Optogenetic Methods to Investigate Brain Alterations in Preclinical Models
Abstract
:1. Introduction
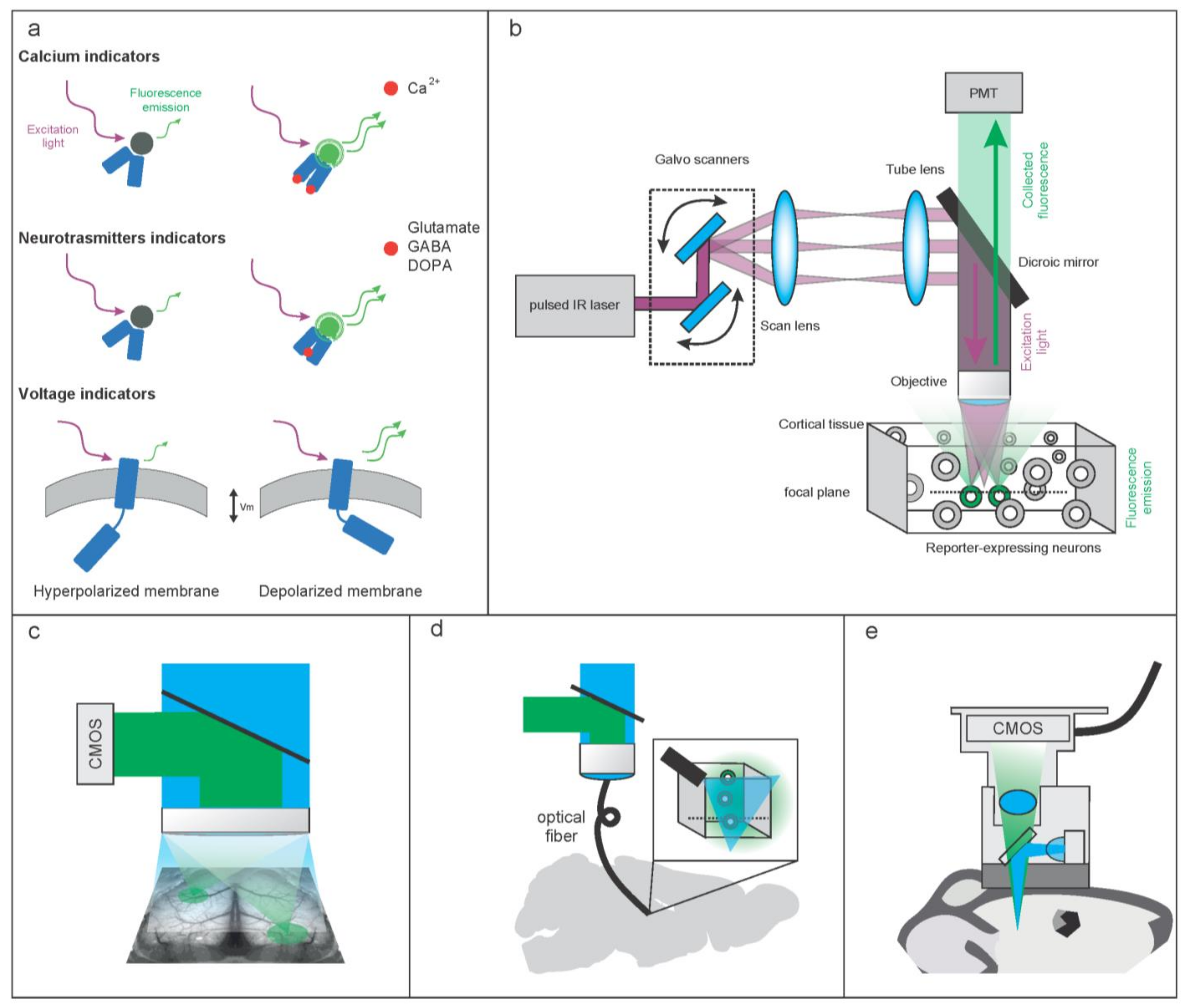
2. Molecular and Hardware Tools to Optically Record Brain Activity
2.1. Molecular Tools to Optically Record Brain Activity
2.1.1. Genetically Encoded Calcium-Based Indicators (GECI)
2.1.2. Genetically Encoded Voltage Indicators (GEVI)
2.1.3. Imaging Neurotransmitters and Neuromodulators
| Reporter | Analyte | Kd (nM) | Koff (ms) | ΔF/F0 Peak % | Available @ | Ref. | |
|---|---|---|---|---|---|---|---|
| GCaMP6s | Calcium | 147 | 1796 | 1680 | Addgene | [9] | |
| GCaMP6f | Calcium | 375 | 400 | 1314 | Addgene | [9] | |
| jGCaMP7s | Calcium | 68 | 1260 | Addgene | [6] | ||
| jGCaMP7f | Calcium | 150 | 270 | 3100 | Addgene | [6] | |
| jGCaMP8f | Calcium | 334 | 27 | 7880 | Addgene | Janelia | |
| jGCaMP8m | Calcium | 108 | 55 | 4570 | Addgene | Janelia | |
| jGCaMP8s | Calcium | 46 | 272 | 4950 | Addgene | Janelia | |
| dLight1.1 | dopamine | 330 | 230 | Addgene | [48] | ||
| dLight1.2 | dopamine | 770 | 90 | 340 | Addgene | [48] | |
| dLight1.3b | dopamine | 1680 | 930 | Addgene | [48] | ||
| GRABDA1m | dopamine | 130 | 700 | 90 | Addgene | [46] | |
| GRABDA2m | dopamine | 90 | 340 | Yu Long Li lab | [47] | ||
| GRABDA1h | dopamine | 10 | 2500 | 90 | Addgene | [46] | |
| GRABDA2h | dopamine | 7 | 280 | Yu Long Li lab | [47] | ||
| GRABNE1h | norepinephrine | 83 | 2000 | 130 | Yu Long Li lab | [50] | |
| GRABNE1m | norepinephrine | 930 | 750 | 250 | Yu Long Li lab | [50] | |
| iGABASnFR | GABA | 9000 | Addgene | [43] | |||
| GRAB5HT1.0 | serotonin | 22 | 3100 | 280 | Yu Long Li lab | [56] | |
| iSeroSnFr | serotonin | 1500 | 250 | Tian lab | [57] | ||
| iGluSnFR | glutamate | 4900 | 92 | 100 | Addgene | [41] | |
| iGlu f | glutamate | 137,000 | 2.1 | Addgene | [40] | ||
| iGlu u | glutamate | 600,000 | 700 | Addgene | [40] | ||
| iACHSnFR | acetylcholine | 1300 | 1200 | Addgene | [58] | ||
| GACh2.0 | acetylcholine | 2000 | 3700 | Yu Long Li lab | [45] | ||
| GRABATP1.0 | ATP | 45 | 9 | 1000 | Yu Long Li lab | [54] | |
| iATPSnFR1 | ATP | 50 | 190 | Addgene | [53] |
2.1.4. Directing Reporters Expression to Neuronal Populations of Interests
2.2. Hardware Tools to Optically Record Brain Activity
2.2.1. Hardware Configurations to Optically Record Brain Activity at High Resolution
2.2.2. Methods for Extending the Imaging in the Third Dimension
2.2.3. Methods for Extending the Imaging Wider and Deeper in the Brain
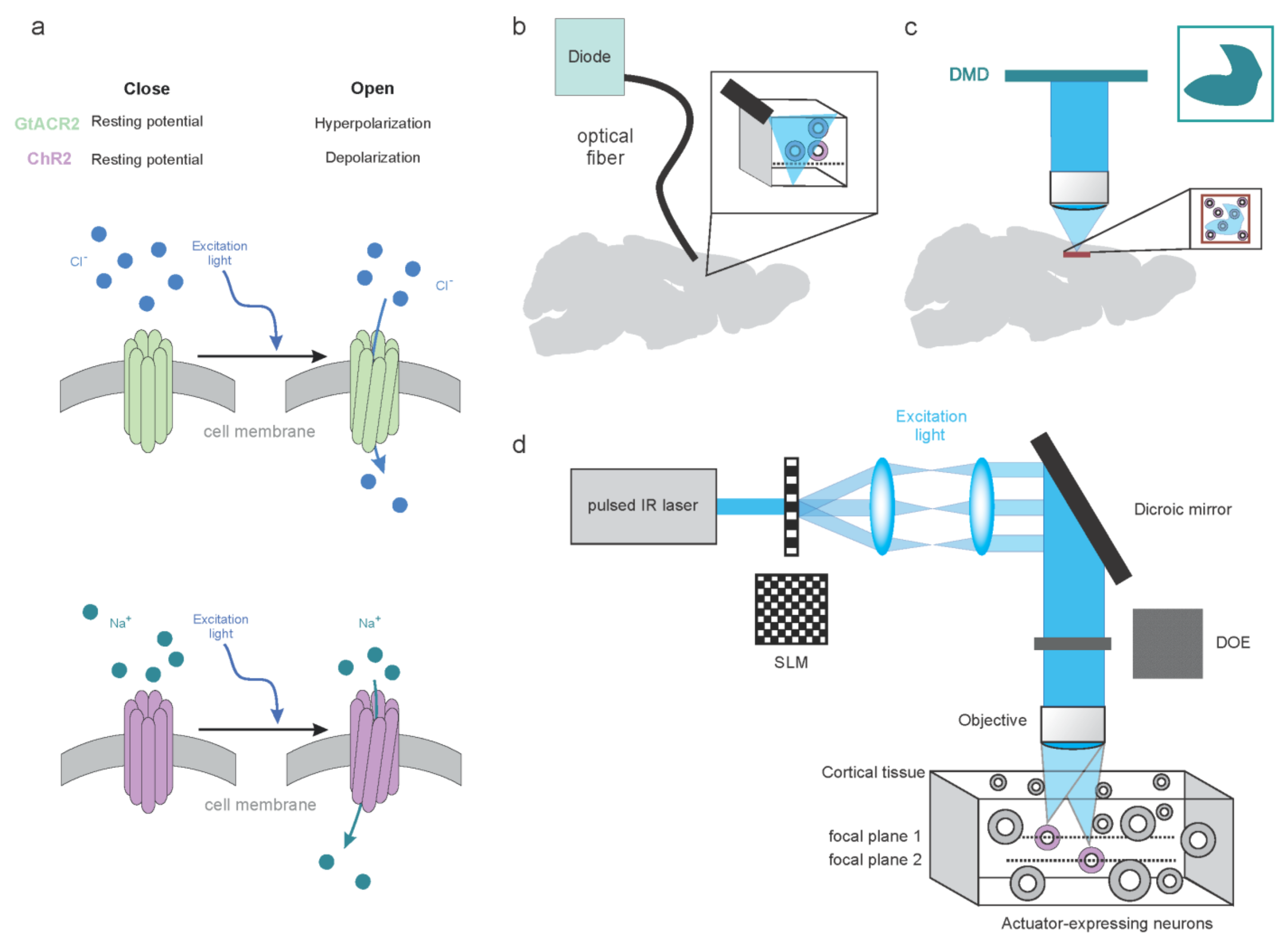
3. Molecular and Hardware Tools to Modulate Brain Activity
3.1. Molecular Tools to Dissect Brain Activity
3.1.1. Synthetic Tools to Dissect Brain Activity
3.1.2. Genetic Tools to Optically Dissect Brain Activity
3.2. Optical Approaches to Dissect Brain Activity
3.2.1. Low-Resolution Modulation of the Neuronal Circuits
3.2.2. High-Resolution Modulation of the Neuronal Circuits
4. Light-Based Brain Circuit Analysis and Modulation in Pathological Conditions
4.1. Neuropsychiatric and Neurological Disorders
Schizophrenia
4.2. Alzheimer Disorder
4.3. Parkinson’s Disease
4.4. Stroke
4.5. Epilepsy
4.6. Autism Spectrum Disorders
4.7. Migraine
4.8. Depression
4.9. Application of Optical Methods to Non-Human Primates
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grienberger, C.; Konnerth, A. Imaging Calcium in Neurons. Neuron 2012, 73, 862–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shemesh, O.A.; Linghu, C.; Piatkevich, K.D.; Goodwin, D.; Celiker, O.T.; Gritton, H.J.; Romano, M.F.; Gao, R.; Yu, C.-C.; Tseng, H.-A.; et al. Precision Calcium Imaging of Dense Neural Populations via a Cell-Body-Targeted Calcium Indicator. Neuron 2020, 107, 470–486.e11. [Google Scholar] [CrossRef]
- Chen, Y.; Jang, H.; Spratt, P.W.E.; Kosar, S.; Taylor, D.E.; Essner, R.A.; Bai, L.; Leib, D.E.; Kuo, T.-W.; Lin, Y.-C.; et al. Soma-Targeted Imaging of Neural Circuits by Ribosome Tethering. Neuron 2020, 107, 454–469.e6. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, X.; Zhang, N.; Gottipati, M.K.; Parpura, V.; Ding, S. Imaging of Mitochondrial Ca2+ Dynamics in Astrocytes Using Cell-Specific Mitochondria-Targeted GCaMP5G/6s: Mitochondrial Ca2+ Uptake and Cytosolic Ca2+ Availability via the Endoplasmic Reticulum Store. Cell Calcium 2014, 56, 457–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakai, J.; Ohkura, M.; Imoto, K. A High Signal-to-Noise Ca2+ Probe Composed of a Single Green Fluorescent Protein. Nat. Biotechnol. 2001, 19, 137–141. [Google Scholar] [CrossRef]
- Dana, H.; Sun, Y.; Mohar, B.; Hulse, B.K.; Kerlin, A.M.; Hasseman, J.P.; Tsegaye, G.; Tsang, A.; Wong, A.; Patel, R.; et al. High-Performance Calcium Sensors for Imaging Activity in Neuronal Populations and Microcompartments. Nat. Methods 2019, 16, 649–657. [Google Scholar] [CrossRef]
- Mohr, M.A.; Bushey, D.; Aggarwal, A.; Marvin, J.S.; Kim, J.J.; Marquez, E.J.; Liang, Y.; Patel, R.; Macklin, J.J.; Lee, C.-Y.; et al. JYCaMP: An Optimized Calcium Indicator for Two-Photon Imaging at Fiber Laser Wavelengths. Nat. Methods 2020, 17, 694–697. [Google Scholar] [CrossRef]
- Akerboom, J.; Chen, T.-W.; Wardill, T.J.; Tian, L.; Marvin, J.S.; Mutlu, S.; Calderón, N.C.; Esposti, F.; Borghuis, B.G.; Sun, X.R.; et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci. 2012, 32, 13819–13840. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive Fluorescent Proteins for Imaging Neuronal Activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.K.; Miri, A.; Leung, L.C.; Berndt, A.; Mourrain, P.; Tank, D.W.; Burdine, R.D. Prolonged, Brain-Wide Expression of Nuclear-Localized GCaMP3 for Functional Circuit Mapping. Front. Neural. Circuits 2014, 8, 138. [Google Scholar] [CrossRef] [Green Version]
- Dal Maschio, M.; Donovan, J.C.; Helmbrecht, T.O.; Baier, H. Linking Neurons to Network Function and Behavior by Two-Photon Holographic Optogenetics and Volumetric Imaging. Neuron 2017, 94, 774–789. [Google Scholar] [CrossRef]
- Ahrens, M.B.; Orger, M.B.; Robson, D.N.; Li, J.M.; Keller, P.J. Whole-Brain Functional Imaging at Cellular Resolution Using Light-Sheet Microscopy. Nat. Methods 2013, 10, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ledochowitsch, P.; Knoblich, U.; Lecoq, J.; Murphy, G.J.; Reid, R.C.; de Vries, S.E.; Koch, C.; Zeng, H.; Buice, M.A.; et al. Relationship between Simultaneously Recorded Spiking Activity and Fluorescence Signal in GCaMP6 Transgenic Mice. Elife 2021, 10, e51675. [Google Scholar] [CrossRef]
- Siegle, J.H.; Ledochowitsch, P.; Jia, X.; Millman, D.J.; Ocker, G.K.; Caldejon, S.; Casal, L.; Cho, A.; Denman, D.J.; Durand, S.; et al. Reconciling Functional Differences in Populations of Neurons Recorded with Two-Photon Imaging and Electrophysiology. Elife 2021, 10, e69068. [Google Scholar] [CrossRef]
- Bennett, C.; Arroyo, S.; Hestrin, S. Subthreshold Mechanisms Underlying State-Dependent Modulation of Visual Responses. Neuron 2013, 80, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Hulse, B.K.; Lubenov, E.V.; Siapas, A.G. Brain State Dependence of Hippocampal Subthreshold Activity in Awake Mice. Cell Rep. 2017, 18, 136–147. [Google Scholar] [CrossRef] [Green Version]
- Pérez Koldenkova, V.; Nagai, T. Genetically Encoded Ca2+ Indicators: Properties and Evaluation. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 1787–1797. [Google Scholar] [CrossRef] [Green Version]
- Knöpfel, T.; Song, C. Optical Voltage Imaging in Neurons: Moving from Technology Development to Practical Tool. Nat. Rev. Neurosci. 2019, 20, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Jares-Erijman, E.A.; Jovin, T.M. FRET Imaging. Nat. Biotechnol. 2003, 21, 1387–1395. [Google Scholar] [CrossRef]
- Mathieu, B.; Villette, V.; Akemann, W.; Evans, S.W.; Chavarha, M.; Chavarha, M.; Bradley, J.; Shi, D.; Shi, D.; Bourdieu, L.; et al. Multiphoton Ultrafast LOcal Volume Excitation (ULOVE) through Acousto-Optic Wavefront Shaping to Record and Control Neuronal Activity. In Proceedings of the Biophotonics Congress: Biomedical Optics Congress 2018 (Microscopy/Translational/Brain/OTS), Fort Lauderdale, FL, USA, 3–6 April 2018; Optical Society of America: Washington, DC, USA, 2018. [Google Scholar]
- Villette, V.; Chavarha, M.; Dimov, I.K.; Bradley, J.; Pradhan, L.; Mathieu, B.; Evans, S.W.; Chamberland, S.; Shi, D.; Yang, R.; et al. Ultrafast Two-Photon Imaging of a High-Gain Voltage Indicator in Awake Behaving Mice. Cell 2019, 179, 1590–1608.e23. [Google Scholar] [CrossRef] [PubMed]
- Hochbaum, D.R.; Zhao, Y.; Farhi, S.L.; Klapoetke, N.; Werley, C.A.; Kapoor, V.; Zou, P.; Kralj, J.M.; Maclaurin, D.; Smedemark-Margulies, N.; et al. All-Optical Electrophysiology in Mammalian Neurons Using Engineered Microbial Rhodopsins. Nat. Methods 2014, 11, 825–833. [Google Scholar] [CrossRef] [Green Version]
- Avery, M.C.; Krichmar, J.L. Neuromodulatory Systems and Their Interactions: A Review of Models, Theories, and Experiments. Front. Neural. Circuits 2017, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Shine, J.M.; Müller, E.J.; Munn, B.; Cabral, J.; Moran, R.J.; Breakspear, M. Computational Models Link Cellular Mechanisms of Neuromodulation to Large-Scale Neural Dynamics. Nat. Neurosci. 2021, 24, 765–776. [Google Scholar] [CrossRef]
- Loued-Khenissi, L.; Preuschoff, K. Apathy and Noradrenaline: Silent Partners to Mild Cognitive Impairment in Parkinson’s Disease? Curr. Opin. Neurol. 2015, 28, 344–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karvat, G.; Kimchi, T. Acetylcholine Elevation Relieves Cognitive Rigidity and Social Deficiency in a Mouse Model of Autism. Neuropsychopharmacology 2014, 39, 831–840. [Google Scholar] [CrossRef]
- Cools, R.; Arnsten, A.F.T. Neuromodulation of Prefrontal Cortex Cognitive Function in Primates: The Powerful Roles of Monoamines and Acetylcholine. Neuropsychopharmacology 2022, 47, 309–328. [Google Scholar] [CrossRef]
- Brisch, R.; Saniotis, A.; Wolf, R.; Bielau, H.; Bernstein, H.-G.; Steiner, J.; Bogerts, B.; Braun, K.; Jankowski, Z.; Kumaratilake, J.; et al. The Role of Dopamine in Schizophrenia from a Neurobiological and Evolutionary Perspective: Old Fashioned, but Still in Vogue. Front. Psychiatry 2014, 5, 47. [Google Scholar] [CrossRef]
- Kesby, J.P.; Eyles, D.W.; McGrath, J.J.; Scott, J.G. Dopamine, Psychosis and Schizophrenia: The Widening Gap between Basic and Clinical Neuroscience. Transl. Psychiatry 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.C.; Li, C.-S.R. Noradrenergic Dysfunction in Alzheimer’s and Parkinson’s Diseases-An Overview of Imaging Studies. Front. Aging Neurosci. 2018, 10, 127. [Google Scholar] [CrossRef]
- Leopold, A.V.; Shcherbakova, D.M.; Verkhusha, V.V. Fluorescent Biosensors for Neurotransmission and Neuromodulation: Engineering and Applications. Front. Cell Neurosci. 2019, 13, 474. [Google Scholar] [CrossRef]
- Ribeiro, L.F.; Amarelle, V.; Ribeiro, L.F.C.; Guazzaroni, M.-E. Converting a Periplasmic Binding Protein into a Synthetic Biosensing Switch through Domain Insertion. Biomed. Res. Int. 2019, 2019, 4798793. [Google Scholar] [CrossRef] [Green Version]
- Haider, R.S.; Godbole, A.; Hoffmann, C. To Sense or Not to Sense—New Insights from GPCR-Based and Arrestin-Based Biosensors. Curr. Opin. Cell Biol. 2019, 57, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Nasu, Y.; Shen, Y.; Kramer, L.; Campbell, R.E. Structure- and Mechanism-Guided Design of Single Fluorescent Protein-Based Biosensors. Nat. Chem. Biol. 2021, 17, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Marvin, J.S.; Borghuis, B.G.; Tian, L.; Cichon, J.; Harnett, M.T.; Akerboom, J.; Gordus, A.; Renninger, S.L.; Chen, T.-W.; Bargmann, C.I.; et al. An Optimized Fluorescent Probe for Visualizing Glutamate Neurotransmission. Nat. Methods 2013, 10, 162–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Truong, T.M.; Ai, H. Illuminating Brain Activities with Fluorescent Protein-Based Biosensors. Chemosensors 2017, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Sabatini, B.L.; Tian, L. Imaging Neurotransmitter and Neuromodulator Dynamics In Vivo with Genetically Encoded Indicators. Neuron 2020, 108, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.P.; Zheng, K.; Cole, N.; Marvin, J.S.; Looger, L.L.; Rusakov, D.A. Multiplex Imaging Relates Quantal Glutamate Release to Presynaptic Ca2+ Homeostasis at Multiple Synapses in Situ. Nat. Commun. 2019, 10, 1414. [Google Scholar] [CrossRef] [Green Version]
- Coates, C.; Kerruth, S.; Helassa, N.; Török, K. Kinetic Mechanisms of Fast Glutamate Sensing by Fluorescent Protein Probes. Biophys. J. 2020, 118, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Helassa, N.; Dürst, C.D.; Coates, C.; Kerruth, S.; Arif, U.; Schulze, C.; Wiegert, J.S.; Geeves, M.; Oertner, T.G.; Török, K. Ultrafast Glutamate Sensors Resolve High-Frequency Release at Schaffer Collateral Synapses. Proc. Natl. Acad. Sci. USA 2018, 115, 5594–5599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marvin, J.S.; Scholl, B.; Wilson, D.E.; Podgorski, K.; Kazemipour, A.; Müller, J.A.; Schoch, S.; Quiroz, F.J.U.; Rebola, N.; Bao, H.; et al. Stability, Affinity, and Chromatic Variants of the Glutamate Sensor IGluSnFR. Nat. Methods 2018, 15, 936–939. [Google Scholar] [CrossRef]
- Wu, J.; Abdelfattah, A.S.; Zhou, H.; Ruangkittisakul, A.; Qian, Y.; Ballanyi, K.; Campbell, R.E. Genetically Encoded Glutamate Indicators with Altered Color and Topology. ACS Chem. Biol. 2018, 13, 1832–1837. [Google Scholar] [CrossRef] [PubMed]
- Marvin, J.S.; Shimoda, Y.; Magloire, V.; Leite, M.; Kawashima, T.; Jensen, T.P.; Kolb, I.; Knott, E.L.; Novak, O.; Podgorski, K.; et al. A Genetically Encoded Fluorescent Sensor for in Vivo Imaging of GABA. Nat. Methods 2019, 16, 763–770. [Google Scholar] [CrossRef] [Green Version]
- Markovic, D.; Holdich, J.; Al-Sabah, S.; Mistry, R.; Krasel, C.; Mahaut-Smith, M.P.; Challiss, R.A.J. FRET-Based Detection of M1 Muscarinic Acetylcholine Receptor Activation by Orthosteric and Allosteric Agonists. PLoS ONE 2012, 7, e29946. [Google Scholar] [CrossRef] [Green Version]
- Jing, M.; Zhang, P.; Wang, G.; Feng, J.; Mesik, L.; Zeng, J.; Jiang, H.; Wang, S.; Looby, J.C.; Guagliardo, N.A.; et al. A Genetically Encoded Fluorescent Acetylcholine Indicator for in Vitro and in Vivo Studies. Nat. Biotechnol 2018, 36, 726–737. [Google Scholar] [CrossRef]
- Sun, F.; Zeng, J.; Jing, M.; Zhou, J.; Feng, J.; Owen, S.F.; Luo, Y.; Li, F.; Wang, H.; Yamaguchi, T.; et al. A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 2018, 174, 481–496.e19. [Google Scholar] [CrossRef]
- Sun, F.; Zhou, J.; Dai, B.; Qian, T.; Zeng, J.; Li, X.; Zhuo, Y.; Zhang, Y.; Wang, Y.; Qian, C.; et al. Next-Generation GRAB Sensors for Monitoring Dopaminergic Activity in Vivo. Nat. Methods 2020, 17, 1156–1166. [Google Scholar] [CrossRef]
- Patriarchi, T.; Cho, J.R.; Merten, K.; Howe, M.W.; Marley, A.; Xiong, W.-H.; Folk, R.W.; Broussard, G.J.; Liang, R.; Jang, M.J.; et al. Ultrafast Neuronal Imaging of Dopamine Dynamics with Designed Genetically Encoded Sensors. Science 2018, 360, eaat4422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patriarchi, T.; Mohebi, A.; Sun, J.; Marley, A.; Liang, R.; Dong, C.; Puhger, K.; Mizuno, G.O.; Davis, C.M.; Wiltgen, B.; et al. An Expanded Palette of Dopamine Sensors for Multiplex Imaging in Vivo. Nat. Methods 2020, 17, 1147–1155. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, C.; Lischinsky, J.E.; Jing, M.; Zhou, J.; Wang, H.; Zhang, Y.; Dong, A.; Wu, Z.; Wu, H.; et al. A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 2019, 102, 745–761.e8. [Google Scholar] [CrossRef] [PubMed]
- Stoeber, M.; Jullié, D.; Lobingier, B.T.; Laeremans, T.; Steyaert, J.; Schiller, P.W.; Manglik, A.; von Zastrow, M. A Genetically Encoded Biosensor Reveals Location Bias of Opioid Drug Action. Neuron 2018, 98, 963–976.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conley, J.M.; Radhakrishnan, S.; Valentino, S.A.; Tantama, M. Imaging Extracellular ATP with a Genetically-Encoded, Ratiometric Fluorescent Sensor. PLoS ONE 2017, 12, e0187481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobas, M.A.; Tao, R.; Nagai, J.; Kronschläger, M.T.; Borden, P.M.; Marvin, J.S.; Looger, L.L.; Khakh, B.S. A Genetically Encoded Single-Wavelength Sensor for Imaging Cytosolic and Cell Surface ATP. Nat. Commun. 2019, 10, 711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; He, K.; Chen, Y.; Li, H.; Pan, S.; Li, B.; Liu, T.; Xi, F.; Deng, F.; Wang, H.; et al. A Sensitive GRAB Sensor for Detecting Extracellular ATP in Vitro and in Vivo. Neuron 2021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.H.; Herde, M.K.; Mitchell, J.A.; Whitfield, J.H.; Wulff, A.B.; Vongsouthi, V.; Sanchez-Romero, I.; Gulakova, P.E.; Minge, D.; Breithausen, B.; et al. Monitoring Hippocampal Glycine with the Computationally Designed Optical Sensor GlyFS. Nat. Chem. Biol. 2018, 14, 861–869. [Google Scholar] [CrossRef]
- Wan, J.; Peng, W.; Li, X.; Qian, T.; Song, K.; Zeng, J.; Deng, F.; Hao, S.; Feng, J.; Zhang, P.; et al. A Genetically Encoded Sensor for Measuring Serotonin Dynamics. Nat. Neurosci. 2021, 24, 746–752. [Google Scholar] [CrossRef]
- Unger, E.K.; Keller, J.P.; Altermatt, M.; Liang, R.; Matsui, A.; Dong, C.; Hon, O.J.; Yao, Z.; Sun, J.; Banala, S.; et al. Directed Evolution of a Selective and Sensitive Serotonin Sensor via Machine Learning. Cell 2020, 183, 1986–2002.e26. [Google Scholar] [CrossRef]
- Borden, P.M.; Zhang, P.; Shivange, A.V.; Marvin, J.S.; Cichon, J.; Dan, C.; Podgorski, K.; Figueiredo, A.; Novak, O.; Tanimoto, M.; et al. A Fast Genetically Encoded Fluorescent Sensor for Faithful in Vivo Acetylcholine Detection in Mice, Fish, Worms and Flies. SSRN 2020. [Google Scholar] [CrossRef]
- Dal Maschio, M.; Ghezzi, D.; Bony, G.; Alabastri, A.; Deidda, G.; Brondi, M.; Sato, S.S.; Zaccaria, R.P.; Di Fabrizio, E.; Ratto, G.M.; et al. High-Performance and Site-Directed in Utero Electroporation by a Triple-Electrode Probe. Nat. Commun. 2012, 3, 960. [Google Scholar] [CrossRef] [PubMed]
- Saito, T. In Vivo Electroporation in the Embryonic Mouse Central Nervous System. Nat. Protoc. 2006, 1, 1552–1558. [Google Scholar] [CrossRef]
- Challis, R.C.; Ravindra Kumar, S.; Chen, X.; Goertsen, D.; Coughlin, G.M.; Hori, A.M.; Chuapoco, M.R.; Otis, T.S.; Miles, T.F.; Gradinaru, V. Adeno-Associated Virus Toolkit to Target Diverse Brain Cells. Annu. Rev. Neurosci. 2022. [Google Scholar] [CrossRef]
- Daigle, T.L.; Madisen, L.; Hage, T.A.; Valley, M.T.; Knoblich, U.; Larsen, R.S.; Takeno, M.M.; Huang, L.; Gu, H.; Larsen, R.; et al. A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain Cell Type Targeting and Functionality. Cell 2018, 174, 465–480.e22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madisen, L.; Garner, A.R.; Shimaoka, D.; Chuong, A.S.; Klapoetke, N.C.; Li, L.; van der Bourg, A.; Niino, Y.; Egolf, L.; Monetti, C.; et al. Transgenic Mice for Intersectional Targeting of Neural Sensors and Effectors with High Specificity and Performance. Neuron 2015, 85, 942–958. [Google Scholar] [CrossRef] [Green Version]
- Steinmetz, N.A.; Buetfering, C.; Lecoq, J.; Lee, C.R.; Peters, A.J.; Jacobs, E.A.K.; Coen, P.; Ollerenshaw, D.R.; Valley, M.T.; de Vries, S.E.J.; et al. Aberrant Cortical Activity in Multiple GCaMP6-Expressing Transgenic Mouse Lines. eNeuro 2017, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Das, A.T.; Tenenbaum, L.; Berkhout, B. Tet-On Systems for Doxycycline-Inducible Gene Expression. Curr. Gene Ther. 2016, 16, 156–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zingg, B.; Peng, B.; Huang, J.; Tao, H.W.; Zhang, L.I. Synaptic Specificity and Application of Anterograde Transsynaptic AAV for Probing Neural Circuitry. J. Neurosci. 2020, 40, 3250–3267. [Google Scholar] [CrossRef]
- De La Crompe, B.; Coulon, P.; Diester, I. Functional Interrogation of Neural Circuits with Virally Transmitted Optogenetic Tools. J. Neurosc. Methods 2020, 345, 108905. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.Y.; Jang, M.J.; Yoo, B.B.; Greenbaum, A.; Ravi, N.; Wu, W.-L.; Sánchez-Guardado, L.; Lois, C.; Mazmanian, S.K.; Deverman, B.E.; et al. Engineered AAVs for Efficient Noninvasive Gene Delivery to the Central and Peripheral Nervous Systems. Nat. Neurosci. 2017, 20, 1172–1179. [Google Scholar] [CrossRef]
- Han, S.; Yang, W.; Yuste, R. Two-Color Volumetric Imaging of Neuronal Activity of Cortical Columns. Cell Rep. 2019, 27, 2229–2240.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curreli, S.; Bonato, J.; Romanzi, S.; Panzeri, S.; Fellin, T. Complementary Encoding of Spatial Information in Hippocampal Astrocytes. PLoS Biol. 2022, 20, e3001530. [Google Scholar] [CrossRef]
- Sakai, C.; Ijaz, S.; Hoffman, E.J. Zebrafish Models of Neurodevelopmental Disorders: Past, Present, and Future. Front. Mol. Neurosci. 2018, 11, 294. [Google Scholar] [CrossRef] [Green Version]
- Feierstein, C.E.; Portugues, R.; Orger, M.B. Seeing the Whole Picture: A Comprehensive Imaging Approach to Functional Mapping of Circuits in Behaving Zebrafish. Neuroscience 2015, 296, 26–38. [Google Scholar] [CrossRef]
- Zipfel, W.R.; Williams, R.M.; Webb, W.W. Nonlinear Magic: Multiphoton Microscopy in the Biosciences. Nat. Biotechnol. 2003, 21, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Mao, H.; Wang, A.; Chen, L. Two-Photon Fluorescence Imaging. In Optical Imaging in Human Disease and Biological Research; Wei, X., Gu, B., Eds.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2021; pp. 45–61. ISBN 9789811576270. [Google Scholar]
- Pallen, S.; Shetty, Y.; Das, S.; Vaz, J.M.; Mazumder, N. Advances in Nonlinear Optical Microscopy Techniques for in Vivo and in Vitro Neuroimaging. Biophys. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- Denk, W.; Svoboda, K. Photon Upmanship: Why Multiphoton Imaging Is More than a Gimmick. Neuron 1997, 18, 351–357. [Google Scholar] [CrossRef] [Green Version]
- Lecoq, J.; Orlova, N.; Grewe, B.F. Wide. Fast. Deep. Recent Advances in Multi-Photon Microscopy of in Vivo Neuronal Activity. J. Neurosci. 2019. [Google Scholar] [CrossRef] [Green Version]
- Schultz, S.R.; Copeland, C.S.; Foust, A.J.; Quicke, P.; Schuck, R. Advances in Two Photon Scanning and Scanless Microscopy Technologies for Functional Neural Circuit Imaging. Proc. IEEE Inst. Electr. Electron. Eng. 2017, 105, 139–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrödel, T.; Prevedel, R.; Aumayr, K.; Zimmer, M.; Vaziri, A. Brain-Wide 3D Imaging of Neuronal Activity in Caenorhabditis Elegans with Sculpted Light. Nat. Methods 2013, 10, 1013–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- May, M.A.; Bawart, M.; Langeslag, M.; Bernet, S.; Kress, M.; Ritsch-Marte, M.; Jesacher, A. High-NA Two-Photon Single Cell Imaging with Remote Focusing Using a Diffractive Tunable Lens. Biomed. Opt. Express 2020, 11, 7183–7191. [Google Scholar] [CrossRef]
- Anselmi, F.; Ventalon, C.; Bègue, A.; Ogden, D.; Emiliani, V. Three-Dimensional Imaging and Photostimulation by Remote-Focusing and Holographic Light Patterning. Proc. Natl. Acad. Sci. USA 2011, 108, 19504–19509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grewe, B.F.; Voigt, F.F.; van ’t Hoff, M.; Helmchen, F. Fast Two-Layer Two-Photon Imaging of Neuronal Cell Populations Using an Electrically Tunable Lens. Biomed. Opt. Express 2011, 2, 2035–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruzzone, M.; Chiarello, E.; Albanesi, M.; Miletto Petrazzini, M.E.; Megighian, A.; Lodovichi, C.; dal Maschio, M. Whole Brain Functional Recordings at Cellular Resolution in Zebrafish Larvae with 3D Scanning Multiphoton Microscopy. Sci Rep. 2021, 11, 11048. [Google Scholar] [CrossRef]
- Duemani Reddy, G.; Kelleher, K.; Fink, R.; Saggau, P. Three-Dimensional Random Access Multiphoton Microscopy for Functional Imaging of Neuronal Activity. Nat. Neurosci. 2008, 11, 713–720. [Google Scholar] [CrossRef]
- Grewe, B.F.; Langer, D.; Kasper, H.; Kampa, B.M.; Helmchen, F. High-Speed in Vivo Calcium Imaging Reveals Neuronal Network Activity with near-Millisecond Precision. Nat. Methods 2010, 7, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Nadella, K.M.N.S.; Roš, H.; Baragli, C.; Griffiths, V.A.; Konstantinou, G.; Koimtzis, T.; Evans, G.J.; Kirkby, P.A.; Silver, R.A. Random-Access Scanning Microscopy for 3D Imaging in Awake Behaving Animals. Nat. Methods 2016, 13, 1001–1004. [Google Scholar] [CrossRef]
- Liu, R.; Liu, R.; Ball, N.; Brockill, J.; Kuan, L.; Millman, D.; White, C.; Leon, A.; Williams, D.; Nishiwaki, S.; et al. Aberration-Free Multi-Plane Imaging of Neural Activity from the Mammalian Brain Using a Fast-Switching Liquid Crystal Spatial Light Modulator. Biomed. Opt. Express 2019, 10, 5059–5080. [Google Scholar] [CrossRef] [PubMed]
- Geng, Q.; Gu, C.; Cheng, J.; Chen, S. Digital Micromirror Device-Based Two-Photon Microscopy for Three-Dimensional and Random-Access Imaging. Optica 2017, 4, 674–677. [Google Scholar] [CrossRef]
- Hopt, A.; Neher, E. Highly Nonlinear Photodamage in Two-Photon Fluorescence Microscopy. Biophys. J. 2001, 80, 2029–2036. [Google Scholar] [CrossRef] [Green Version]
- Tauer, U. Advantages and Risks of Multiphoton Microscopy in Physiology. Exp. Physiol. 2002, 87, 709–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picot, A.; Dominguez, S.; Liu, C.; Chen, I.-W.; Tanese, D.; Ronzitti, E.; Berto, P.; Papagiakoumou, E.; Oron, D.; Tessier, G.; et al. Temperature Rise under Two-Photon Optogenetic Brain Stimulation. Cell Rep. 2018, 24, 1243–1253.e5. [Google Scholar] [CrossRef] [Green Version]
- Theer, P.; Denk, W. On the Fundamental Imaging-Depth Limit in Two-Photon Microscopy. J. Opt. Soc. Am. A 2006, 23, 3139–3149. [Google Scholar] [CrossRef]
- Wang, T.; Ouzounov, D.G.; Wu, C.; Horton, N.G.; Zhang, B.; Wu, C.-H.; Zhang, Y.; Schnitzer, M.J.; Xu, C. Three-Photon Imaging of Mouse Brain Structure and Function through the Intact Skull. Nat. Methods 2018, 15, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Hontani, Y.; Xia, F.; Xu, C. Multicolor Three-Photon Fluorescence Imaging with Single-Wavelength Excitation Deep in Mouse Brain. Sci. Adv. 2021, 7, eabf3531. [Google Scholar] [CrossRef]
- Meng, G.; Liang, Y.; Sarsfield, S.; Jiang, W.; Lu, R.; Dudman, J.T.; Aponte, Y.; Ji, N. High-Throughput Synapse-Resolving Two-Photon Fluorescence Microendoscopy for Deep-Brain Volumetric Imaging in Vivo. eLife 2019, 8, e40805. [Google Scholar] [CrossRef]
- Antonini, A.; Sattin, A.; Moroni, M.; Bovetti, S.; Moretti, C.; Succol, F.; Forli, A.; Vecchia, D.; Rajamanickam, V.P.; Bertoncini, A.; et al. Extended Field-of-View Ultrathin Microendoscopes for High-Resolution Two-Photon Imaging with Minimal Invasiveness. eLife 2020, 9, e58882. [Google Scholar] [CrossRef]
- Tang, Q.; Tsytsarev, V.; Liang, C.-P.; Akkentli, F.; Erzurumlu, R.S.; Chen, Y. In Vivo Voltage-Sensitive Dye Imaging of Subcortical Brain Function. Sci. Rep. 2015, 5, 17325. [Google Scholar] [CrossRef] [Green Version]
- Pernici, C.D.; Kemp, B.S.; Murray, T.A. Time Course Images of Cellular Injury and Recovery in Murine Brain with High-Resolution GRIN Lens System. Sci. Rep. 2019, 9, 7946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Groot, A.; van den Boom, B.J.; van Genderen, R.M.; Coppens, J.; van Veldhuijzen, J.; Bos, J.; Hoedemaker, H.; Negrello, M.; Willuhn, I.; De Zeeuw, C.I.; et al. NINscope, a Versatile Miniscope for Multi-Region Circuit Investigations. eLife 2020, 9, e49987. [Google Scholar] [CrossRef]
- Barbera, G.; Liang, B.; Zhang, L.; Li, Y.; Lin, D.-T. A Wireless MiniScope for Deep Brain Imaging in Freely Moving Mice. J. Neurosci. Methods 2019, 323, 56–60. [Google Scholar] [CrossRef]
- Skocek, O.; Nöbauer, T.; Weilguny, L.; Martínez Traub, F.; Xia, C.N.; Molodtsov, M.I.; Grama, A.; Yamagata, M.; Aharoni, D.; Cox, D.D.; et al. High-Speed Volumetric Imaging of Neuronal Activity in Freely Moving Rodents. Nat. Methods 2018, 15, 429–432. [Google Scholar] [CrossRef]
- Patel, A.A.; McAlinden, N.; Mathieson, K.; Sakata, S. Simultaneous Electrophysiology and Fiber Photometry in Freely Behaving Mice. Front. Neurosci. 2020, 14, 148. [Google Scholar] [CrossRef] [Green Version]
- Pisano, F.; Pisanello, M.; Lee, S.J.; Lee, J.; Maglie, E.; Balena, A.; Sileo, L.; Spagnolo, B.; Bianco, M.; Hyun, M.; et al. Depth-Resolved Fiber Photometry with a Single Tapered Optical Fiber Implant. Nat. Methods 2019, 16, 1185–1192. [Google Scholar] [CrossRef]
- Sofroniew, N.J.; Flickinger, D.; King, J.; Svoboda, K. A Large Field of View Two-Photon Mesoscope with Subcellular Resolution for in Vivo Imaging. eLife 2016, 5, e14472. [Google Scholar] [CrossRef] [PubMed]
- Janiak, F.K.; Bartel, P.; Bale, M.R.; Yoshimatsu, T.; Komulainen, E.; Zhou, M.; Staras, K.; Prieto-Godino, L.L.; Euler, T.; Maravall, M.; et al. Non-Telecentric Two-Photon Microscopy for 3D Random Access Mesoscale Imaging. Nat. Commun. 2022, 13, 544. [Google Scholar] [CrossRef]
- Yu, C.-H.; Stirman, J.N.; Yu, Y.; Hira, R.; Smith, S.L. Diesel2p Mesoscope with Dual Independent Scan Engines for Flexible Capture of Dynamics in Distributed Neural Circuitry. Nat. Commun. 2020, 12, 1–8. [Google Scholar] [CrossRef]
- Resta, F.; Mascaro, A.L.A.; Montagni, E.; de Vito, G.; Scaglione, A.; Pavone, F.S. Mesoscale Imaging of Neuronal Activity Coupled with Light-Evoked Motor Mapping Reveal Movement-Specific Spatiotemporal Patterns of Cortical Activation in Awake Mice. In Proceedings of the Neural Imaging and Sensing 2020, San Francisco, CA, USA, 3–5 February 2020; SPIE: Bellingham, WA, USA, 2020; Volume 11226, pp. 31–34. [Google Scholar]
- Bermudez-Contreras, E.; Chekhov, S.; Tarnowsky, J.; McNaughton, B.L.; Mohajerani, M.H. High-Performance, Inexpensive Setup for Simultaneous Multisite Recording of Electrophysiological Signals and Mesoscale Voltage Imaging in the Mouse Cortex. NPh 2018, 5, 025005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, C.; Komiyama, T. Characterizing Cortex-Wide Dynamics with Wide-Field Calcium Imaging. J. Neurosci. 2021, 41, 4160–4168. [Google Scholar] [CrossRef]
- Xie, Y.; Chan, A.W.; McGirr, A.; Xue, S.; Xiao, D.; Zeng, H.; Murphy, T.H. Resolution of High-Frequency Mesoscale Intracortical Maps Using the Genetically Encoded Glutamate Sensor IGluSnFR. J. Neurosci. 2016, 36, 1261–1272. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Li, Q.; Jiang, J.; Cable, A.; Chen, Y. Three-Dimensional Coregistered Optical Coherence Tomography and Line-Scanning Fluorescence Laminar Optical Tomography. Opt. Lett. 2009, 34, 1615–1617. [Google Scholar] [CrossRef]
- Hillman, E.M.C.; Bernus, O.; Pease, E.; Bouchard, M.B.; Pertsov, A. Depth-Resolved Optical Imaging of Transmural Electrical Propagation in Perfused Heart. Opt. Express 2007, 15, 17827–17841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Q.; Tsytsarev, V.; Frank, A.; Wu, Y.; Chen, C.; Erzurumlu, R.S.; Chen, Y. In Vivo Mesoscopic Voltage-Sensitive Dye Imaging of Brain Activation. Sci. Rep. 2016, 6, 25269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Packer, A.M.; Russell, L.E.; Dalgleish, H.W.P.; Häusser, M. Simultaneous All-Optical Manipulation and Recording of Neural Circuit Activity with Cellular Resolution in Vivo. Nat. Methods 2015, 12, 140–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emiliani, V.; Cohen, A.E.; Deisseroth, K.; Häusser, M. All-Optical Interrogation of Neural Circuits. J. Neurosci. 2015, 35, 13917–13926. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Z.-F.; Shang, C.-F.; Wang, Y.-F.; Yang, Z.; Yang, C.; Li, F.-N.; Xie, J.-Z.; Pan, J.-W.; Fu, L.; Du, J.-L. All-Optical Imaging and Manipulation of Whole-Brain Neuronal Activities in Behaving Larval Zebrafish. Biomed. Opt. Express 2018, 9, 6154–6169. [Google Scholar] [CrossRef] [PubMed]
- Huckvale, R.; Mortensen, M.; Pryde, D.; Smart, T.G.; Baker, J.R. Azogabazine; a Photochromic Antagonist of the GABAA Receptor. Org. Biomol. Chem. 2016, 14, 6676–6678. [Google Scholar] [CrossRef] [Green Version]
- Pittolo, S.; Gómez-Santacana, X.; Eckelt, K.; Rovira, X.; Dalton, J.; Goudet, C.; Pin, J.-P.; Llobet, A.; Giraldo, J.; Llebaria, A.; et al. An Allosteric Modulator to Control Endogenous G Protein-Coupled Receptors with Light. Nat. Chem. Biol. 2014, 10, 813–815. [Google Scholar] [CrossRef]
- Donthamsetti, P.C.; Winter, N.; Schönberger, M.; Levitz, J.; Stanley, C.; Javitch, J.A.; Isacoff, E.Y.; Trauner, D. Optical Control of Dopamine Receptors Using a Photoswitchable Tethered Inverse Agonist. J. Am. Chem. Soc. 2017, 139, 18522–18535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donthamsetti, P.; Winter, N.; Hoagland, A.; Stanley, C.; Visel, M.; Lammel, S.; Trauner, D.; Isacoff, E. Cell Specific Photoswitchable Agonist for Reversible Control of Endogenous Dopamine Receptors. Nat. Commun. 2021, 12, 4775. [Google Scholar] [CrossRef]
- Maleeva, G.; Nin-Hill, A.; Rustler, K.; Petukhova, E.; Ponomareva, D.; Mukhametova, E.; Gomila, A.M.; Wutz, D.; Alfonso-Prieto, M.; König, B.; et al. Subunit-Specific Photocontrol of Glycine Receptors by Azobenzene-Nitrazepam Photoswitcher. eNeuro 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Maleeva, G.; Wutz, D.; Rustler, K.; Nin-Hill, A.; Rovira, C.; Petukhova, E.; Bautista-Barrufet, A.; Gomila-Juaneda, A.; Scholze, P.; Peiretti, F.; et al. A Photoswitchable GABA Receptor Channel Blocker. Br. J. Pharmacol. 2019, 176, 2661–2677. [Google Scholar] [CrossRef] [Green Version]
- Lemoine, D.; Mondoloni, S.; Tange, J.; Lambolez, B.; Faure, P.; Taly, A.; Tricoire, L.; Mourot, A. Probing the Ionotropic Activity of Glutamate GluD2 Receptor in HEK Cells with Genetically-Engineered Photopharmacology. Elife 2020, 9, e59026. [Google Scholar] [CrossRef] [PubMed]
- Berlin, S.; Szobota, S.; Reiner, A.; Carroll, E.C.; Kienzler, M.A.; Guyon, A.; Xiao, T.; Trauner, D.; Isacoff, E.Y. A Family of Photoswitchable NMDA Receptors. eLife 2016, 5, e12040. [Google Scholar] [CrossRef] [PubMed]
- Levitz, J.; Broichhagen, J.; Leippe, P.; Konrad, D.; Trauner, D.; Isacoff, E.Y. Dual Optical Control and Mechanistic Insights into Photoswitchable Group II and III Metabotropic Glutamate Receptors. Proc. Natl. Acad. Sci. USA 2017, 114, E3546–E3554. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.Y.; Lin, M.Z.; Steinbach, P.; Tsien, R.Y. Characterization of Engineered Channelrhodopsin Variants with Improved Properties and Kinetics. Biophys. J. 2009, 96, 1803–1814. [Google Scholar] [CrossRef] [Green Version]
- Klapoetke, N.C.; Murata, Y.; Kim, S.S.; Pulver, S.R.; Birdsey-Benson, A.; Cho, Y.K.; Morimoto, T.K.; Chuong, A.S.; Carpenter, E.J.; Tian, Z. Independent Optical Excitation of Distinct Neural Populations. Nat. Methods 2014, 11, 338–346. [Google Scholar] [CrossRef] [Green Version]
- Mardinly, A.R.; Oldenburg, I.A.; Pégard, N.C.; Sridharan, S.; Lyall, E.H.; Chesnov, K.; Brohawn, S.G.; Waller, L.; Adesnik, H. Precise Multimodal Optical Control of Neural Ensemble Activity. Nat. Neurosci. 2018, 21, 881–893. [Google Scholar] [CrossRef]
- Sridharan, S.; Gajowa, M.A.; Ogando, M.B.; Jagadisan, U.K.; Abdeladim, L.; Sadahiro, M.; Bounds, H.A.; Hendricks, W.D.; Turney, T.S.; Tayler, I.; et al. High-Performance Microbial Opsins for Spatially and Temporally Precise Perturbations of Large Neuronal Networks. Neuron 2022. [Google Scholar] [CrossRef]
- Marshel, J.H.; Kim, Y.S.; Machado, T.A.; Quirin, S.; Benson, B.; Kadmon, J.; Raja, C.; Chibukhchyan, A.; Ramakrishnan, C.; Inoue, M.; et al. Cortical Layer–Specific Critical Dynamics Triggering Perception. Science 2019, 365, eaaw5202. [Google Scholar] [CrossRef]
- Mager, T.; Lopez de la Morena, D.; Senn, V.; Schlotte, J.; D´Errico, A.; Feldbauer, K.; Wrobel, C.; Jung, S.; Bodensiek, K.; Rankovic, V.; et al. High Frequency Neural Spiking and Auditory Signaling by Ultrafast Red-Shifted Optogenetics. Nat. Commun. 2018, 9, 1750. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Janz, R.; Liu, X.; Spudich, J.L. Natural Light-Gated Anion Channels: A Family of Microbial Rhodopsins for Advanced Optogenetics. Science 2015, 349, 647–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, B.Y.; Han, X.; Dobry, A.S.; Qian, X.; Chuong, A.S.; Li, M.; Henninger, M.A.; Belfort, G.M.; Lin, Y.; Monahan, P.E. High-Performance Genetically Targetable Optical Neural Silencing by Light-Driven Proton Pumps. Nature 2010, 463, 98–102. [Google Scholar] [CrossRef] [Green Version]
- Gradinaru, V.; Zhang, F.; Ramakrishnan, C.; Mattis, J.; Prakash, R.; Diester, I.; Goshen, I.; Thompson, K.R.; Deisseroth, K. Molecular and Cellular Approaches for Diversifying and Extending Optogenetics. Cell 2010, 141, 154–165. [Google Scholar] [CrossRef] [Green Version]
- Govorunova, E.G.; Sineshchekov, O.A.; Li, H.; Wang, Y.; Brown, L.S.; Spudich, J.L. RubyACRs, Nonalgal Anion Channelrhodopsins with Highly Red-Shifted Absorption. Proc. Natl. Acad. Sci. USA 2020, 117, 22833. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a Directly Light-Gated Cation-Selective Membrane Channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.Y.; Knutsen, P.M.; Muller, A.; Kleinfeld, D.; Tsien, R.Y. ReaChR: A Red-Shifted Variant of Channelrhodopsin Enables Deep Transcranial Optogenetic Excitation. Nat. Neurosci. 2013, 16, 1499–1508. [Google Scholar] [CrossRef] [Green Version]
- Mattis, J.; Tye, K.M.; Ferenczi, E.A.; Ramakrishnan, C.; O’shea, D.J.; Prakash, R.; Gunaydin, L.A.; Hyun, M.; Fenno, L.E.; Gradinaru, V. Principles for Applying Optogenetic Tools Derived from Direct Comparative Analysis of Microbial Opsins. Nat. Methods 2012, 9, 159–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahn, M.; Prigge, M.; Ron, S.; Levy, R.; Yizhar, O. Biophysical Constraints of Optogenetic Inhibition at Presynaptic Terminals. Nat. Neurosci. 2016, 19, 554–556. [Google Scholar] [CrossRef] [PubMed]
- Ferenczi, E.A.; Tan, X.; Huang, C.L.-H. Principles of Optogenetic Methods and Their Application to Cardiac Experimental Systems. Front. Physiol. 2019, 10, 1096. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, L.-P.; Brauner, M.; Liewald, J.F.; Kay, K.; Watzke, N.; Wood, P.G.; Bamberg, E.; Nagel, G.; Gottschalk, A. Multimodal Fast Optical Interrogation of Neural Circuitry. Nature 2007, 446, 633–639. [Google Scholar] [CrossRef]
- Berndt, A.; Lee, S.Y.; Ramakrishnan, C.; Deisseroth, K. Structure-Guided Transformation of Channelrhodopsin into a Light-Activated Chloride Channel. Science 2014, 344, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Wietek, J.; Beltramo, R.; Scanziani, M.; Hegemann, P.; Oertner, T.G.; Wiegert, J.S. An Improved Chloride-Conducting Channelrhodopsin for Light-Induced Inhibition of Neuronal Activity in Vivo. Sci. Rep. 2015, 5, 14807. [Google Scholar] [CrossRef] [Green Version]
- Wietek, J.; Broser, M.; Krause, B.S.; Hegemann, P. Identification of a Natural Green Light Absorbing Chloride Conducting Channelrhodopsin from Proteomonas Sulcata*. J. Biol. Chem. 2016, 291, 4121–4127. [Google Scholar] [CrossRef] [Green Version]
- Wietek, J.; Rodriguez-Rozada, S.; Tutas, J.; Tenedini, F.; Grimm, C.; Oertner, T.G.; Soba, P.; Hegemann, P.; Wiegert, J.S. Anion-Conducting Channelrhodopsins with Tuned Spectra and Modified Kinetics Engineered for Optogenetic Manipulation of Behavior. Sci. Rep. 2017, 7, 14957. [Google Scholar] [CrossRef] [Green Version]
- Berndt, A.; Lee, S.Y.; Wietek, J.; Ramakrishnan, C.; Steinberg, E.E.; Rashid, A.J.; Kim, H.; Park, S.; Santoro, A.; Frankland, P.W.; et al. Structural Foundations of Optogenetics: Determinants of Channelrhodopsin Ion Selectivity. Proc. Natl. Acad. Sci. USA 2016, 113, 822–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oppermann, J.; Fischer, P.; Silapetere, A.; Liepe, B.; Rodriguez-Rozada, S.; Flores-Uribe, J.; Peter, E.; Keidel, A.; Vierock, J.; Kaufmann, J.; et al. MerMAIDs: A Family of Metagenomically Discovered Marine Anion-Conducting and Intensely Desensitizing Channelrhodopsins. Nat. Commun. 2019, 10, 3315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimondo, J.V.; Kay, L.; Ellender, T.J.; Akerman, C.J. Optogenetic Silencing Strategies Differ in Their Effects on Inhibitory Synaptic Transmission. Nat. Neurosci. 2012, 15, 1102–1104. [Google Scholar] [CrossRef] [Green Version]
- Messier, J.E.; Chen, H.; Cai, Z.-L.; Xue, M. Targeting Light-Gated Chloride Channels to Neuronal Somatodendritic Domain Reduces Their Excitatory Effect in the Axon. Elife 2018, 7, e38506. [Google Scholar] [CrossRef]
- Lewis, D.A. Inhibitory Neurons in Human Cortical Circuits: Substrate for Cognitive Dysfunction in Schizophrenia. Curr. Opin. Neurobiol. 2014, 26, 22–26. [Google Scholar] [CrossRef] [Green Version]
- Uhlhaas, P.J.; Singer, W. Abnormal Neural Oscillations and Synchrony in Schizophrenia. Nat. Rev. Neurosci. 2010, 11, 100–113. [Google Scholar] [CrossRef]
- Hirano, Y.; Uhlhaas, P.J. Current Findings and Perspectives on Aberrant Neural Oscillations in Schizophrenia. Psychiatry Clin. Neurosci. 2021, 75, 358–368. [Google Scholar] [CrossRef]
- Rolls, E.T.; Loh, M.; Deco, G.; Winterer, G. Computational Models of Schizophrenia and Dopamine Modulation in the Prefrontal Cortex. Nat. Rev. Neurosci. 2008, 9, 696–709. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Reid, L.; Yuste, R. Playing the Piano with the Cortex: Role of Neuronal Ensembles and Pattern Completion in Perception and Behavior. Curr. Opin. Neurobiol. 2020, 64, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Reid, L.; Yang, W.; Bando, Y.; Peterka, D.S.; Yuste, R. Imprinting and Recalling Cortical Ensembles. Science 2016, 353, 691–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamm, J.P.; Peterka, D.S.; Gogos, J.A.; Yuste, R. Altered Cortical Ensembles in Mouse Models of Schizophrenia. Neuron 2017, 94, 153–167.e8. [Google Scholar] [CrossRef] [Green Version]
- Hamm, J.P.; Shymkiv, Y.; Mukai, J.; Gogos, J.A.; Yuste, R. Aberrant Cortical Ensembles and Schizophrenia-like Sensory Phenotypes in Setd1a+/- Mice. Biol. Psychiatry 2020, 88, 215–223. [Google Scholar] [CrossRef]
- Hopfield, J.J. Neural Networks and Physical Systems with Emergent Collective Computational Abilities. Proc. Natl. Acad. Sci. USA 1982, 79, 2554–2558. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zheng, L.; Fang, T.; Li, R.; Ma, X.; Sun, Y.; Wang, L.; Tian, H.; Jiang, D.; Zhuo, C. Exploration of the Cortical Pathophysiology Underlying Visual Disturbances in Schizophrenia Comorbid with Depressive Disorder—An Evidence from Mouse Model. Brain Behav. 2021, 11, e02113. [Google Scholar] [CrossRef]
- Michaiel, A.M.; Parker, P.R.L.; Niell, C.M. A Hallucinogenic Serotonin-2A Receptor Agonist Reduces Visual Response Gain and Alters Temporal Dynamics in Mouse V1. Cell Rep. 2019, 26, 3475–3483.e4. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Kong, D.; Xue, R.; Chen, M.; Li, G.; Xu, Y.; Liu, S.; Tian, H.; Zhuo, C. Metformin Enhances Antidepressant/Antipsychotic Combination Therapy of Schizophrenia with Comorbid Depression in a Murine Model. Front. Neurosci. 2020, 14. [Google Scholar] [CrossRef]
- McNally, J.M.; Aguilar, D.D.; Katsuki, F.; Radzik, L.K.; Schiffino, F.L.; Uygun, D.S.; McKenna, J.T.; Strecker, R.E.; Deisseroth, K.; Spencer, K.M.; et al. Optogenetic Manipulation of an Ascending Arousal System Tunes Cortical Broadband Gamma Power and Reveals Functional Deficits Relevant to Schizophrenia. Mol. Psychiatry 2021, 26, 3461–3475. [Google Scholar] [CrossRef]
- Wolff, A.R.; Bygrave, A.M.; Sanderson, D.J.; Boyden, E.S.; Bannerman, D.M.; Kullmann, D.M.; Kätzel, D. Optogenetic Induction of the Schizophrenia-Related Endophenotype of Ventral Hippocampal Hyperactivity Causes Rodent Correlates of Positive and Cognitive Symptoms. Sci. Rep. 2018, 8, 12871. [Google Scholar] [CrossRef] [PubMed]
- Duan, A.R.; Varela, C.; Zhang, Y.; Shen, Y.; Xiong, L.; Wilson, M.A.; Lisman, J. Delta Frequency Optogenetic Stimulation of the Thalamic Nucleus Reuniens Is Sufficient to Produce Working Memory Deficits: Relevance to Schizophrenia. Biol. Psychiatry 2015, 77, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Palop, J.J.; Mucke, L. Network Abnormalities and Interneuron Dysfunction in Alzheimer Disease. Nat. Rev. Neurosci. 2016, 17, 777–792. [Google Scholar] [CrossRef] [PubMed]
- Verret, L.; Mann, E.O.; Hang, G.B.; Barth, A.M.I.; Cobos, I.; Ho, K.; Devidze, N.; Masliah, E.; Kreitzer, A.C.; Mody, I.; et al. Inhibitory Interneuron Deficit Links Altered Network Activity and Cognitive Dysfunction in Alzheimer Model. Cell 2012, 149, 708–721. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, A.K.; Jones, E.A.; Lin, Y.-H.; Karlsson, M.P.; Kay, K.; Yoon, S.Y.; Tong, L.M.; Nova, P.; Carr, J.S.; Frank, L.M.; et al. Apolipoprotein E4 Causes Age-Dependent Disruption of Slow Gamma Oscillations during Hippocampal Sharp-Wave Ripples. Neuron 2016, 90, 740–751. [Google Scholar] [CrossRef] [Green Version]
- Iaccarino, H.F.; Singer, A.C.; Martorell, A.J.; Rudenko, A.; Gao, F.; Gillingham, T.Z.; Mathys, H.; Seo, J.; Kritskiy, O.; Abdurrob, F.; et al. Gamma Frequency Entrainment Attenuates Amyloid Load and Modifies Microglia. Nature 2016, 540, 230–235. [Google Scholar] [CrossRef] [Green Version]
- Bero, A.W.; Yan, P.; Roh, J.H.; Cirrito, J.R.; Stewart, F.R.; Raichle, M.E.; Lee, J.-M.; Holtzman, D.M. Neuronal Activity Regulates the Regional Vulnerability to Amyloid-β Deposition. Nat. Neurosci. 2011, 14, 750–756. [Google Scholar] [CrossRef] [Green Version]
- Adaikkan, C.; Middleton, S.J.; Marco, A.; Pao, P.-C.; Mathys, H.; Kim, D.N.-W.; Gao, F.; Young, J.Z.; Suk, H.-J.; Boyden, E.S.; et al. Gamma Entrainment Binds Higher-Order Brain Regions and Offers Neuroprotection. Neuron 2019, 102, 929–943.e8. [Google Scholar] [CrossRef]
- Hawkes, C.H.; Del Tredici, K.; Braak, H. A Timeline for Parkinson’s Disease. Parkinsonism Relat. Disord. 2010, 16, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Doty, R.L. Olfactory Dysfunction in Parkinson Disease. Nat. Rev. Neurol. 2012, 8, 329–339. [Google Scholar] [CrossRef]
- Rey, N.L.; Steiner, J.A.; Maroof, N.; Luk, K.C.; Madaj, Z.; Trojanowski, J.Q.; Lee, V.M.-Y.; Brundin, P. Widespread Transneuronal Propagation of α-Synucleinopathy Triggered in Olfactory Bulb Mimics Prodromal Parkinson’s Disease. J. Exp. Med. 2016, 213, 1759–1778. [Google Scholar] [CrossRef] [PubMed]
- Burré, J.; Sharma, M.; Tsetsenis, T.; Buchman, V.; Etherton, M.R.; Südhof, T.C. Alpha-Synuclein Promotes SNARE-Complex Assembly in Vivo and in Vitro. Science 2010, 329, 1663–1667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouke, K.E.; Wegman, M.E.; Weber, S.A.; Brady, E.B.; Román-Vendrell, C.; Morgan, J.R. Synuclein Regulates Synaptic Vesicle Clustering and Docking at a Vertebrate Synapse. F. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Soukup, S.-F.; Vanhauwaert, R.; Verstreken, P. Parkinson’s Disease: Convergence on Synaptic Homeostasis. EMBO J. 2018, 37, e98960. [Google Scholar] [CrossRef]
- Maset, A.; Albanesi, M.; di Soccio, A.; Canova, M.; dal Maschio, M.; Lodovichi, C. Aberrant Patterns of Sensory-Evoked Activity in the Olfactory Bulb of LRRK2 Knockout Mice. Cells 2021, 10, 3212. [Google Scholar] [CrossRef]
- Blumenstock, S.; Sun, F.; Klaus, C.; Marinković, P.; Sgobio, C.; Paeger, L.; Liebscher, S.; Herms, J. Cortical Circuit Dysfunction in a Mouse Model of Alpha-Synucleinopathy in Vivo. Brain Commun. 2021, 3, fcab273. [Google Scholar] [CrossRef] [PubMed]
- Benabid, A.L.; Pollak, P.; Gross, C.; Hoffmann, D.; Benazzouz, A.; Gao, D.M.; Laurent, A.; Gentil, M.; Perret, J. Acute and Long-Term Effects of Subthalamic Nucleus Stimulation in Parkinson’s Disease. SFN 1994, 62, 76–84. [Google Scholar] [CrossRef]
- Yates, D. Targeting Circuits with DBS. Nat. Rev. Neurosci. 2021, 22, 721. [Google Scholar] [CrossRef]
- Kuncel, A.M.; Cooper, S.E.; Grill, W.M. A Method to Estimate the Spatial Extent of Activation in Thalamic Deep Brain Stimulation. Electroencephal. Clin. Neurophysiol. Electromyogr. Motor Control. 2008, 119, 2148–2158. [Google Scholar] [CrossRef] [Green Version]
- Gradinaru, V.; Mogri, M.; Thompson, K.R.; Henderson, J.M.; Deisseroth, K. Optical Deconstruction of Parkinsonian Neural Circuitry. Science 2009, 324, 354–359. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.H.; Park, J.H.; Kim, Y.H.; Min, J.; Hwang, E.; Lee, C.J.; Suh, J.-K.F.; Hwang, O.; Jeon, S.R. Optogenetic Inactivation of the Subthalamic Nucleus Improves Forelimb Akinesia in a Rat Model of Parkinson Disease. Neurosurgery 2014, 74, 533–540. [Google Scholar] [CrossRef]
- Seeger-Armbruster, S.; Bosch-Bouju, C.; Little, S.T.C.; Smither, R.A.; Hughes, S.M.; Hyland, B.I.; Parr-Brownlie, L.C. Patterned, But Not Tonic, Optogenetic Stimulation in Motor Thalamus Improves Reaching in Acute Drug-Induced Parkinsonian Rats. J. Neurosci. 2015, 35, 1211–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magno, L.A.V.; Tenza-Ferrer, H.; Collodetti, M.; Aguiar, M.F.G.; Rodrigues, A.P.C.; da Silva, R.S.; do Silva, J.P.; Nicolau, N.F.; Rosa, D.V.F.; Birbrair, A.; et al. Optogenetic Stimulation of the M2 Cortex Reverts Motor Dysfunction in a Mouse Model of Parkinson’s Disease. J. Neurosci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bordia, T.; Perez, X.A.; Heiss, J.; Zhang, D.; Quik, M. Optogenetic Activation of Striatal Cholinergic Interneurons Regulates L-Dopa-Induced Dyskinesias. Neurobiol. Dis. 2016, 91, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Balbi, M.; Vanni, M.P.; Silasi, G.; Sekino, Y.; Bolanos, L.; LeDue, J.M.; Murphy, T.H. Targeted Ischemic Stroke Induction and Mesoscopic Imaging Assessment of Blood Flow and Ischemic Depolarization in Awake Mice. NPh 2017, 4, 035001. [Google Scholar] [CrossRef]
- Jiang, L.; Li, W.; Mamtilahun, M.; Song, Y.; Ma, Y.; Qu, M.; Lu, Y.; He, X.; Zheng, J.; Fu, Z.; et al. Optogenetic Inhibition of Striatal GABAergic Neuronal Activity Improves Outcomes After Ischemic Brain Injury. Stroke 2017, 48, 3375–3383. [Google Scholar] [CrossRef]
- He, X.; Lu, Y.; Lin, X.; Jiang, L.; Tang, Y.; Tang, G.; Chen, X.; Zhang, Z.; Wang, Y.; Yang, G.-Y. Optical Inhibition of Striatal Neurons Promotes Focal Neurogenesis and Neurobehavioral Recovery in Mice after Middle Cerebral Artery Occlusion. J. Cereb. Blood Flow Metab. 2017, 37, 837–847. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.Y.; Wang, E.H.; Woodson, W.J.; Wang, S.; Sun, G.; Lee, A.G.; Arac, A.; Fenno, L.E.; Deisseroth, K.; Steinberg, G.K. Optogenetic Neuronal Stimulation Promotes Functional Recovery after Stroke. Proc. Natl. Acad. Sci. USA 2014, 111, 12913–12918. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Jiang, L.; Li, W.; Qu, M.; Song, Y.; He, X.; Zhang, Z.; Yang, G.-Y.; Wang, Y. Optogenetic Inhibition of Striatal Neuronal Activity Improves the Survival of Transplanted Neural Stem Cells and Neurological Outcomes after Ischemic Stroke in Mice. Stem Cells Int. 2017, 2017, e4364302. [Google Scholar] [CrossRef] [Green Version]
- Tennant, K.A.; Taylor, S.L.; White, E.R.; Brown, C.E. Optogenetic Rewiring of Thalamocortical Circuits to Restore Function in the Stroke Injured Brain. Nat. Commun. 2017, 8, 15879. [Google Scholar] [CrossRef] [Green Version]
- Balbi, M.; Xiao, D.; Jativa Vega, M.; Hu, H.; Vanni, M.P.; Bernier, L.-P.; LeDue, J.; MacVicar, B.; Murphy, T.H. Gamma Frequency Activation of Inhibitory Neurons in the Acute Phase after Stroke Attenuates Vascular and Behavioral Dysfunction. Cell Rep. 2021, 34, 108696. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.S.; Lowenstein, D.H. Epilepsy. N. Engl. J. Med. 2003, 349, 1257–1266. [Google Scholar] [CrossRef]
- Tønnesen, J.; Sørensen, A.T.; Deisseroth, K.; Lundberg, C.; Kokaia, M. Optogenetic Control of Epileptiform Activity. Proc. Natl. Acad. Sci. USA 2009, 106, 12162–12167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paz, J.T.; Davidson, T.J.; Frechette, E.S.; Delord, B.; Parada, I.; Peng, K.; Deisseroth, K.; Huguenard, J.R. Closed-Loop Optogenetic Control of Thalamus as a Tool for Interrupting Seizures after Cortical Injury. Nat. Neurosci. 2013, 16, 64–70. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Harangozó, M.; Pedraza, L.; Földi, T.; Kozák, G.; Li, Q.; Berényi, A. Closed-Loop Stimulation of the Medial Septum Terminates Epileptic Seizures. Brain 2021, 144, 885–908. [Google Scholar] [CrossRef] [PubMed]
- Berényi, A.; Belluscio, M.; Mao, D.; Buzsáki, G. Closed-Loop Control of Epilepsy by Transcranial Electrical Stimulation. Science 2012, 337, 735–737. [Google Scholar] [CrossRef] [Green Version]
- Krook-Magnuson, E.; Armstrong, C.; Oijala, M.; Soltesz, I. On-Demand Optogenetic Control of Spontaneous Seizures in Temporal Lobe Epilepsy. Nat. Commun. 2013, 4, 1376. [Google Scholar] [CrossRef] [Green Version]
- Kozák, G.; Berényi, A. Sustained Efficacy of Closed Loop Electrical Stimulation for Long-Term Treatment of Absence Epilepsy in Rats. Sci. Rep. 2017, 7, 6300. [Google Scholar] [CrossRef] [Green Version]
- Farrell, J.S.; Nguyen, Q.-A.; Soltesz, I. Resolving the Micro-Macro Disconnect to Address Core Features of Seizure Networks. Neuron 2019, 101, 1016–1028. [Google Scholar] [CrossRef] [Green Version]
- Hadjiabadi, D.; Lovett-Barron, M.; Raikov, I.G.; Sparks, F.T.; Liao, Z.; Baraban, S.C.; Leskovec, J.; Losonczy, A.; Deisseroth, K.; Soltesz, I. Maximally Selective Single-Cell Target for Circuit Control in Epilepsy Models. Neuron 2021, 109, 2556–2572.e6. [Google Scholar] [CrossRef] [PubMed]
- Cohen, I.; Navarro, V.; Clemenceau, S.; Baulac, M.; Miles, R. On the Origin of Interictal Activity in Human Temporal Lobe Epilepsy in Vitro. Science 2002, 298, 1418–1421. [Google Scholar] [CrossRef] [Green Version]
- Huberfeld, G.; Wittner, L.; Clemenceau, S.; Baulac, M.; Kaila, K.; Miles, R.; Rivera, C. Perturbed Chloride Homeostasis and GABAergic Signaling in Human Temporal Lobe Epilepsy. J. Neurosci. 2007, 27, 9866–9873. [Google Scholar] [CrossRef] [Green Version]
- Pallud, J.; Le Van Quyen, M.; Bielle, F.; Pellegrino, C.; Varlet, P.; Cresto, N.; Baulac, M.; Duyckaerts, C.; Kourdougli, N.; Chazal, G.; et al. Cortical GABAergic Excitation Contributes to Epileptic Activities around Human Glioma. Sci. Transl. Med. 2014, 6, 244ra89. [Google Scholar] [CrossRef] [Green Version]
- Ellender, T.J.; Raimondo, J.V.; Irkle, A.; Lamsa, K.P.; Akerman, C.J. Excitatory Effects of Parvalbumin-Expressing Interneurons Maintain Hippocampal Epileptiform Activity via Synchronous Afterdischarges. J. Neurosci. 2014, 34, 15208–15222. [Google Scholar] [CrossRef] [Green Version]
- Sulis Sato, S.; Artoni, P.; Landi, S.; Cozzolino, O.; Parra, R.; Pracucci, E.; Trovato, F.; Szczurkowska, J.; Luin, S.; Arosio, D.; et al. Simultaneous Two-Photon Imaging of Intracellular Chloride Concentration and PH in Mouse Pyramidal Neurons in Vivo. Proc. Natl. Acad. Sci. USA 2017, 114, E8770–E8779. [Google Scholar] [CrossRef] [Green Version]
- Maset, A.; Galla, L.; Francia, S.; Cozzolino, O.; Capasso, P.; Goisis, R.C.; Losi, G.; Lombardo, A.; Ratto, G.M.; Lodovichi, C. Altered Cl- Homeostasis Hinders Forebrain GABAergic Interneuron Migration in a Mouse Model of Intellectual Disability. Proc. Natl. Acad. Sci. USA 2021, 118, e2016034118. [Google Scholar] [CrossRef]
- Lodovichi, C.; Ratto, G.M.; Trevelyan, A.J.; Arosio, D. Genetically Encoded Sensors for Chloride Concentration. J. Neurosci. Methods 2022, 368, 109455. [Google Scholar] [CrossRef] [PubMed]
- Frith, U.; Happé, F. Autism Spectrum Disorder. Curr. Biol. 2005, 15, 786–790. [Google Scholar] [CrossRef] [Green Version]
- Amso, D.; Haas, S.; Tenenbaum, E.; Markant, J.; Sheinkopf, S.J. Bottom-up Attention Orienting in Young Children with Autism. J. Autism Dev. Disord. 2014, 44, 664–673. [Google Scholar] [CrossRef]
- Chen, Q.; Deister, C.A.; Gao, X.; Guo, B.; Lynn-Jones, T.; Chen, N.; Wells, M.F.; Liu, R.; Goard, M.J.; Dimidschstein, J.; et al. Dysfunction of Cortical GABAergic Neurons Leads to Sensory Hyper-Reactivity in a Shank3 Mouse Model of ASD. Nat. Neurosci. 2020, 23, 520–532. [Google Scholar] [CrossRef]
- Gogolla, N.; Leblanc, J.J.; Quast, K.B.; Südhof, T.C.; Fagiolini, M.; Hensch, T.K. Common Circuit Defect of Excitatory-Inhibitory Balance in Mouse Models of Autism. J. Neurodev. Disord. 2009, 1, 172–181. [Google Scholar] [CrossRef] [Green Version]
- Marín, O. Interneuron Dysfunction in Psychiatric Disorders. Nat. Rev. Neurosci. 2012, 13, 107–120. [Google Scholar] [CrossRef]
- Chao, H.-T.; Chen, H.; Samaco, R.C.; Xue, M.; Chahrour, M.; Yoo, J.; Neul, J.L.; Gong, S.; Lu, H.-C.; Heintz, N.; et al. Dysfunction in GABA Signalling Mediates Autism-like Stereotypies and Rett Syndrome Phenotypes. Nature 2010, 468, 263–269. [Google Scholar] [CrossRef]
- Constantin, L.; Poulsen, R.E.; Scholz, L.A.; Favre-Bulle, I.A.; Taylor, M.A.; Sun, B.; Goodhill, G.J.; Vanwalleghem, G.C.; Scott, E.K. Altered Brain-Wide Auditory Networks in a Zebrafish Model of Fragile X Syndrome. BMC Biol. 2020, 18, 125. [Google Scholar] [CrossRef]
- May, A.; Schulte, L.H. Chronic Migraine: Risk Factors, Mechanisms and Treatment. Nat. Rev. Neurol 2016, 12, 455–464. [Google Scholar] [CrossRef]
- Burstein, R.; Noseda, R.; Borsook, D. Migraine: Multiple Processes, Complex Pathophysiology. J. Neurosci. 2015, 35, 6619–6629. [Google Scholar] [CrossRef]
- Capuani, C.; Melone, M.; Tottene, A.; Bragina, L.; Crivellaro, G.; Santello, M.; Casari, G.; Conti, F.; Pietrobon, D. Defective Glutamate and K+ Clearance by Cortical Astrocytes in Familial Hemiplegic Migraine Type 2. EMBO Mol. Med. 2016, 8, 967–986. [Google Scholar] [CrossRef]
- Parker, P.D.; Suryavanshi, P.; Melone, M.; Sawant-Pokam, P.A.; Reinhart, K.M.; Kaufmann, D.; Theriot, J.J.; Pugliese, A.; Conti, F.; Shuttleworth, C.W.; et al. Non-Canonical Glutamate Signaling in a Genetic Model of Migraine with Aura. Neuron 2021, 109, 611–628.e8. [Google Scholar] [CrossRef]
- Kendler, K.S.; Karkowski, L.M.; Prescott, C.A. Causal Relationship between Stressful Life Events and the Onset of Major Depression. Am. J. Psychiatry 1999, 156, 837–841. [Google Scholar] [CrossRef]
- Kessler, R.C. The Effects of Stressful Life Events on Depression. Annu. Rev. Psychol. 1997, 48, 191–214. [Google Scholar] [CrossRef]
- Slattery, D.A.; Cryan, J.F. Modelling Depression in Animals: At the Interface of Reward and Stress Pathways. Psychopharmacology 2017, 234, 1451–1465. [Google Scholar] [CrossRef]
- Grace, A.A. Phasic versus Tonic Dopamine Release and the Modulation of Dopamine System Responsivity: A Hypothesis for the Etiology of Schizophrenia. Neuroscience 1991, 41, 1–24. [Google Scholar] [CrossRef]
- Cao, J.-L.; Covington, H.E.; Friedman, A.K.; Wilkinson, M.B.; Walsh, J.J.; Cooper, D.C.; Nestler, E.J.; Han, M.-H. Mesolimbic Dopamine Neurons in the Brain Reward Circuit Mediate Susceptibility to Social Defeat and Antidepressant Action. J. Neurosci. 2010, 30, 16453–16458. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, V.; Han, M.-H.; Graham, D.L.; Berton, O.; Renthal, W.; Russo, S.J.; Laplant, Q.; Graham, A.; Lutter, M.; Lagace, D.C.; et al. Molecular Adaptations Underlying Susceptibility and Resistance to Social Defeat in Brain Reward Regions. Cell 2007, 131, 391–404. [Google Scholar] [CrossRef] [Green Version]
- Chaudhury, D.; Walsh, J.J.; Friedman, A.K.; Juarez, B.; Ku, S.M.; Koo, J.W.; Ferguson, D.; Tsai, H.-C.; Pomeranz, L.; Christoffel, D.J.; et al. Rapid Regulation of Depression-Related Behaviours by Control of Midbrain Dopamine Neurons. Nature 2013, 493, 532–536. [Google Scholar] [CrossRef]
- Tye, K.M.; Mirzabekov, J.J.; Warden, M.R.; Ferenczi, E.A.; Tsai, H.-C.; Finkelstein, J.; Kim, S.-Y.; Adhikari, A.; Thompson, K.R.; Andalman, A.S.; et al. Dopamine Neurons Modulate Neural Encoding and Expression of Depression-Related Behaviour. Nature 2013, 493, 537–541. [Google Scholar] [CrossRef] [Green Version]
- Lowes, D.C.; Chamberlin, L.A.; Kretsge, L.N.; Holt, E.S.; Abbas, A.I.; Park, A.J.; Yusufova, L.; Bretton, Z.H.; Firdous, A.; Enikolopov, A.G.; et al. Ventral Tegmental Area GABA Neurons Mediate Stress-Induced Blunted Reward-Seeking in Mice. Nat. Commun. 2021, 12, 3539. [Google Scholar] [CrossRef]
- Bagot, R.C.; Parise, E.M.; Peña, C.J.; Zhang, H.-X.; Maze, I.; Chaudhury, D.; Persaud, B.; Cachope, R.; Bolaños-Guzmán, C.A.; Cheer, J.F.; et al. Ventral Hippocampal Afferents to the Nucleus Accumbens Regulate Susceptibility to Depression. Nat. Commun. 2015, 6, 7062. [Google Scholar] [CrossRef]
- Ramirez, S.; Liu, X.; Lin, P.-A.; Suh, J.; Pignatelli, M.; Redondo, R.L.; Ryan, T.J.; Tonegawa, S. Creating a False Memory in the Hippocampus. Science 2013, 341, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ramirez, S.; Pang, P.T.; Puryear, C.B.; Govindarajan, A.; Deisseroth, K.; Tonegawa, S. Optogenetic Stimulation of a Hippocampal Engram Activates Fear Memory Recall. Nature 2012, 484, 381–385. [Google Scholar] [CrossRef] [Green Version]
- Covington, H.E.; Lobo, M.K.; Maze, I.; Vialou, V.; Hyman, J.M.; Zaman, S.; LaPlant, Q.; Mouzon, E.; Ghose, S.; Tamminga, C.A.; et al. Antidepressant Effect of Optogenetic Stimulation of the Medial Prefrontal Cortex. J. Neurosci. 2010, 30, 16082–16090. [Google Scholar] [CrossRef] [PubMed]
- Treadway, M.T.; Pizzagalli, D.A. Imaging the Pathophysiology of Major Depressive Disorder - from Localist Models to Circuit-Based Analysis. Biol. Mood Anxiety Disord. 2014, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Muir, J.; Lopez, J.; Bagot, R.C. Wiring the Depressed Brain: Optogenetic and Chemogenetic Circuit Interrogation in Animal Models of Depression. Neuropsychopharmacology 2019, 44, 1013–1026. [Google Scholar] [CrossRef]
- Challis, C.; Boulden, J.; Veerakumar, A.; Espallergues, J.; Vassoler, F.M.; Pierce, R.C.; Beck, S.G.; Berton, O. Raphe GABAergic Neurons Mediate the Acquisition of Avoidance after Social Defeat. J. Neurosci. 2013, 33, 13978–13988. [Google Scholar] [CrossRef]
- Challis, C.; Beck, S.; Berton, O. Optogenetic Modulation of Descending Prefrontocortical Inputs to the Dorsal Raphe Bidirectionally Bias Socioaffective Choices after Social Defeat. Fron. Behav. Neurosci. 2014, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares-Cunha, C.; Coimbra, B.; David-Pereira, A.; Borges, S.; Pinto, L.; Costa, P.; Sousa, N.; Rodrigues, A.J. Activation of D2 Dopamine Receptor-Expressing Neurons in the Nucleus Accumbens Increases Motivation. Nat. Commun. 2016, 7, 11829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGirr, A.; LeDue, J.; Chan, A.W.; Xie, Y.; Murphy, T.H. Cortical Functional Hyperconnectivity in a Mouse Model of Depression and Selective Network Effects of Ketamine. Brain 2017, 140, 2210–2225. [Google Scholar] [CrossRef] [PubMed]
- Desimone, R.; Albright, T.D.; Gross, C.G.; Bruce, C. Stimulus-Selective Properties of Inferior Temporal Neurons in the Macaque. J. Neurosci. 1984, 4, 2051–2062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, C.G.; Rocha-Miranda, C.E.; Bender, D.B. Visual Properties of Neurons in Inferotemporal Cortex of the Macaque. J. Neurophysiol. 1972, 35, 96–111. [Google Scholar] [CrossRef]
- Tanaka, K.; Saito, H.; Fukada, Y.; Moriya, M. Coding Visual Images of Objects in the Inferotemporal Cortex of the Macaque Monkey. J. Neurophysiol. 1991, 66, 170–189. [Google Scholar] [CrossRef] [Green Version]
- Stauffer, W.R.; Lak, A.; Yang, A.; Borel, M.; Paulsen, O.; Boyden, E.S.; Schultz, W. Dopamine Neuron-Specific Optogenetic Stimulation in Rhesus Macaques. Cell 2016, 166, 1564–1571.e6. [Google Scholar] [CrossRef] [Green Version]
- Gerits, A.; Farivar, R.; Rosen, B.R.; Wald, L.L.; Boyden, E.S.; Vanduffel, W. Optogenetically Induced Behavioral and Functional Network Changes in Primates. Curr. Biol. 2012, 22, 1722–1726. [Google Scholar] [CrossRef] [Green Version]
- Galvan, A.; Stauffer, W.R.; Acker, L.; El-Shamayleh, Y.; Inoue, K.-I.; Ohayon, S.; Schmid, M.C. Nonhuman Primate Optogenetics: Recent Advances and Future Directions. J. Neurosci. 2017, 37, 10894–10903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heider, B.; Nathanson, J.L.; Isacoff, E.Y.; Callaway, E.M.; Siegel, R.M. Two-Photon Imaging of Calcium in Virally Transfected Striate Cortical Neurons of Behaving Monkey. PLoS ONE 2010, 5, e13829. [Google Scholar] [CrossRef] [PubMed]
- Sadakane, O.; Masamizu, Y.; Watakabe, A.; Terada, S.-I.; Ohtsuka, M.; Takaji, M.; Mizukami, H.; Ozawa, K.; Kawasaki, H.; Matsuzaki, M.; et al. Long-Term Two-Photon Calcium Imaging of Neuronal Populations with Subcellular Resolution in Adult Non-Human Primates. Cell Rep. 2015, 13, 1989–1999. [Google Scholar] [CrossRef] [Green Version]
- Santisakultarm, T.P.; Kersbergen, C.J.; Bandy, D.K.; Ide, D.C.; Choi, S.-H.; Silva, A.C. Two-Photon Imaging of Cerebral Hemodynamics and Neural Activity in Awake and Anesthetized Marmosets. J. Neurosci. Methods 2016, 271, 55–64. [Google Scholar] [CrossRef] [Green Version]
- Yamahachi, H.; Marik, S.A.; McManus, J.N.J.; Denk, W.; Gilbert, C.D. Rapid Axonal Sprouting and Pruning Accompany Functional Reorganization in Primary Visual Cortex. Neuron 2009, 64, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, O.; Lustig, B.R.; Nassi, J.J.; Cetin, A.; Reynolds, J.H.; Albright, T.D.; Callaway, E.M.; Stoner, G.R.; Roe, A.W. Optogenetics through Windows on the Brain in the Nonhuman Primate. J. Neurophysiol. 2013, 110, 1455–1467. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, F.; Jiang, H.; Lee, T.S.; Tang, S. Long-Term Two-Photon Imaging in Awake Macaque Monkey. Neuron 2017, 93, 1049–1057.e3. [Google Scholar] [CrossRef]
- Ju, N.; Jiang, R.; Macknik, S.L.; Martinez-Conde, S.; Tang, S. Long-Term All-Optical Interrogation of Cortical Neurons in Awake-Behaving Nonhuman Primates. PLoS Biol. 2018, 16, e2005839. [Google Scholar] [CrossRef]
- Park, J.E.; Zhang, X.F.; Choi, S.-H.; Okahara, J.; Sasaki, E.; Silva, A.C. Generation of Transgenic Marmosets Expressing Genetically Encoded Calcium Indicators. Sci. Rep. 2016, 6, 34931. [Google Scholar] [CrossRef] [PubMed]
- Seidemann, E.; Chen, Y.; Bai, Y.; Chen, S.C.; Mehta, P.; Kajs, B.L.; Geisler, W.S.; Zemelman, B.V. Calcium Imaging with Genetically Encoded Indicators in Behaving Primates. eLife 2016, 5, e16178. [Google Scholar] [CrossRef] [PubMed]
| Opsins | Ions | Spectral Peak (nm) | Tau Off (ms) | References | ||
|---|---|---|---|---|---|---|
| Influx | Efflux | |||||
| Depolarizing | ChR2 | Na+ | - | 470 | 10 | [126] |
| CoChR | Na+ | - | 470 | 30 | [127] | |
| Chronos | Na+ | - | 530 | 3.6 | [127] | |
| ChroME | Na+ | - | 530 | 3 | [128] | |
| ChroMEs | Na+ | - | 530 | 13 | [129] | |
| ChroMEf | Na+ | - | 530 | 9.6 | [129] | |
| ChRmine | Na+ | - | 585 | 2 | [130] | |
| ChrimsonR | Na+ | - | 590 | 15.8 | [127] | |
| f-Crimson | Na+ | - | 590 | 5.7 | [131] | |
| vf-Crimson | Na+ | - | 590 | 2.7 | [131] | |
| Hyperpolarizing | GtACR2 | Cl- | - | 480 | 40 | [132] |
| GtACR1 | Cl- | - | 520 | 15 | [128,132] | |
| Arch | - | H+ | 570 | 20 | [133] | |
| eNpHr3.0 | Cl- | - | 590 | 40.5 | [134] | |
| AIACR1 | Cl- | - | 590 | 90 | [135] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brondi, M.; Bruzzone, M.; Lodovichi, C.; dal Maschio, M. Optogenetic Methods to Investigate Brain Alterations in Preclinical Models. Cells 2022, 11, 1848. https://doi.org/10.3390/cells11111848
Brondi M, Bruzzone M, Lodovichi C, dal Maschio M. Optogenetic Methods to Investigate Brain Alterations in Preclinical Models. Cells. 2022; 11(11):1848. https://doi.org/10.3390/cells11111848
Chicago/Turabian StyleBrondi, Marco, Matteo Bruzzone, Claudia Lodovichi, and Marco dal Maschio. 2022. "Optogenetic Methods to Investigate Brain Alterations in Preclinical Models" Cells 11, no. 11: 1848. https://doi.org/10.3390/cells11111848






