hepatic Stellate Cells through PI3K p110 Alpha Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Primary Hepatic Stellate Cells and LX-2 Cells
2.2. Cell Culture
2.3. siRNA Protein Suppression
2.4. LDH Assay
2.5. WST Assay
2.6. BrdU Assay
2.7. DNA Quantification Assay
2.8. Collagen Quantification In Vitro
2.9. Cell Counting
2.10. Immunoblotting
2.11. Statistical Analysis
3. Results
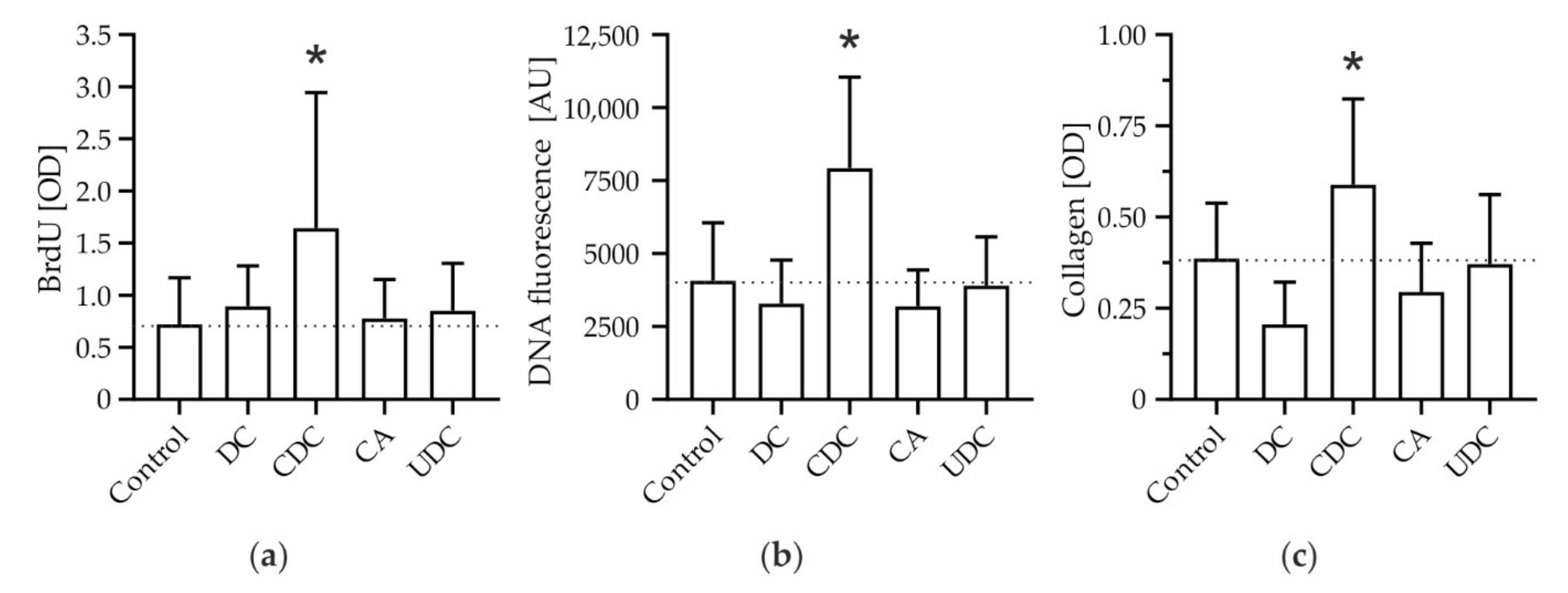
3.1. The Hydrophobic Bile Salt CDC Specifically Promotes Proliferation and Expansion of HSCs with Subsequent Deposition of Collagen
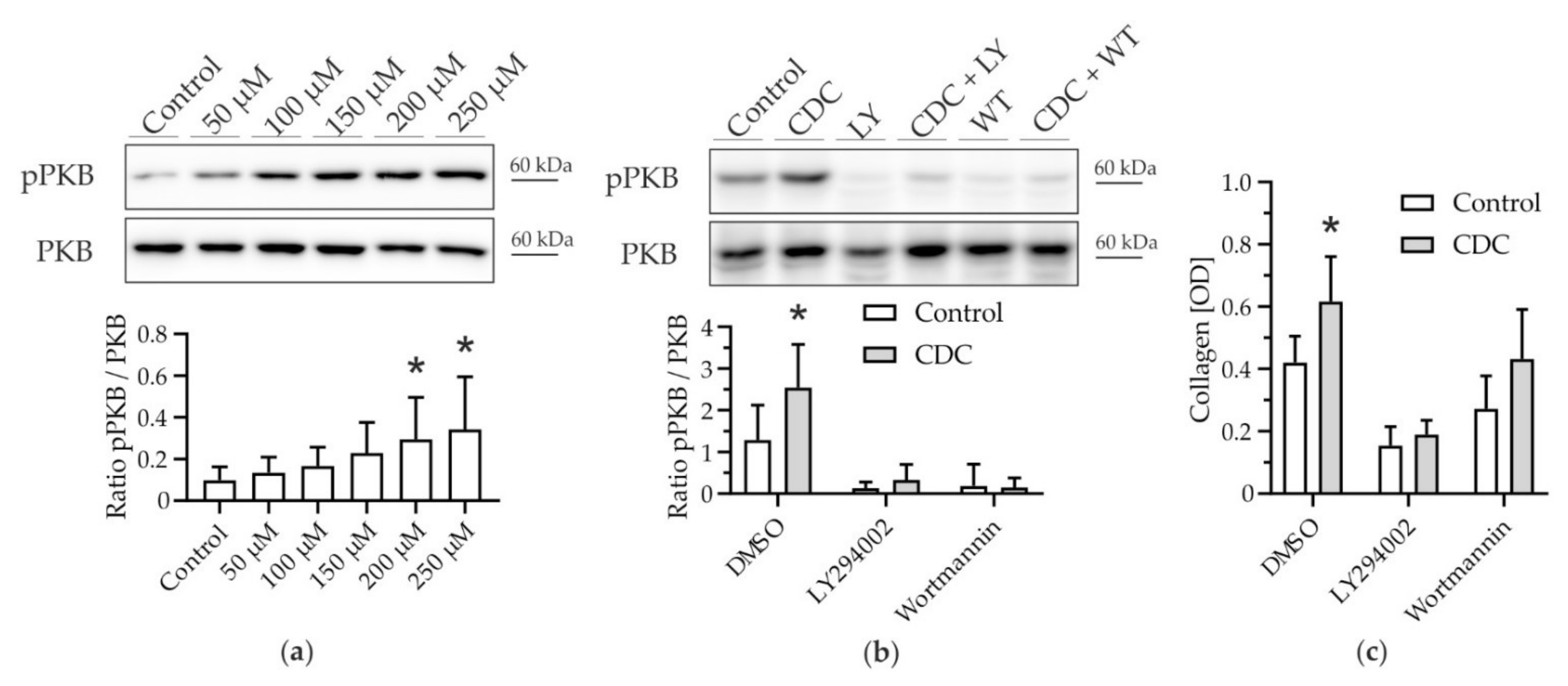
3.2. CDC-Induced Pro-Fibrotic Effects in HSC Engage PI3K-PKB Signaling
3.3. p110α Is the Catalytic PI3K Isoform Predominantly Mediating Pro-Fibrotic Effects of CDC
3.4. Activation of the Human HSC Cell Line LX-2 Depends on PI3K p110α Signaling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Strautnieks, S.S.; Bull, L.N.; Knisely, A.S.; Kocoshis, S.A.; Dahl, N.; Arnell, H.; Sokal, E.; Dahan, K.; Childs, S.; Ling, V.; et al. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat. Genet. 1998, 20, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.; Karam, V.; Cailliez, V.; Grady, J.G.O.; Mirza, D.; Cherqui, D.; Klempnauer, J.; Salizzoni, M.; Pratschke, J.; Jamieson, N.; et al. 2018 Annual Report of the European Liver Transplant Registry (ELTR)-50-year evolution of liver transplantation. Transpl. Int. 2018, 31, 1293–1317. [Google Scholar] [CrossRef] [PubMed]
- Dilger, K.; Hohenester, S.; Winkler-Budenhofer, U.; Bastiaansen, B.A.; Schaap, F.; Rust, C.; Beuers, U. Effect of ursodeoxycholic acid on bile acid profiles and intestinal detoxification machinery in primary biliary cirrhosis and health. J. Hepatol. 2012, 57, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Greim, H.; Trülzsch, D.; Czygan, P.; Rudick, J.; Hutterer, F.; Schaffner, F.; Popper, H. Mechanism of cholestasis. 6. Bile acids in human livers with or without biliary obstruction. Gastroenterology 1972, 63, 846–850. [Google Scholar] [CrossRef]
- Greim, H.; Trülzsch, D.; Roboz, J.; Dressler, K.; Czygan, P.; Hutterer, F.; Schaffner, F.; Popper, H. Mechanism of cholestasis. 5. Bile acids in normal rat livers and in those after bile duct ligation. Gastroenterology 1972, 63, 837–845. [Google Scholar] [CrossRef]
- Murphy, G.M.; Ross, A.; Billing, B.H. Serum bile acids in primary biliary cirrhosis. Gut 1972, 13, 201–206. [Google Scholar] [CrossRef]
- Dyrszka, H.; Salen, G.; Zaki, F.G.; Chen, T.; Mosbach, E. Hepatic Toxicity in the Rhesus Monkey Treated with Chenodeoxycholic Acid for 6 Months: Biochemical and Ultrastructural Studies. Gastroenterology 1976, 70, 93–104. [Google Scholar] [CrossRef]
- Malhi, H.; Gores, G.J. Cellular and Molecular Mechanisms of Liver Injury. Gastroenterology 2008, 134, 1641–1654. [Google Scholar] [CrossRef]
- Friedman, S.L. Mechanisms of Hepatic Fibrogenesis. Gastroenterology 2008, 134, 1655–1669. [Google Scholar] [CrossRef]
- Guicciardi, M.; Gores, G. Bile acid-mediated hepatocyte apoptosis and cholestatic liver disease. Dig. Liver Dis. 2002, 34, 387–392. [Google Scholar] [CrossRef]
- Fickert, P.; Wagner, M. Biliary bile acids in hepatobiliary injury—What is the link. J. Hepatol. 2017, 67, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Hohenester, S.; Kanitz, V.; Kremer, A.E.; Paulusma, C.C.; Wimmer, R.; Kuehn, H.; Denk, G.; Horst, D.; Elferink, R.O.; Beuers, U. Glycochenodeoxycholate Promotes Liver Fibrosis in Mice with Hepatocellular Cholestasis. Cells 2020, 9, 281. [Google Scholar] [CrossRef]
- Fuchs, C.D.; Paumgartner, G.; Wahlström, A.; Schwabl, P.; Reiberger, T.; Leditznig, N.; Stojakovic, T.; Rohr-Udilova, N.; Chiba, P.; Marschall, H.U.; et al. Metabolic preconditioning protects BSEP/ABCB11(−/−) mice against cholestatic liver injury. J. Hepatol. 2017, 66, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Salem, M.; Yousef, I.M.; Tuchweber, B.; Lam, P.; Childs, S.J.; Helgason, C.D.; Ackerley, C.; Phillips, M.J.; Ling, V. Targeted inactivation of sister of P-glycoprotein gene (spgp) in mice results in nonprogressive but persistent intrahepatic cholestasis. Proc. Natl. Acad. Sci. USA 2001, 98, 2011–2016. [Google Scholar] [CrossRef] [PubMed]
- Hohenester, S.; Vennegeerts, T.; Wagner, M.; Wimmer, R.; Drolle, H.; Rieger, C.; Denk, G.U.; Rust, C.; Fiegl, M. Physiological hypoxia prevents bile salt-induced apoptosis in human and rat hepatocytes. Liver Int. 2013, 34, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Hohenester, S.; Gates, A.; Wimmer, R.; Beuers, U.; Anwer, M.S.; Rust, C.; Webster, C.R. Phosphatidylinositol-3-kinase p110γ contributes to bile salt-induced apoptosis in primary rat hepatocytes and human hepatoma cells. J. Hepatol. 2010, 53, 918–926. [Google Scholar] [CrossRef]
- Myung, S.J.; Yoon, J.-H.; Gwak, G.-Y.; Kim, W.; Yang, J.I.; Lee, S.H.; Jang, J.J.; Lee, H.-S. Bile acid-mediated thrombospondin-1 induction in hepatocytes leads to transforming growth factor-β-dependent hepatic stellate cell activation. Biochem. Biophys. Res. Commun. 2007, 353, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Roll, F. Isolation and culture of hepatic lipocytes, Kupffer cells, and sinusoidal endothelial cells by density gradient centrifugation with Stractan. Anal. Biochem. 1987, 161, 207–218. [Google Scholar] [CrossRef]
- Wimmer, R.; Hohenester, S.; Pusl, T.; Denk, G.U.; Rust, C.; Beuers, U. Tauroursodeoxycholic acid exerts anticholestatic effects by a cooperative cPKC alpha-/PKA-dependent mechanism in rat liver. Gut 2008, 57, 1448–1454. [Google Scholar] [CrossRef] [PubMed]
- Denk, G.U.; Hohenester, S.; Wimmer, R.; Böhland, C.; Rust, C.; Beuers, U. Role of mitogen-activated protein kinases in tauroursodeoxycholic acid-induced bile formation in cholestatic rat liver. Hepatol. Res. 2008, 38, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Gates, A.; Hohenester, S.; Anwer, M.S.; Webster, C.R. cAMP-GEF cytoprotection by Src tyrosine kinase activation of phosphoinositide -3 -kinase p110β/α in rat hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G764–G774. [Google Scholar] [CrossRef] [PubMed]
- Hohenester, S.; Maillette de Buy Wenniger, L.; Paulusma, C.C.; van Vliet, S.J.; Jefferson, D.M.; Oude Elferink, R.P.; Beuers, U. A biliary HCO3- umbrella constitutes a protective mechanism against bile acid-induced injury in human cholangiocytes. Hepatology 2012, 55, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Schonhoff, C.M.; Yamazaki, A.; Hohenester, S.; Webster, C.R.; Bouscarel, B.; Anwer, M.S. PKCε-dependent and -independent effects of taurolithocholate on PI3K/PKB pathway and taurocholate uptake in HuH-NTCP cell line. American journal of physiology. Gastrointest. Liver Physiol. 2009, 297, G1259–G1267. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. [Google Scholar] [CrossRef]
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Mol. Biol. Rep. 2020, 47, 4587–4629. [Google Scholar] [CrossRef]
- Arcaro, A.; Wymann, M. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: The role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem. J. 1993, 296, 297–301. [Google Scholar] [CrossRef]
- Vlahos, C.J.; Matter, W.F.; Hui, K.Y.; Brown, R.F. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 1994, 269, P5241–P5248. [Google Scholar] [CrossRef]
- Fritsch, C.; Huang, A.; Chatenay-Rivauday, C.; Schnell, C.; Reddy, A.; Liu, M.; Kauffmann, A.; Guthy, D.; Erdmann, D.; De Pover, A.; et al. Characterization of the Novel and Specific PI3Kα Inhibitor NVP-BYL719 and Development of the Patient Stratification Strategy for Clinical Trials. Mol. Cancer Ther. 2014, 13, 1117–1129. [Google Scholar] [CrossRef]
- André, F. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef]
- Reiter, F.P.; Ye, L.; Bösch, F.; Wimmer, R.; Artmann, R.; Ziesch, A.; Kanitz, V.; Mayr, D.; Steib, C.J.; Trauner, M.; et al. Antifibrotic effects of hypocalcemic vitamin D analogs in murine and human hepatic stellate cells and in the CCl4 mouse model. Lab. Investig. 2019, 99, 1906–1917. [Google Scholar] [CrossRef] [PubMed]
- Simoes, I.C.; Janikiewicz, J.; Bauer, J.; Karkucinska-Wieckowska, A.; Kalinowski, P.; Dobrzyń, A.; Wolski, A.; Pronicki, M.; Zieniewicz, K.; Dobrzyń, P.; et al. Fat and Sugar—A Dangerous Duet. A Comparative Review on Metabolic Remodeling in Rodent Models of Nonalcoholic Fatty Liver Disease. Nutrients 2019, 11, 2871. [Google Scholar] [CrossRef] [PubMed]
- Weiskirchen, S.; Tag, C.G.; Sauer-Lehnen, S.; Tacke, F.; Weiskirchen, R. Isolation and Culture of Primary Murine Hepatic Stellate Cells. Fibrosis 2017, 1627, 165–191. [Google Scholar] [CrossRef]
- Reiter, F.P.; Ye, L.; Ofner, A.; Schiergens, T.S.; Ziesch, A.; Brandl, L.; Ben Khaled, N.; Hohenester, S.; Wimmer, R.; Artmann, R.; et al. p70 Ribosomal Protein S6 Kinase Is a Checkpoint of Human Hepatic Stellate Cell Activation and Liver Fibrosis in Mice. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Camps, M.; Rückle, T.; Ji, H.; Ardissone, V.; Rintelen, F.; Shaw, J.; Ferrandi, C.; Chabert, C.; Gillieron, C.; Françon, B.; et al. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat. Med. 2005, 11, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Hui, A.Y.; Albanis, E.; Arthur, M.J.; O’Byrne, S.M.; Blaner, W.S.; Mukherjee, P.; Friedman, S.L.; Eng, F.J. Human hepatic stellate cell lines, LX-1 and LX-2: New tools for analysis of hepatic fibrosis. Gut 2005, 54, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Dewidar, B.; Meyer, C.; Dooley, S.; Meindl-Beinker, A.N. TGF-β in Hepatic Stellate Cell Activation and Liver Fibrogenesis—Updated 2019. Cells 2019, 8, 1419. [Google Scholar] [CrossRef] [PubMed]
- Schon, H.-T.; Weiskirchen, R. Immunomodulatory effects of transforming growth factor-β in the liver. Hepatobiliary Surg. Nutr. 2014, 3, 386–406. [Google Scholar] [CrossRef] [PubMed]
- Svegliati-Baroni, G.; Ridolfi, F.; Hannivoort, R.; Saccomanno, S.; Homan, M.; de Minicis, S.; Jansen, P.L.; Candelaresi, C.; Benedetti, A.; Moshage, H. Bile acids induce hepatic stellate cell proliferation via activation of the epidermal growth factor receptor. Gastroenterology 2005, 128, 1042–1055. [Google Scholar] [CrossRef]
- Sommerfeld, A.; Reinehr, R.; Häussinger, D. Bile Acid-induced Epidermal Growth Factor Receptor Activation in Quiescent Rat Hepatic Stellate Cells Can Trigger Both Proliferation and Apoptosis. J. Biol. Chem. 2009, 284, 22173–22183. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y. Recent advances in understanding bile acid homeostasis. F1000Research 2017, 6, 2029. [Google Scholar] [CrossRef] [PubMed]
- Rust, C.; Bauchmüller, K.; Fickert, P.; Fuchsbichler, A.; Beuers, U. Phosphatidylinositol 3-kinase-dependent signaling modulates taurochenodeoxycholic acid-induced liver injury and cholestasis in perfused rat livers. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 289, G88–G94. [Google Scholar] [CrossRef]
- Nakhaeirad, S.; Nakhaeizadeh, H.; Götze, S.; Kordes, C.; Sawitza, I.; Hoffmann, M.J.; Franke, M.; Schulz, W.; Scheller, J.; Piekorz, R.P.; et al. The Role of Embryonic Stem Cell-expressed RAS (ERAS) in the Maintenance of Quiescent Hepatic Stellate Cells. J. Biol. Chem. 2016, 291, 8399–8413. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Nölting, S.; Rentsch, J.; Freitag, H.; Detjen, K.; Briest, F.; Möbs, M.; Weissmann, V.; Siegmund, B.; Auernhammer, C.J.; Prada, E.T.A.; et al. The selective PI3Kα inhibitor BYL719 as a novel therapeutic option for neuroendocrine tumors: Results from multiple cell line models. PLoS ONE 2017, 12, e0182852. [Google Scholar] [CrossRef] [PubMed]
- Son, G.; Hines, I.N.; Lindquist, J.; Schrum, L.W.; Rippe, R.A. Inhibition of phosphatidylinositol 3-kinase signaling in hepatic stellate cells blocks the progression of hepatic fibrosis. Hepatology 2009, 50, 1512–1523. [Google Scholar] [CrossRef] [PubMed]
- Son, M.K.; Ryu, Y.-L.; Jung, K.H.; Lee, H.; Lee, H.S.; Yan, H.H.; Park, H.J.; Ryu, J.-K.; Suh, J.; Hong, S.; et al. HS-173, a Novel PI3K Inhibitor, Attenuates the Activation of Hepatic Stellate Cells in Liver Fibrosis. Sci. Rep. 2013, 3, 3470. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zimny, S.; Koob, D.; Li, J.; Wimmer, R.; Schiergens, T.; Nagel, J.; Reiter, F.P.; Denk, G.; Hohenester, S. Hydrophobic Bile Salts Induce Pro-Fibrogenic Proliferation of Hepatic Stellate Cells through PI3K p110 Alpha Signaling. Cells 2022, 11, 2344. https://doi.org/10.3390/cells11152344
Zimny S, Koob D, Li J, Wimmer R, Schiergens T, Nagel J, Reiter FP, Denk G, Hohenester S. Hydrophobic Bile Salts Induce Pro-Fibrogenic Proliferation of Hepatic Stellate Cells through PI3K p110 Alpha Signaling. Cells. 2022; 11(15):2344. https://doi.org/10.3390/cells11152344
Chicago/Turabian StyleZimny, Sebastian, Dennis Koob, Jingguo Li, Ralf Wimmer, Tobias Schiergens, Jutta Nagel, Florian Paul Reiter, Gerald Denk, and Simon Hohenester. 2022. "Hydrophobic Bile Salts Induce Pro-Fibrogenic Proliferation of Hepatic Stellate Cells through PI3K p110 Alpha Signaling" Cells 11, no. 15: 2344. https://doi.org/10.3390/cells11152344
APA StyleZimny, S., Koob, D., Li, J., Wimmer, R., Schiergens, T., Nagel, J., Reiter, F. P., Denk, G., & Hohenester, S. (2022). Hydrophobic Bile Salts Induce Pro-Fibrogenic Proliferation of Hepatic Stellate Cells through PI3K p110 Alpha Signaling. Cells, 11(15), 2344. https://doi.org/10.3390/cells11152344





