Human Sperm Morphology as a Marker of Its Nuclear Quality and Epigenetic Pattern
Abstract
:1. Introduction
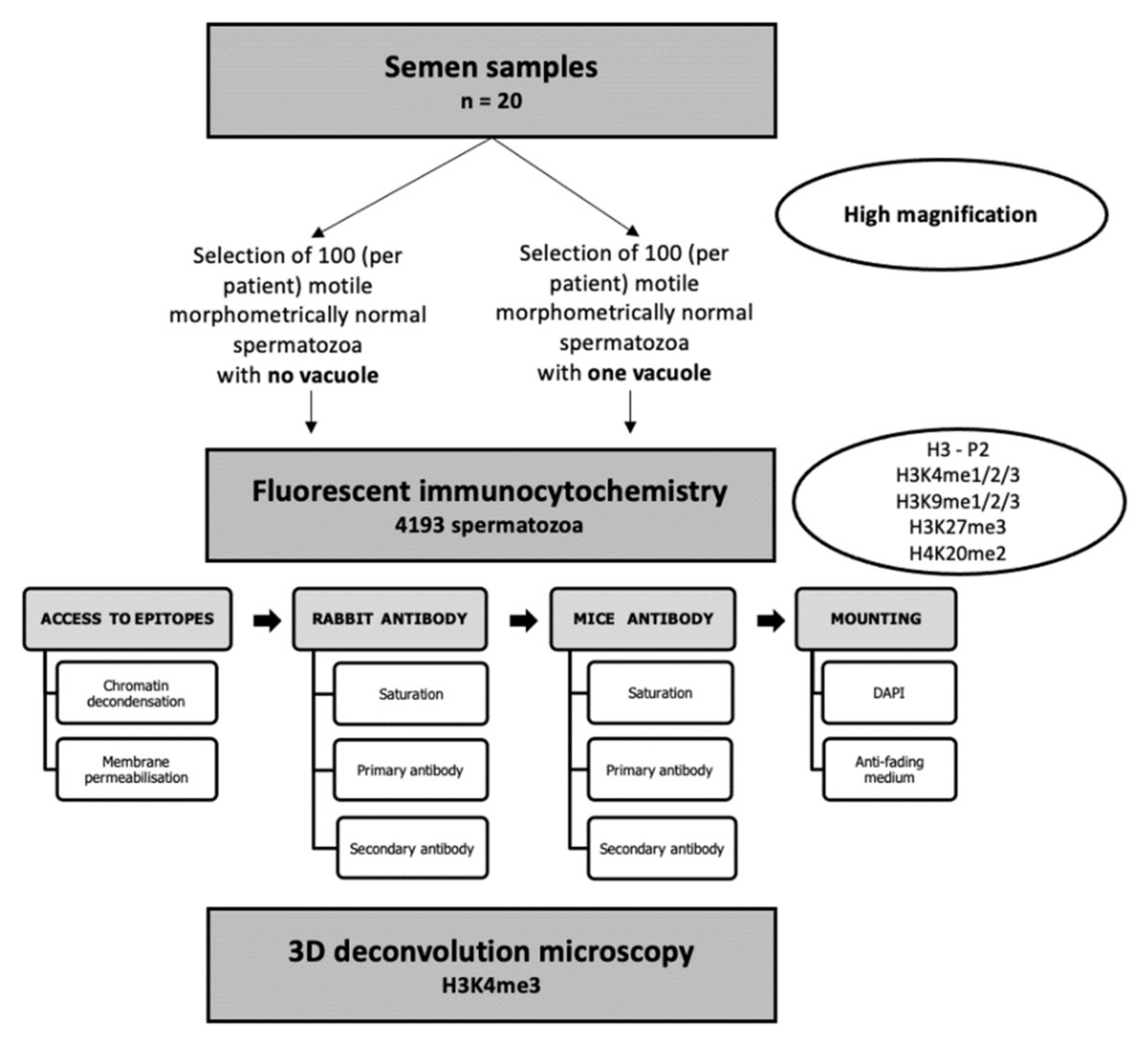
2. Materials and Methods
2.1. Patients
- -
- alpha (the type I error, i.e., the probability of wrongly rejecting H0 and detecting a statistically significant difference when the groups are not actually different) at 0.05 and
- -
- beta (the type II error, i.e., the probability of wrongly accepting H0 and not detecting a statistically significant difference when a specified difference between the groups in reality exists) at 0.1, the power of our analyses reached 90% (1-beta: 0.9) with 12 patients included. As we had already included 20 patients, we analyzed the results obtained in these 20 patients, thus exceeding the required number of patients.
2.2. Sperm Preparation
2.3. Sperm Selection under High-Magnification Microscopy (IMSI-like Methodology)
2.4. Selection of Epigenetic Marks of Interest in Male Fertility: Nuclear Proteins and PTMs
2.5. Fluorescent Immunocytochemistry and Two-Dimensional (2D) Microscopy
2.6. Three-Dimensional (3D) Microscopy
3. Results
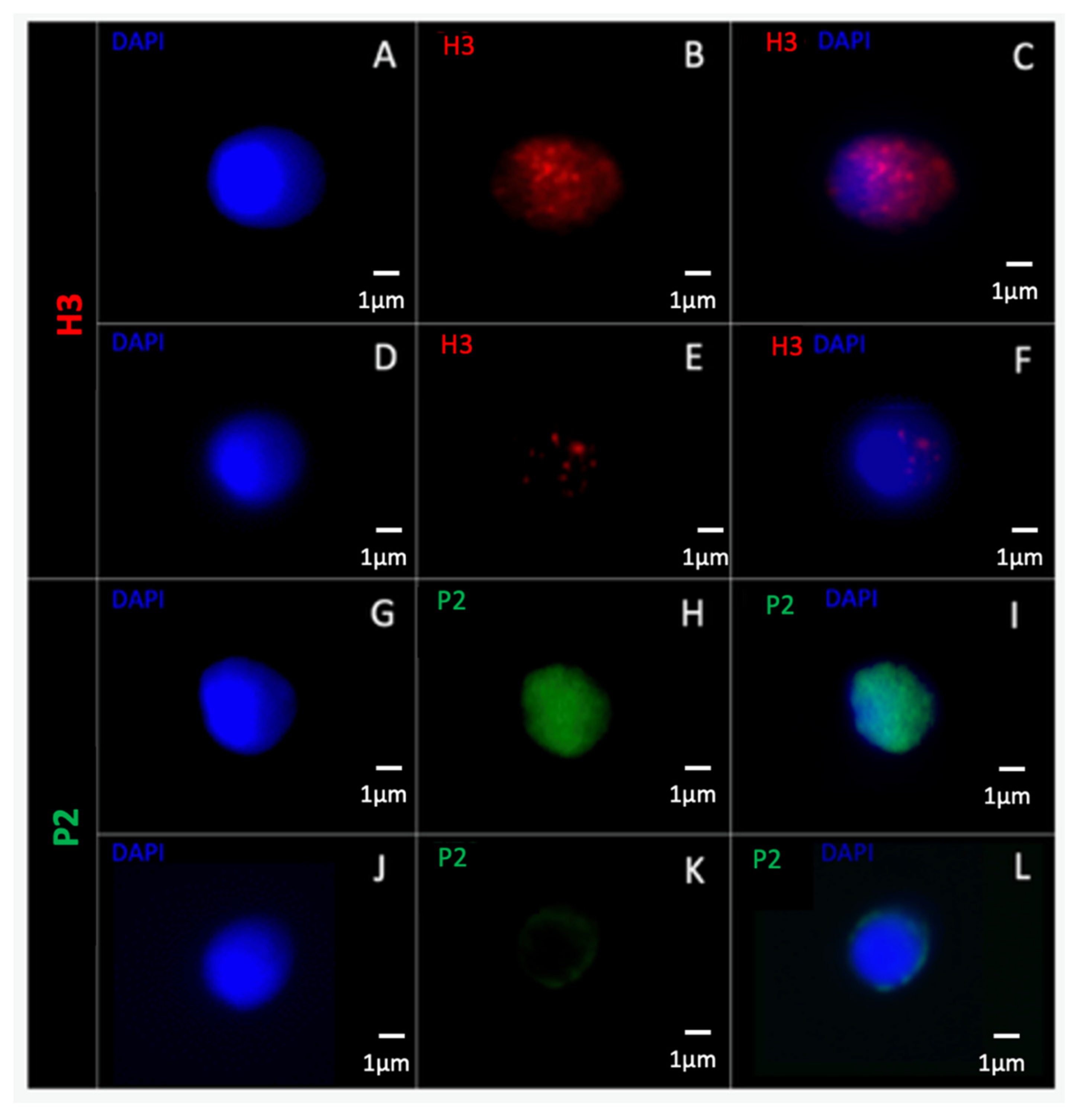
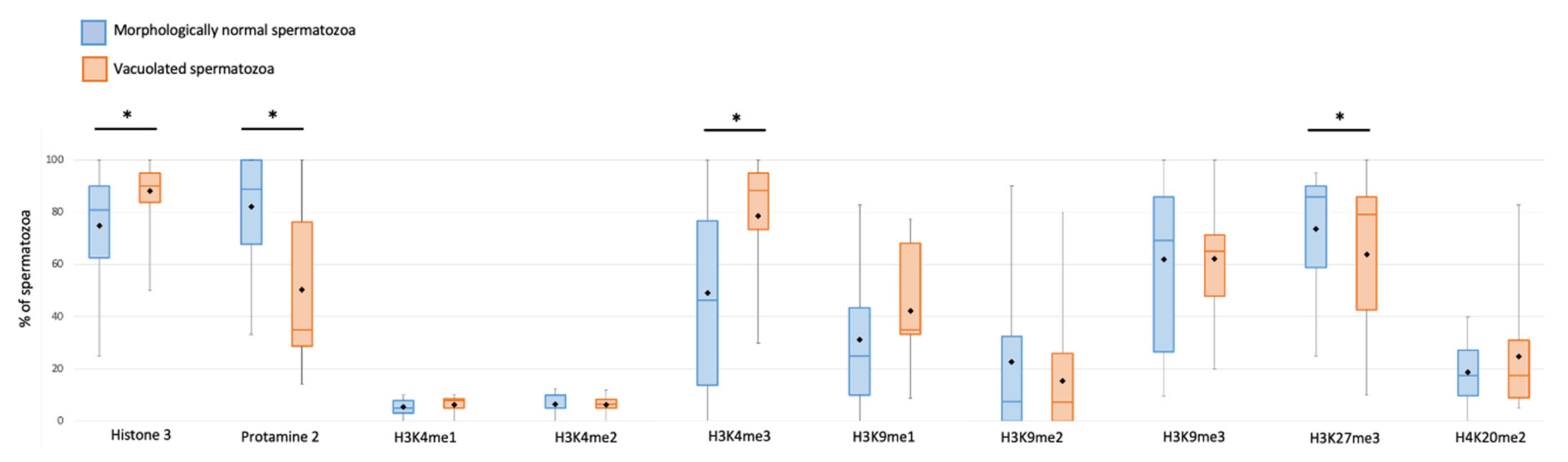
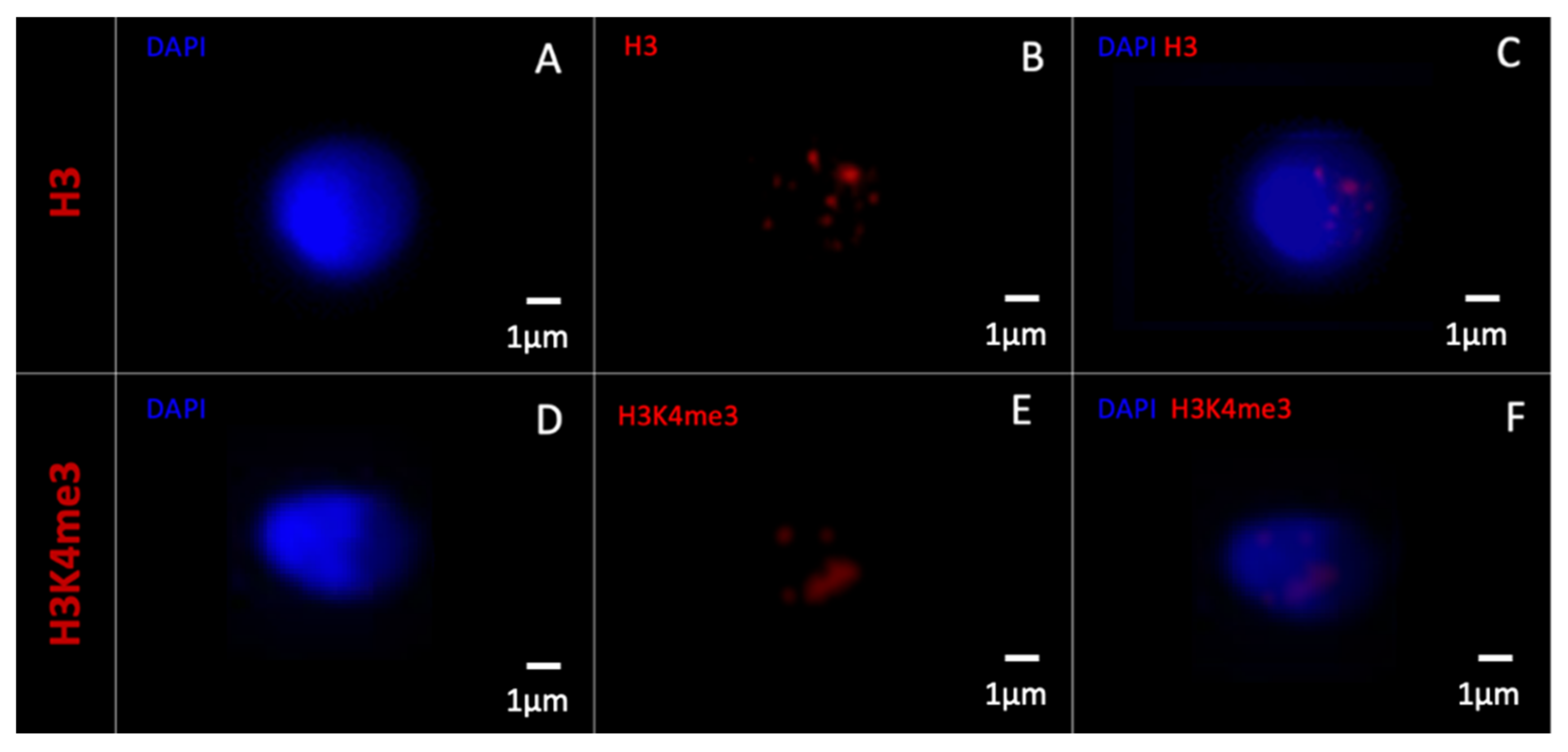
3.1. Sperm Epigenetic Marks and Sperm Morphology under High-Magnification Microscopy
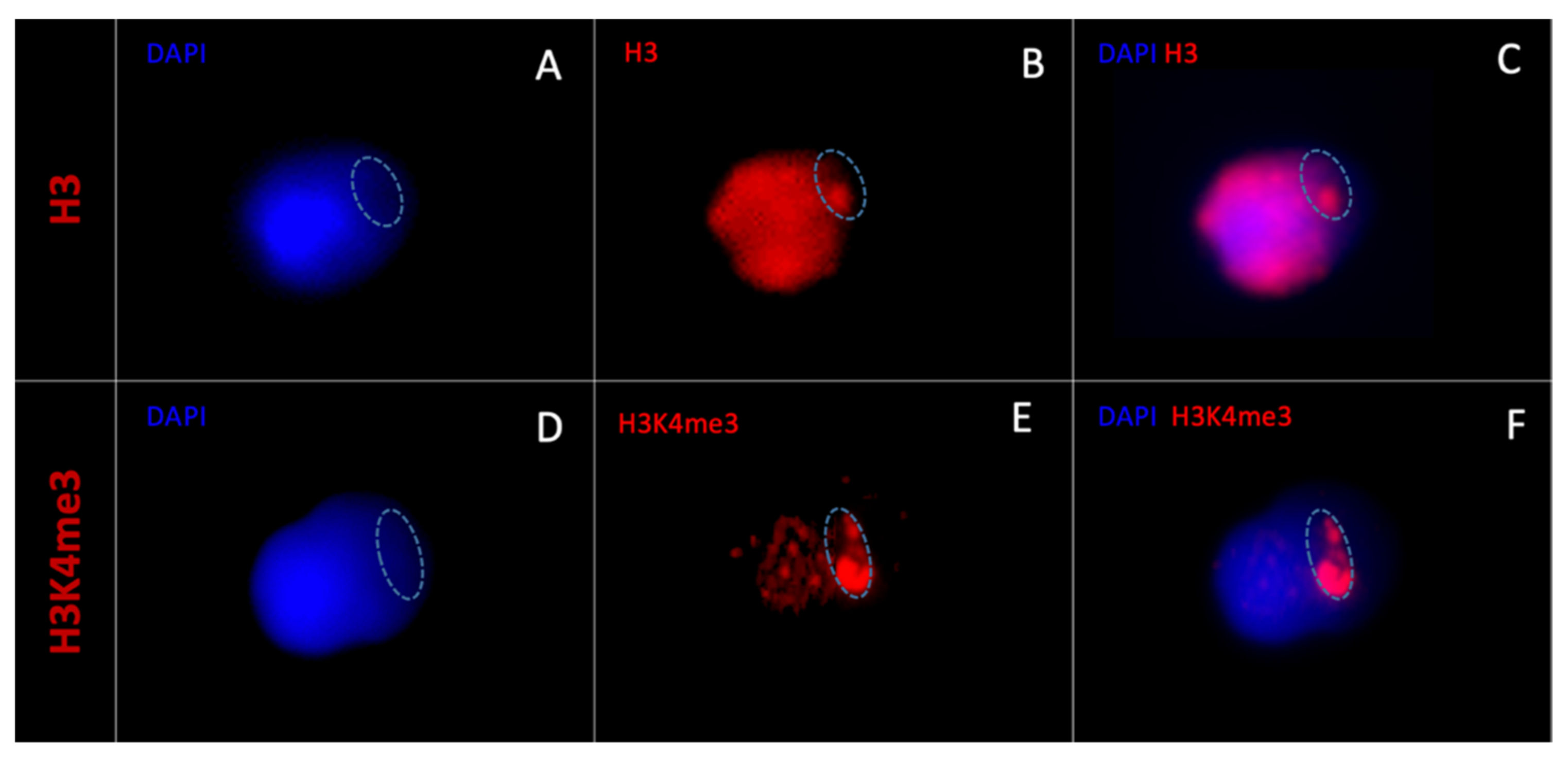
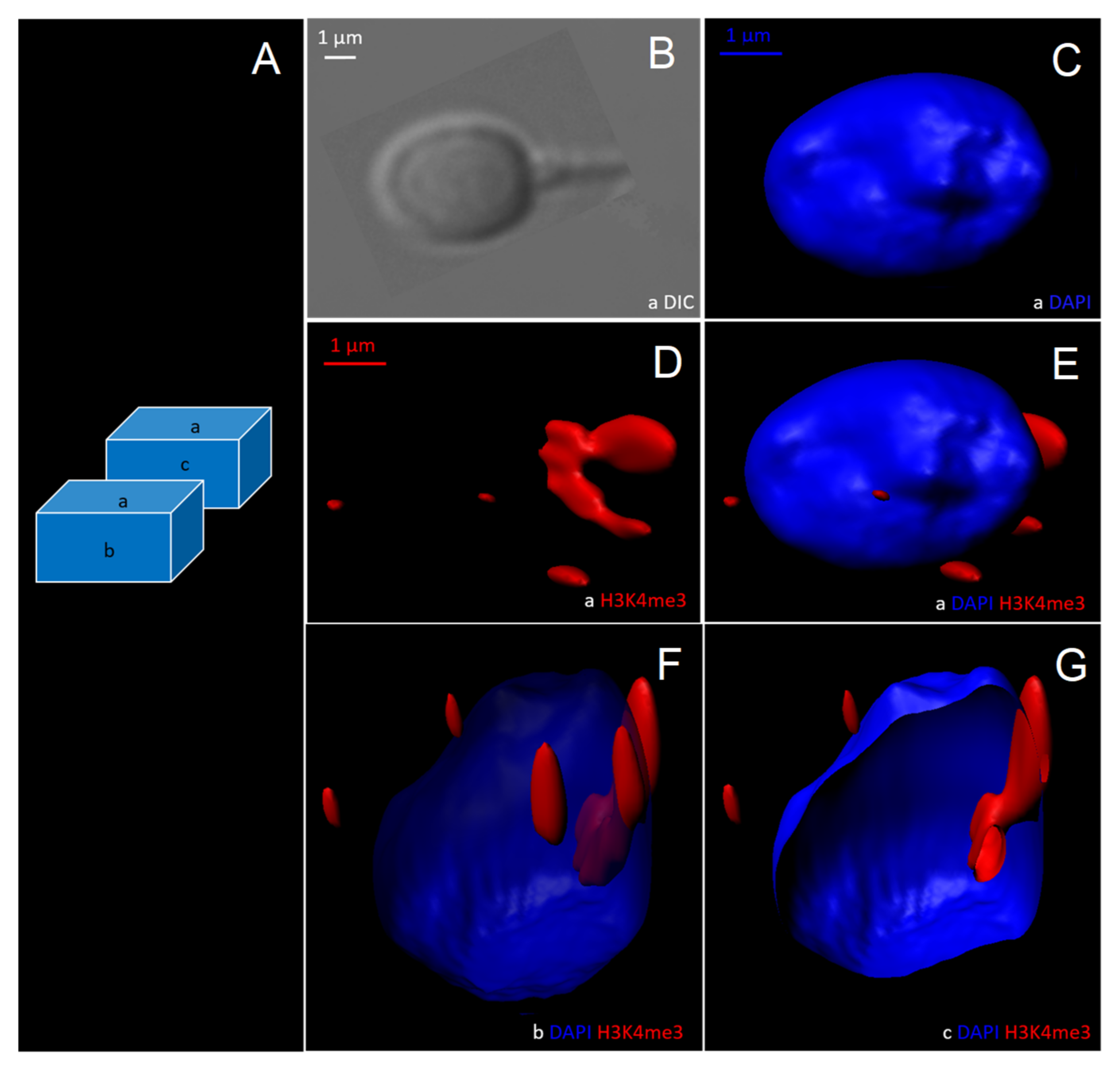
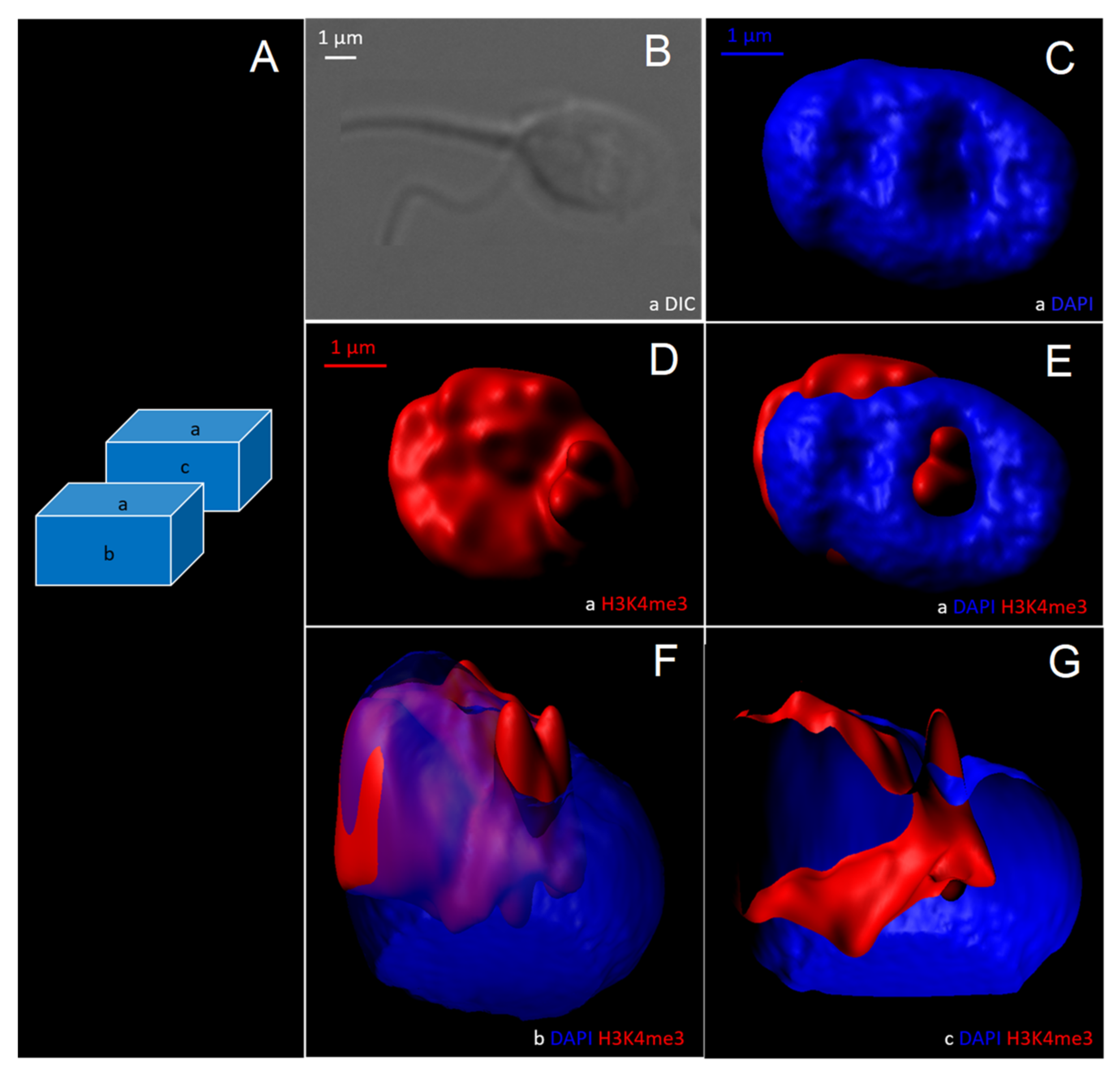
3.2. Anatomical Relationships between Vacuoles and Epigenetic Marks
3.2.1. 2D Microscopy
3.2.2. 3D- Deconvolution Microscopy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hao, S.L.; Ni, F.D.; Yang, W.X. The dynamics and regulation of chromatin remodeling during spermiogenesis. Gene 2019, 706, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.G.; Ketchum, C.C.; Meyer-Ficca, M.L. Heritable sperm chromatin epigenetics: A break to remember. Biol. Reprod. 2017, 97, 784–797. [Google Scholar] [CrossRef]
- Kimmins, S.; Sassone-Corsi, P. Chromatin remodelling and epigenetic features of germ cells. Nature 2005, 434, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.P.; Krawetz, S.A. Decondensing the protamine domain for transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 8340–8345. [Google Scholar] [CrossRef] [Green Version]
- Rooney, A.P.; Zhang, J.; Nei, M. An unusual form of purifying selection in a sperm protein. Mol. Biol. Evol. 2000, 17, 278–283. [Google Scholar] [CrossRef]
- Gatewood, J.M.; Schroth, G.P.; Schmid, C.W.; Bradbury, E.M. Zinc-induced secondary structure transitions in human sperm protamines. J. Biol. Chem. 1990, 265, 20667–20672. [Google Scholar] [CrossRef]
- Gusse, M.; Sautiere, P.; Belaiche, D.; Martinage, A.; Roux, C.; Dadoune, J.P.; Chevaillier, P. Purification and characterization of nuclear basic proteins of human sperm. Biochim. Biophys. Acta 1986, 884, 124–134. [Google Scholar] [CrossRef]
- Dominguez, K.; Arca, C.D.R.; Ward, W.S. The Relationship between Chromatin Structure and DNA Damage in Mammalian Spermatozoa. In Sperm Chromatin: Biological and Clinical Applications in Male Infertility and Assisted Reprod; Zini, A., Agarwal, A., Eds.; Springer: New York, NY, USA, 2011; pp. 61–68. [Google Scholar]
- Ward, W.S. Function of sperm chromatin structural elements in fertilization and development. Mol. Hum. Reprod. 2010, 16, 30–36. [Google Scholar] [CrossRef] [Green Version]
- Arpanahi, A.; Brinkworth, M.; Iles, D.; Krawetz, S.A.; Paradowska, A.; Platts, A.E.; Saida, M.; Steger, K.; Tedder, P.; Miller, D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009, 19, 1338–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunner, A.M.; Nanni, P.; Mansuy, I.M. Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics Chromatin 2014, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Brykczynska, U.; Hisano, M.; Erkek, S.; Ramos, L.; Oakeley, E.J.; Roloff, T.C.; Beisel, C.; Schübeler, D.; Stadler, M.B.; Peters, A.H. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 2010, 17, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Carrell, D.T. The Sperm Epigenome: Implications for Assisted Reproductive Technologies. Adv. Exp. Med. Biol. 2019, 1166, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, S.S.; Nix, D.A.; Zhang, H.; Purwar, J.; Carrell, D.T.; Cairns, B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009, 460, 473–478. [Google Scholar] [CrossRef] [Green Version]
- Erkek, S.; Hisano, M.; Liang, C.Y.; Gill, M.; Murr, R.; Dieker, J.; Schübeler, D.; Van Der Vlag, J.; Stadler, M.B.; Peters, A.H. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol. 2013, 20, 868–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammoud, S.S.; Nix, D.A.; Hammoud, A.O.; Gibson, M.; Cairns, B.R.; Carrell, D.T. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum. Reprod. 2011, 26, 2558–2569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihara, M.; Meyer-Ficca, M.L.; Leu, N.A.; Rao, S.; Li, F.; Gregory, B.D.; Zalenskaya, I.A.; Schultz, R.M.; Meyer, R.G. Paternal poly (ADP-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS Genet. 2014, 10, e1004317. [Google Scholar] [CrossRef] [Green Version]
- Schagdarsurengin, U.; Paradowska, A.; Steger, K. Analysing the sperm epigenome: Roles in early embryogenesis and assisted reproduction. Nat. Rev. Urol. 2012, 9, 609–619. [Google Scholar] [CrossRef]
- Castillo, J.; Amaral, A.; Oliva, R. Sperm nuclear proteome and its epigenetic potential. Andrology 2014, 2, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Blanco, M.; Cocquet, J. Genetic Factors Affecting Sperm Chromatin Structure. Adv. Exp. Med. Biol. 2019, 1166, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Castillo, J.; Estanyol, J.M.; Ballescà, J.L. Human sperm chromatin epigenetic potential: Genomics, proteomics, and male infertility. Asian J. Androl. 2015, 17, 601–609. [Google Scholar] [CrossRef]
- Luense, L.J.; Wang, X.; Schon, S.; Weller, A.H.; Shiao, E.L.; Bryant, J.; Bartolomei, M.; Coutifaris, C.; Garcia, B.A.; Berger, S.L. Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenetics Chromatin 2016, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciel, V.L., Jr.; Tamashiro, L.K.; Bertolla, R.P. Post-translational modifications of seminal proteins and their importance in male fertility potential. Expert Rev. Proteom. 2019, 16, 941–950. [Google Scholar] [CrossRef]
- Steilmann, C.; Paradowska, A.; Bartkuhn, M.; Vieweg, M.; Schuppe, H.C.; Bergmann, M.; Kliesch, S.; Weidner, W.; Steger, K. Presence of histone H3 acetylated at lysine 9 in male germ cells and its distribution pattern in the genome of human spermatozoa. Reprod. Fertil. Dev. 2011, 23, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gao, H.; Li, W.; Liu, C. Essential Role of Histone Replacement and Modifications in Male Fertility. Front. Genet. 2019, 10, 962. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Q.; Tang, F.; Yan, L.; Qiao, J. Epigenetic Regulation and Risk Factors During the Development of Human Gametes and Early Embryos. Annu. Rev. Genom. Hum. Genet. 2019, 20, 21–40. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.; Jodar, M.; Oliva, R. The contribution of human sperm proteins to the development and epigenome of the preimplantation embryo. Hum. Reprod. Update 2018, 24, 535–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bichara, C.; Berby, B.; Rives, A.; Jumeau, F.; Letailleur, M.; Setif, V.; Sibert, L.; Rondanino, C.; Rives, N. Sperm chromatin condensation defects, but neither DNA fragmentation nor aneuploidy, are an independent predictor of clinical pregnancy after intracytoplasmic sperm injection. J. Assist. Reprod. Genet. 2019, 36, 1387–1399. [Google Scholar] [CrossRef]
- Boitrelle, F.; Ferfouri, F.; Petit, J.M.; Segretain, D.; Tourain, C.; Bergere, M.; Bailly, M.; Vialard, F.; Albert, M.; Selva, J. Large human sperm vacuoles observed in motile spermatozoa under high magnification: Nuclear thumbprints linked to failure of chromatin condensation. Hum. Reprod. 2011, 26, 1650–1658. [Google Scholar] [CrossRef] [Green Version]
- Boitrelle, F.; Pagnier, M.; Athiel, Y.; Swierkowski-Blanchard, N.; Torre, A.; Alter, L.; Muratorio, C.; Vialard, F.; Albert, M.; Selva, J. A human morphologically normal spermatozoon may have noncondensed chromatin. Andrologia 2014, 47, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Perdrix, A.; Travers, A.; Chelli, M.H.; Escalier, D.; Rego, J.L.D.; Milazzo, J.P.; Mousset-Simeon, N.; Mace, B.; Rives, N. Assessment of acrosome and nuclear abnormalities in human spermatozoa with large vacuoles. Hum. Reprod. 2010, 26, 47–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldini, D.; Ferri, D.; Baldini, G.M.; Lot, D.; Catino, A.; Vizziello, D.; Vizziello, G. Sperm Selection for ICSI: Do We Have a Winner? Cells 2021, 10, 3566. [Google Scholar] [CrossRef]
- Ogle, R.A.; Netherton, J.; Schneider, E.; Velkov, T.; Zhang, H.; Cole, N.; Hetherington, L.; Villaverde, A.I.S.B.; Baker, M.A. Nuclear heterogeneity is prevalent in high-quality fractionated human sperm cells typically used for assisted conception. Hum. Reprod. 2021, 36, 2073–2082. [Google Scholar] [CrossRef] [PubMed]
- Boitrelle, F.; Albert, M.; Petit, J.-M.; Ferfouri, F.; Wainer, R.; Bergere, M.; Bailly, M.; Vialard, F.; Selva, J. Small human sperm vacuoles observed under high magnification are pocket-like nuclear concavities linked to chromatin condensation failure. Reprod. Biomed. Online 2013, 27, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Franco, J.G., Jr.; Mauri, A.L.; Petersen, C.G.; Massaro, F.C.; Silva, L.F.I.; Felipe, V.; Cavagna, M.; Pontes, A.; Baruffi, R.L.R.; Oliveira, J.B.A.; et al. Large nuclear vacuoles are indicative of abnormal chromatin packaging in human spermatozoa. Int. J. Androl. 2011, 35, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Utsuno, H.; Miyamoto, T.; Oka, K.; Shiozawa, T. Morphological alterations in protamine-deficient spermatozoa. Hum. Reprod. 2014, 29, 2374–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, V.W.; Liu, L.; Jones, K.P.; Hatasaka, H.H.; Gibson, M.; Peterson, C.M.; Carrell, D.T. Sperm protamine 1/protamine 2 ratios are related to in vitro fertilization pregnancy rates and predictive of fertilization ability. Fertil. Steril. 2006, 86, 1408–1415. [Google Scholar] [CrossRef] [PubMed]
- Asmarinah; Syauqy, A.; Umar, L.A.; Lestari, S.W.; Mansyur, E.; Hestiantoro, A.; Paradowszka-Dogan, A. Sperm chromatin maturity and integrity correlated to zygote development in ICSI program. Syst. Biol. Reprod. Med. 2016, 62, 309–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boitrelle, F.; Guthauser, B.; Alter, L.; Bailly, M.; Bergere, M.; Wainer, R.; Vialard, F.; Albert, M.; Selva, J. High-magnification selection of spermatozoa prior to oocyte injection: Confirmed and potential indications. Reprod. Biomed. Online 2014, 28, 6–13. [Google Scholar] [CrossRef] [Green Version]
- Iranpour, F.G. Impact of sperm chromatin evaluation on fertilization rate in intracytoplasmic sperm injection. Adv. Biomed. Res. 2014, 3, 229. [Google Scholar] [CrossRef] [PubMed]
- Nasr-Esfahani, M.H.; Salehi, M.; Razavi, S.; Mardani, M.; Bahramian, H.; Steger, K.; Oreizi, F. Effect of protamine-2 deficiency on ICSI outcome. Reprod. Biomed. Online 2004, 9, 652–658. [Google Scholar] [CrossRef]
- Ribas-Maynou, J.; Abad, C.; García-Segura, S.; Oliver-Bonet, M.; Prada, E.; Amengual, M.J.; Navarro, J.; Benet, J. Sperm chromatin condensation and single- and double-stranded DNA damage as important parameters to define male factor related recurrent miscarriage. Mol. Reprod. Dev. 2020, 87, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Liu, L.; Murphy, K.; Ge, S.; Hotaling, J.; Aston, K.I.; Emery, B.; Carrell, D.T. Comparative analysis of three sperm DNA damage assays and sperm nuclear protein content in couples undergoing assisted reproduction treatment. Hum. Reprod. 2014, 29, 904–917. [Google Scholar] [CrossRef] [PubMed]
- Talebi, A.R.; Vahidi, S.; Aflatoonian, A.; Ghasemi, N.; Ghasemzadeh, J.; Firoozabadi, R.D.; Moein, M.R. Cytochemical evaluation of sperm chromatin and DNA integrity in couples with unexplained recurrent spontaneous abortions. Andrologia 2011, 44, 462–470. [Google Scholar] [CrossRef]
- Aoki, V.W.; Emery, B.R.; Carrell, U.T. Global sperm deoxyribonucleic acid methylation is unaffected in protamine-deficient infertile males. Fertil. Steril. 2006, 86, 1541–1543. [Google Scholar] [CrossRef]
- World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th ed.; WHO Press: Geneva, Switzerland, 2010. [Google Scholar]
- Auger, J.; Jouannet, P.; Eustache, F. Another look at human sperm morphology. Hum. Reprod. 2016, 31, 10–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Björndahl, L.; Barratt, C.L.; Mortimer, D.; Jouannet, P. ‘How to count sperm properly’: Checklist for acceptability of studies based on human semen analysis. Hum. Reprod. 2015, 31, 227–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukaszuk, K.; Jakiel, G.; Potocka, I.W.; Kiewisz, J.; Olszewska, J.; Sieg, W.; Podolak, A.; Pastuszek, E.; Wdowiak, A. IMSI—Guidelines for Sperm Quality Assessment. Diagnostics 2022, 12, 192. [Google Scholar] [CrossRef]
- Boitrelle, F.; Guthauser, B.; Alter, L.; Bailly, M.; Wainer, R.; Vialard, F.; Albert, M.; Selva, J. The nature of human sperm head vacuoles: A systematic literature review. Basic Clin. Androl. 2013, 23, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Vanderzwalmen, P.; Hiemer, A.; Rubner, P.; Bach, M.; Neyer, A.; Stecher, A.; Uher, P.; Zintz, M.; Lejeune, B.; Vanderzwalmen, S.; et al. Blastocyst development after sperm selection at high magnification is associated with size and number of nuclear vacuoles. Reprod. Biomed. Online 2008, 17, 617–627. [Google Scholar] [CrossRef]
- Bartoov, B.; Berkovitz, A.; Eltes, F.; Kogosowski, A.; Menezo, Y.; Barak, Y. Real-Time Fine Morphology of Motile Human Sperm Cells is Associated with IVF-ICSI Outcome. J. Androl. 2002, 23, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Peer, S.; Eltes, F.; Berkovitz, A.; Yehuda, R.; Itsykson, P.; Bartoov, B. Is fine morphology of the human sperm nuclei affected by in vitro incubation at 37 °C? Fertil. Steril. 2007, 88, 1589–1594. [Google Scholar] [CrossRef]
- Krejčí, J.; Stixová, L.; Pagáčová, E.; Legartová, S.; Kozubek, S.; Lochmanová, G.; Zdráhal, Z.; Sehnalová, P.; Dabravolski, S.; Hejátko, J.; et al. Post-Translational Modifications of Histones in Human Sperm. J. Cell. Biochem. 2015, 116, 2195–2209. [Google Scholar] [CrossRef] [PubMed]
- La Spina, F.A.; Romanato, M.; Brugo-Olmedo, S.; De Vincentiis, S.; Julianelli, V.; Rivera, R.M.; Buffone, M.G. Heterogeneous distribution of histone methylation in mature human sperm. J. Assist. Reprod. Genet. 2013, 31, 45–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Lalancette, C.; Miller, D.; Krawetz, S.A. Characterization of nucleohistone and nucleoprotamine components in the mature human sperm nucleus. Asian J. Androl. 2008, 10, 535–541. [Google Scholar] [CrossRef]
- Ramos, L.; Van Der Heijden, G.; Derijck, A.; Berden, J.; Kremer, J.; Van Der Vlag, J.; De Boer, P. Incomplete nuclear transformation of human spermatozoa in oligo-astheno-teratospermia: Characterization by indirect immunofluorescence of chromatin and thiol status. Hum. Reprod. 2007, 23, 259–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuen, B.T.; Bush, K.M.; Barrilleaux, B.L.; Cotterman, R.; Knoepfler, P.S. Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development 2014, 141, 3483–3494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.K.; Jee, B.C.; Kim, S.H. Histone methylation and acetylation in ejaculated human sperm: Effects of swim-up and smoking. Fertil. Steril. 2015, 103, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Jee, B.C.; Lee, J.M.; Suh, C.S.; Kim, S.H. Histone Acetylation Level and Histone Acetyltransferase/Deacetylase Activity in Ejaculated Sperm from Normozoospermic Men. Yonsei Med. J. 2014, 55, 1333–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biggs, D.S. 3D deconvolution microscopy. Curr. Protoc. Cytom. 2010, 52, 12–19. [Google Scholar] [CrossRef]
- Ghasemian, F.; Bahadori, M.H.; Hosseini Kolkooh, S.Z.; Esmaeili, M. Using Deep Learning Algorithm: The Study of Sperm Head Vacuoles and Its Correlation with Protamine mRNA Ratio. Cell J. 2022, 24, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, P.G.; Manicardi, G.C.; Bizzaro, D.; Bianchi, U.; Sakkas, D. Effect of Deoxyribonucleic Acid Protamination on Fluorochrome Staining and in Situ Nick-Translation of Murine and Human Mature Spermatozoa1. Biol. Reprod. 1993, 49, 1083–1088. [Google Scholar] [CrossRef]
- De Iuliis, G.N.; Thomson, L.K.; Mitchell, L.A.; Finnie, J.M.; Koppers, A.J.; Hedges, A.; Nixon, B.; Aitken, R.J. DNA Damage in Human Spermatozoa Is Highly Correlated with the Efficiency of Chromatin Remodeling and the Formation of 8-Hydroxy-2′-Deoxyguanosine, a Marker of Oxidative Stress1. Biol. Reprod. 2009, 81, 517–524. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Ghanghas, P.; Kaushal, N.; Kaur, J.; Kaur, P. Epigenetics and oxidative stress: A twin-edged sword in spermatogenesis. Andrologia 2019, 51, e13432. [Google Scholar] [CrossRef] [PubMed]
- Balhorn, R. Sperm Chromatin: An Overview. In A Clinician’s Guide to Sperm DNA and Chromatin Damage; Zini, A., Agarwal, A., Eds.; Springer: New York, NY, USA, 2011; pp. 3–18. [Google Scholar]
- Garolla, A.; Fortini, D.; Menegazzo, M.; De Toni, L.; Nicoletti, V.; Moretti, A.; Selice, R.; Engl, B.; Foresta, C. High-power microscopy for selecting spermatozoa for ICSI by physiological status. Reprod. Biomed. Online 2008, 17, 610–616. [Google Scholar] [CrossRef]
- Hammoud, I.; Boitrelle, F.; Ferfouri, F.; Vialard, F.; Bergere, M.; Wainer, B.; Bailly, M.; Albert, M.; Selva, J. Selection of normal spermatozoa with a vacuole-free head (x6300) improves selection of spermatozoa with intact DNA in patients with high sperm DNA fragmentation rates. Andrologia 2012, 45, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Hosseinifar, H.; Yazdanikhah, S.; Modarresi, T.; Totonchi, M.; Gilani, M.A.S.; Sabbaghian, M. Correlation between sperm DNA fragmentation index and CMA3 positive spermatozoa in globozoospermic patients. Andrology 2015, 3, 526–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López, G.; Lafuente, R.; Checa, M.A.; Carreras, R.; Brassesco, M. Diagnostic value of sperm DNA fragmentation and sperm high-magnification for predicting outcome of assisted reproduction treatment. Asian J. Androl. 2013, 15, 790–794. [Google Scholar] [CrossRef] [Green Version]
- Pastuszek, E.; Kiewisz, J.; Skowronska, P.; Liss, J.; Lukaszuk, M.; Bruszczynska, A.; Jakiel, G.; Lukaszuk, K. An investigation of the potential effect of sperm nuclear vacuoles in human spermatozoa on DNA fragmentation using a neutral and alkaline Comet assay. Andrology 2017, 5, 392–398. [Google Scholar] [CrossRef] [Green Version]
- Braun, R.E. Packaging paternal chromosomes with protamine. Nat. Genet. 2001, 28, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Teperek, M.; Simeone, A.; Gaggioli, V.; Miyamoto, K.; Allen, G.E.; Erkek, S.; Kwon, T.; Marcotte, E.M.; Zegerman, P.; Bradshaw, C.R.; et al. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016, 26, 1034–1046. [Google Scholar] [CrossRef] [Green Version]
- Carrell, D.T.; Hammoud, S.S. The human sperm epigenome and its potential role in embryonic development. Mol. Hum. Reprod. 2009, 16, 37–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, K.; Hada, M.; Fukuda, Y.; Inoue, E.; Makino, Y.; Katou, Y.; Shirahige, K.; Okada, Y. Re-evaluating the Localization of Sperm-Retained Histones Revealed the Modification-Dependent Accumulation in Specific Genome Regions. Cell Rep. 2018, 23, 3920–3932. [Google Scholar] [CrossRef]
- Xu, Q.; Xie, W. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 2018, 28, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Lambrot, R.; Chan, D.; Shao, X.; Aarabi, M.; Kwan, T.; Bourque, G.; Moskovtsev, S.; Librach, C.; Trasler, J.; Dumeaux, V.; et al. Whole-genome sequencing of H3K4me3 and DNA methylation in human sperm reveals regions of overlap linked to fertility and development. Cell Rep. 2021, 36, 109418. [Google Scholar] [CrossRef]
- Oikawa, M.; Simeone, A.; Hormanseder, E.; Teperek, M.; Gaggioli, V.; O’Doherty, A.; Falk, E.; Sporniak, M.; D’Santos, C.; Franklin, V.N.R.; et al. Epigenetic homogeneity in histone methylation underlies sperm programming for embryonic transcription. Nat. Commun. 2020, 11, 3491. [Google Scholar] [CrossRef] [PubMed]
- Lismer, A.; Siklenka, K.; LaFleur, C.; Dumeaux, V.; Kimmins, S. Sperm histone H3 lysine 4 trimethylation is altered in a genetic mouse model of transgenerational epigenetic inheritance. Nucleic Acids Res. 2020, 48, 11380–11393. [Google Scholar] [CrossRef] [PubMed]
- Pepin, A.-S.; Lafleur, C.; Lambrot, R.; Dumeaux, V.; Kimmins, S. Sperm histone H3 lysine 4 tri-methylation serves as a metabolic sensor of paternal obesity and is associated with the inheritance of metabolic dysfunction. Mol. Metab. 2022, 59, 101463. [Google Scholar] [CrossRef]
- Siklenka, K.; Erkek, S.; Godmann, M.; Lambrot, R.; McGraw, S.; Lafleur, C.; Cohen, T.; Xia, J.; Suderman, M.; Hallett, M.; et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015, 350, aab2006. [Google Scholar] [CrossRef] [PubMed]
- Blanco Rodríguez, J.; Camprubí Sánchez, C. Epigenetic Transgenerational Inheritance. Adv. Exp. Med. Biol. 2019, 1166, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Champroux, A.; Cocquet, J.; Henry-Berger, J.; Drevet, J.; Kocer, A. A Decade of Exploring the Mammalian Sperm Epigenome: Paternal Epigenetic and Transgenerational Inheritance. Front. Cell Dev. Biol. 2018, 6, 50. [Google Scholar] [CrossRef] [Green Version]
- Hart, K.; Tadros, N.N. The role of environmental factors and lifestyle on male reproductive health, the epigenome, and resulting offspring. Panminerva Med. 2019, 61, 187–195. [Google Scholar] [CrossRef]
- King, S.E.; McBirney, M.; Beck, D.; Sadler-Riggleman, I.; Nilsson, E.; Skinner, M.K. Sperm epimutation biomarkers of obesity and pathologies following DDT induced epigenetic transgenerational inheritance of disease. Environ. Epigenetics 2019, 5, dvz008. [Google Scholar] [CrossRef] [Green Version]
- Lewens, T. Blurring the germline: Genome editing and transgenerational epigenetic inheritance. Bioethics 2019, 34, 7–15. [Google Scholar] [CrossRef]
- Shukla, A.; Bunkar, N.; Kumar, R.; Bhargava, A.; Tiwari, R.; Chaudhury, K.; Goryacheva, I.Y.; Mishra, P.K. Air pollution associated epigenetic modifications: Transgenerational inheritance and underlying molecular mechanisms. Sci. Total Environ. 2018, 656, 760–777. [Google Scholar] [CrossRef]
- Kaufman, P.D.; Rando, O.J. Chromatin as a potential carrier of heritable information. Curr. Opin. Cell Biol. 2010, 22, 284–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legoff, L.; D’Cruz, S.C.; Tevosian, S.; Primig, M.; Smagulova, F. Transgenerational Inheritance of Environmentally Induced Epigenetic Alterations during Mammalian Development. Cells 2019, 8, 1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkies, P. Molecular mechanisms of epigenetic inheritance: Possible evolutionary implications. Semin. Cell Dev. Biol. 2019, 97, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Torres-Flores, U.; Hernández-Hernández, A. The Interplay Between Replacement and Retention of Histones in the Sperm Genome. Front. Genet. 2020, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- Ben Maamar, M.; Sadler-Riggleman, I.; Beck, D.; McBirney, M.; Nilsson, E.; Klukovich, R.; Xie, Y.; Tang, C.; Yan, W.; Skinner, M.K. Alterations in sperm DNA methylation, non-coding RNA expression, and histone retention mediate vinclozolin-induced epigenetic transgenerational inheritance of disease. Environ. Epigenetics 2018, 4, dvy010. [Google Scholar] [CrossRef] [PubMed]
- Schagdarsurengin, U.; Steger, K. Epigenetics in male reproduction: Effect of paternal diet on sperm quality and offspring health. Nat. Rev. Urol. 2016, 13, 584–595. [Google Scholar] [CrossRef]
- Skinner, M.K.; Ben Maamar, M.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; McBirney, M.; Klukovich, R.; Xie, Y.; Tang, C.; Yan, W. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin 2018, 11, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Terashima, M.; Barbour, S.; Ren, J.; Yu, W.; Han, Y.; Muegge, K. Effect of high fat diet on paternal sperm histone distribution and male offspring liver gene expression. Epigenetics 2015, 10, 861–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lismer, A.; Dumeaux, V.; Lafleur, C.; Lambrot, R.; Brind’Amour, J.; Lorincz, M.C.; Kimmins, S. Histone H3 lysine 4 trimethylation in sperm is transmitted to the embryo and associated with diet-induced phenotypes in the offspring. Dev. Cell 2021, 56, 671–686.e6. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K. Sure, Fathers Give Birth, Too!: Postnatal paternal folate deficiency increases congenital disabilities through H3K4me3 histone methylation changes in sperm and embryos. Mol. Cells 2021, 44, 696–698. [Google Scholar] [CrossRef] [PubMed]

| Patients | Age | Etiology | Abstinence Time (Days) | Semen Volume (mL) | Sperm Concentration (106/mL) | Total Sperm Count (106/ Ejaculate) | Sperm Vitality (%) | Sperm Total Motility (%) | Sperm Progressive Motility (%) | Sperm Typical Forms 1 (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 34 | Exclusively female | 2 | 3.2 | 34.3 | 109.8 | 78 | 60 | 40 | 25 |
| 2 | 42 | Idiopathic | 2 | 4.3 | 21.6 | 92.9 | 83 | 45 | 35 | 31 |
| 3 | 32 | Exclusively female | 3 | 2.6 | 42.7 | 111.0 | 65 | 50 | 40 | 24 |
| 4 | 38 | Idiopathic | 2 | 2.8 | 57.2 | 160.2 | 71 | 60 | 45 | 36 |
| 5 | 29 | Idiopathic | 4 | 5.9 | 19.9 | 117.4 | 81 | 60 | 50 | 39 |
| 6 | 26 | Exclusively female | 3 | 3.3 | 33.2 | 109.6 | 82 | 50 | 40 | 33 |
| 7 | 44 | Idiopathic | 5 | 4.1 | 48.3 | 198.0 | 76 | 60 | 45 | 27 |
| 8 | 48 | Idiopathic | 4 | 2.3 | 29.0 | 66.7 | 79 | 60 | 55 | 29 |
| 9 | 31 | Idiopathic | 3 | 2.6 | 28.3 | 73.6 | 64 | 45 | 35 | 32 |
| 10 | 32 | Exclusively female | 4 | 2.7 | 46.1 | 124.5 | 69 | 50 | 35 | 25 |
| 11 | 28 | Exclusively male | 2 | 6.4 | 2.2 | 14.1 | 64 | 30 | 20 | 13 |
| 12 | 34 | Combined male and female | 5 | 1.9 | 8.6 | 16.3 | 76 | 50 | 30 | 21 |
| 13 | 26 | Exclusively male | 2 | 5.8 | 1.9 | 11.0 | 45 | 20 | 15 | 5 |
| 14 | 47 | Exclusively male | 2 | 3.1 | 5.7 | 17.7 | 77 | 40 | 30 | 12 |
| 15 | 39 | Combined male and female | 3 | 4.2 | 6.3 | 26.5 | 71 | 40 | 30 | 17 |
| 16 | 44 | Combined male and female | 3 | 1.7 | 7.2 | 12.2 | 48 | 20 | 10 | 2 |
| 17 | 32 | Combined male and female | 2 | 2.5 | 10.3 | 25.8 | 52 | 30 | 15 | 13 |
| 18 | 38 | Exclusively male | 4 | 2.1 | 11.9 | 25.0 | 56 | 30 | 20 | 18 |
| 19 | 27 | Exclusively male | 2 | 3.0 | 9.4 | 28.2 | 82 | 50 | 30 | 20 |
| 20 | 33 | Combined male and female | 4 | 3.2 | 10.1 | 32.3 | 64 | 40 | 30 | 8 |
| Nuclear Proteins (n = 7) | |
|---|---|
| Protamine (2) | protamine 1, protamine 2 |
| Core histones (5) | H1, H2A, H2B, H3, H4 |
| Protamine epigenetic marks (n = 3) | |
| P1 (3) | ph1, ph27ac1, ph3 |
| P2 (0) | |
| Histone variants (n = 23) | |
| H1 (3) | H1.4, H1.t, H1.t2 |
| H2A (11) | H2A.1a, H2A.2b, H2A.2c, H2A.3, H2A.J, H2A.V, H2A.X, H2A-bbd 2/3, H2A.Z, macroH2A.1, macroH2A.2 |
| H2B (6) | tH2B., H2B.1b, H2B.1c/e/f, H2B.1d, H2B.1 l, H2B.2f |
| H3 (3) | H3, H3.3, H3.1t |
| H4 (0) | |
| Histones PTMs (n = 104) | |
| H1 (4) | K43ac, R50me1, K62me1, K63ac |
| H1t (13) | K112me1, K113ac, K122ac, K124me1, K170ac, K173me1, K183ac, K183me3, R185me1, S180ph, S187ph, K188me1, K190ac |
| H2A (3) | K5ac, R11me2, R29me1 |
| H2AV/Z (21) | K4ac, K4me2/3, K7ac, K7cr, K7me1/2/3, S9ph, K11cr, K11me1/2/3, K13cr, K13me2/3, K15ac, R19me1, K27ac, K37cr, K37me1 |
| H2B (2) | K16me1, K20me3 |
| tH2B (15) | K6ac, T9ph, K12ac, K12me1/3, K13me1/3, K16me1/3, K17me1, K28ac, K29ac, R30me2, K86ac, R87me1 |
| H3 (31) | T3ph, K4me1/2/3, K9ac, K9me1/2/3, K14ac, K18ac, K18me1, K23ac, K23me1/3, R26me1/2 K27me1/2/3, K36cr, K36me1/2/3, K37ac, K37me2/3, R53me1, K56ac, K79me1/2, K12Ox |
| H4 (15) | S1ph, R3me1, K5ac, K8ac, K12ac, K16ac, K9ac, K20me1/2/3, K31ac, R35me1, M84Ox, K91ac, R92me1 |
| Pairing | Rabbit Antibody Cy3 (Red) | Mouse Antibody FITC (Green) |
|---|---|---|
| 1 | Pan H3 Millipore 06-755 | Protamine 2 Briar Patch Biosciences Hup-2B |
| 2 | H3K27me3 Millipore 07-449 | H3K9me2 Active motif 39683 clone MABI 0307 |
| 3 | H3K9me1 Millipore ABE101 | H3K4me2 Millipore 05-1338 clone CMA303 |
| 4 | H3K4me3 Abcam ab8580 | H4K20me2 GeneTex GTX630545 clone GT1851 |
| 5 | H3K9me3 Active motif 39765 | H3K4me1 GeneTex GTX50902 clone MABI0302 |
| Non-Vacuolated Sperm (%) | Vacuolated Sperm (%) | ||||||
|---|---|---|---|---|---|---|---|
| Nuclear Proteins and PTMs | Mean ± SEM | Median | Q3–Q1 | Mean ± SEM | Median | Q3–Q1 | p-Value |
| H3 | 74.8 ± 4.8 | 80.9 | 62.5–90 | 88.1 ± 2.7 | 90 | 83.7–95 | 0.009 * |
| P2 | 82.1 ± 4.4 | 88.8 | 67.8–100 | 50.2 ± 6.2 | 35 | 28.8–76.3 | <0.001 * |
| H3K4me1 | 5.4 ± 0.8 | 5.0 | 3.1–8 | 6.2 ± 0.8 | 8 | 5–8.5 | 0.325 |
| H3K4me2 | 6.4 ± 1.0 | 5.0 | 5.0–10 | 6.3 ± 0.9 | 6.5 | 5– 8.3 | 0.71 |
| H3K4me3 | 49.1 ± 7.4 | 46.3 | 13.8–76.7 | 78.5 ± 5.2 | 88.2 | 73.3–95 | <0.001 * |
| H3K9me1 | 31.1 ± 6.3 | 25 | 10–43.5 | 42.3 ± 4.8 | 35 | 33.3–68.2 | 0.165 |
| H3K9me2 | 22.6 ± 6.5 | 7.5 | 0.0–32.5 | 15.4 ± 4.7 | 73 | 0.0–25.9 | 0.13 |
| H3K9me3 | 61.8 ± 6.9 | 69.1 | 26.7–85.7 | 62.2 ± 4.9 | 65 | 47.7–71.2 | 0.911 |
| H3K27me3 | 73.6 ± 5.1 | 85.7 | 58.8–90 | 63.9 ± 6.3 | 79.2 | 42.5–85.9 | 0.028 * |
| H4K20me2 | 18.7 ± 2.6 | 17.4 | 9.9–27.3 | 24.7 ± 4.5 | 17.4 | 9.1–31 | 0.121 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bendayan, M.; Caceres, L.; Saïs, E.; Swierkowski-Blanchard, N.; Alter, L.; Bonnet-Garnier, A.; Boitrelle, F. Human Sperm Morphology as a Marker of Its Nuclear Quality and Epigenetic Pattern. Cells 2022, 11, 1788. https://doi.org/10.3390/cells11111788
Bendayan M, Caceres L, Saïs E, Swierkowski-Blanchard N, Alter L, Bonnet-Garnier A, Boitrelle F. Human Sperm Morphology as a Marker of Its Nuclear Quality and Epigenetic Pattern. Cells. 2022; 11(11):1788. https://doi.org/10.3390/cells11111788
Chicago/Turabian StyleBendayan, Marion, Liliana Caceres, Emine Saïs, Nelly Swierkowski-Blanchard, Laura Alter, Amélie Bonnet-Garnier, and Florence Boitrelle. 2022. "Human Sperm Morphology as a Marker of Its Nuclear Quality and Epigenetic Pattern" Cells 11, no. 11: 1788. https://doi.org/10.3390/cells11111788






