Dysregulation of Immune Response Mediators and Pain-Related Ion Channels Is Associated with Pain-like Behavior in the GLA KO Mouse Model of Fabry Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Groups
2.2. Tissue Collection
2.3. Immunohistochemistry
2.4. Bright Field and Fluorescence Microscopy
2.5. Image Analysis
2.6. Gene Expression Analysis
2.7. Behavioral Testing
2.8. Statistics
3. Results
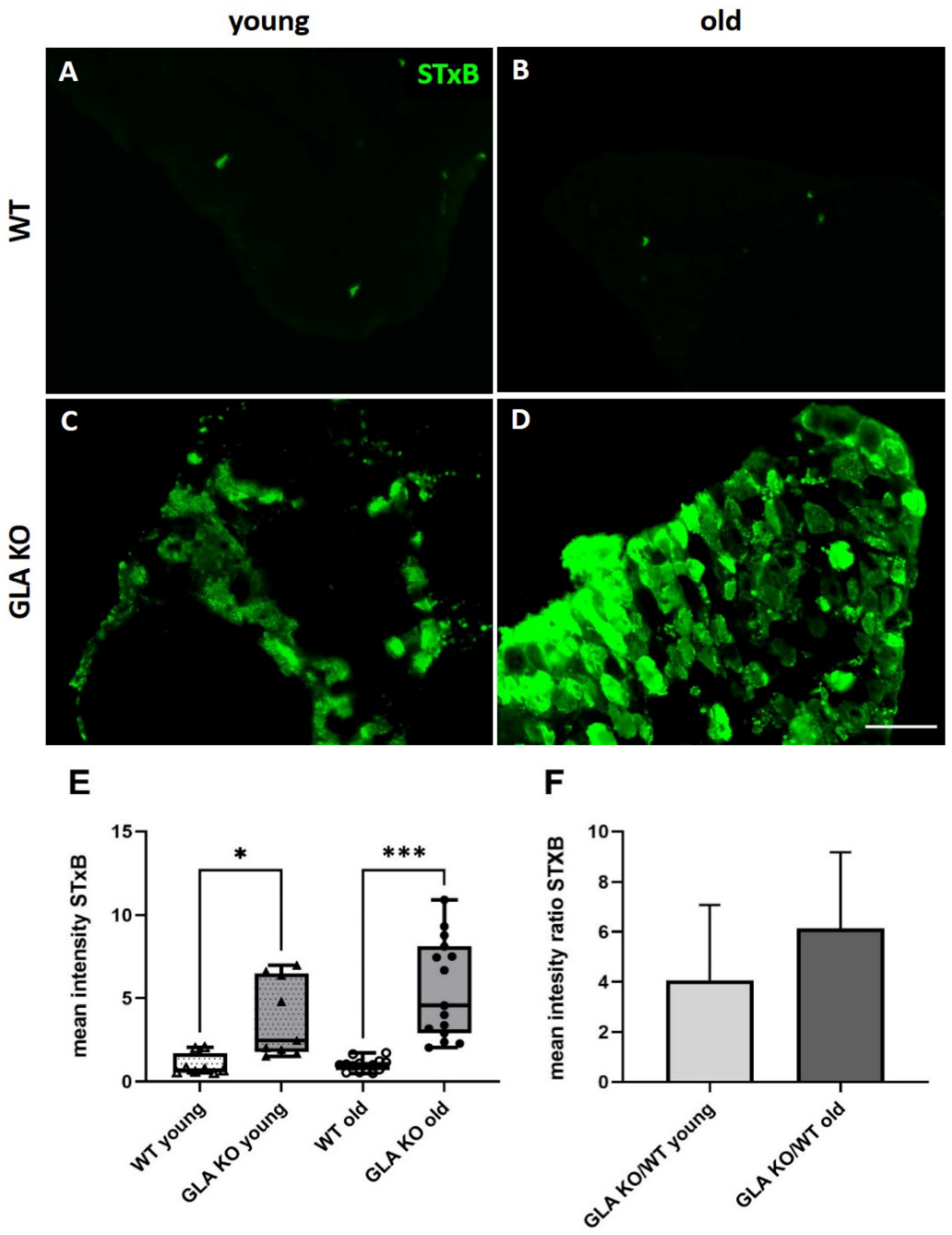
3.1. Higher Gb3 Load in DRG of Young and Old GLA KO Compared to WT Mice
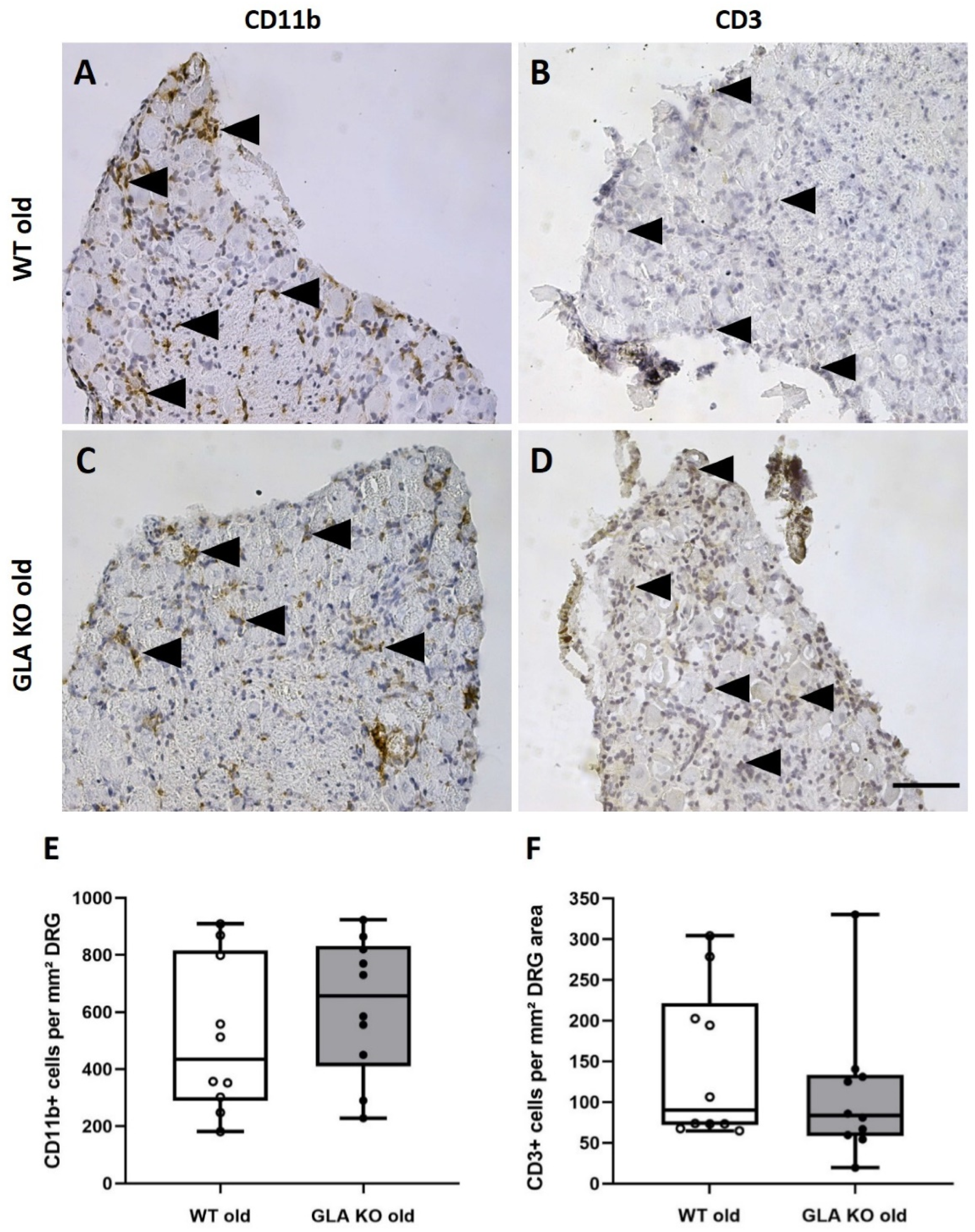
3.2. No Macrophage or T-Cell Infiltration in DRG of Old GLA KO Mice
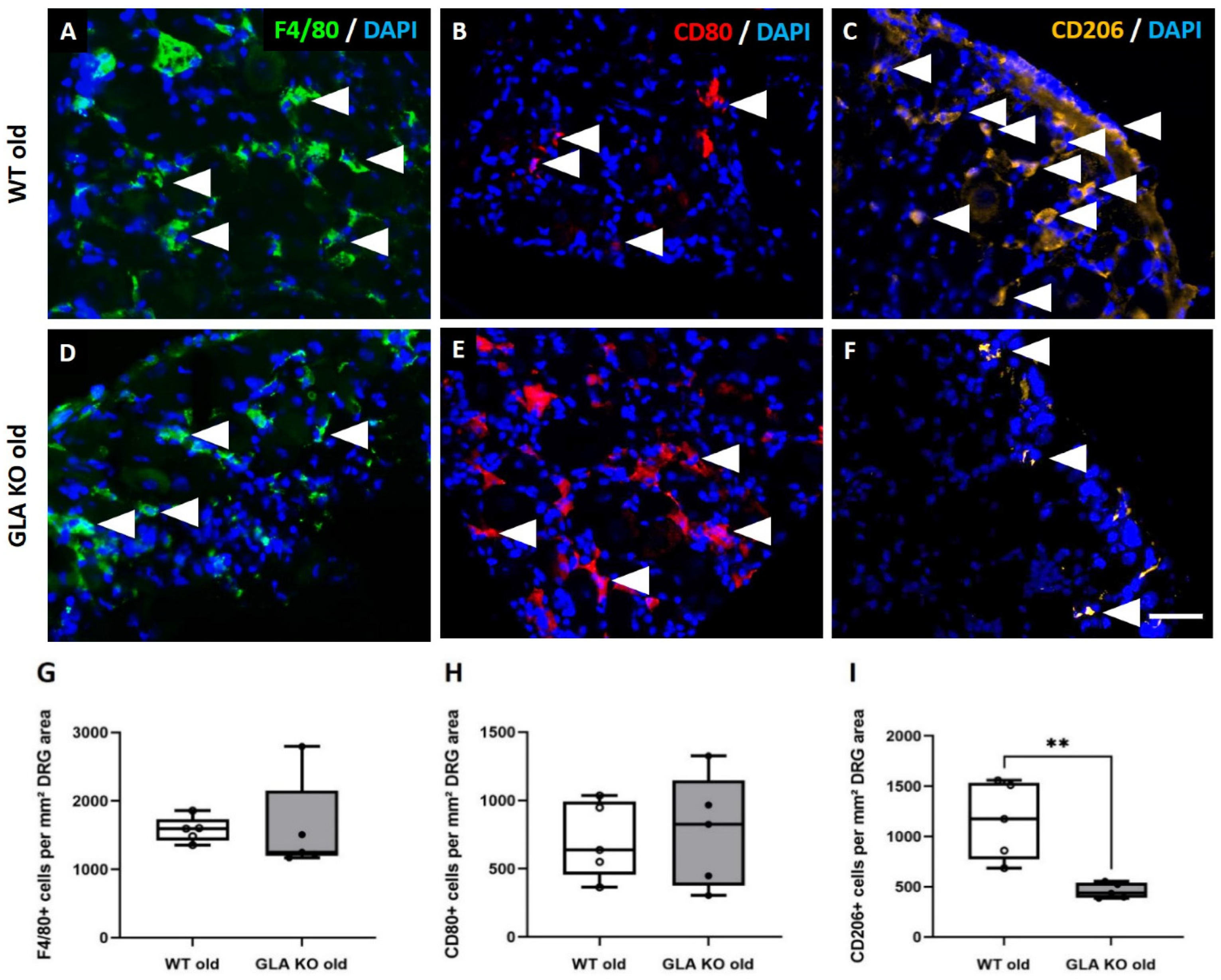
3.3. Lower M2 Phenotype Macrophage Differentiation in DRG of Old GLA KO Compared to Old WT Mice
3.4. Lower Expression of Inflammation-Associated Mediators IL1b, IL10, GFAP, and LRG1 in DRG of Old GLA KO Compared to Old WT Mice
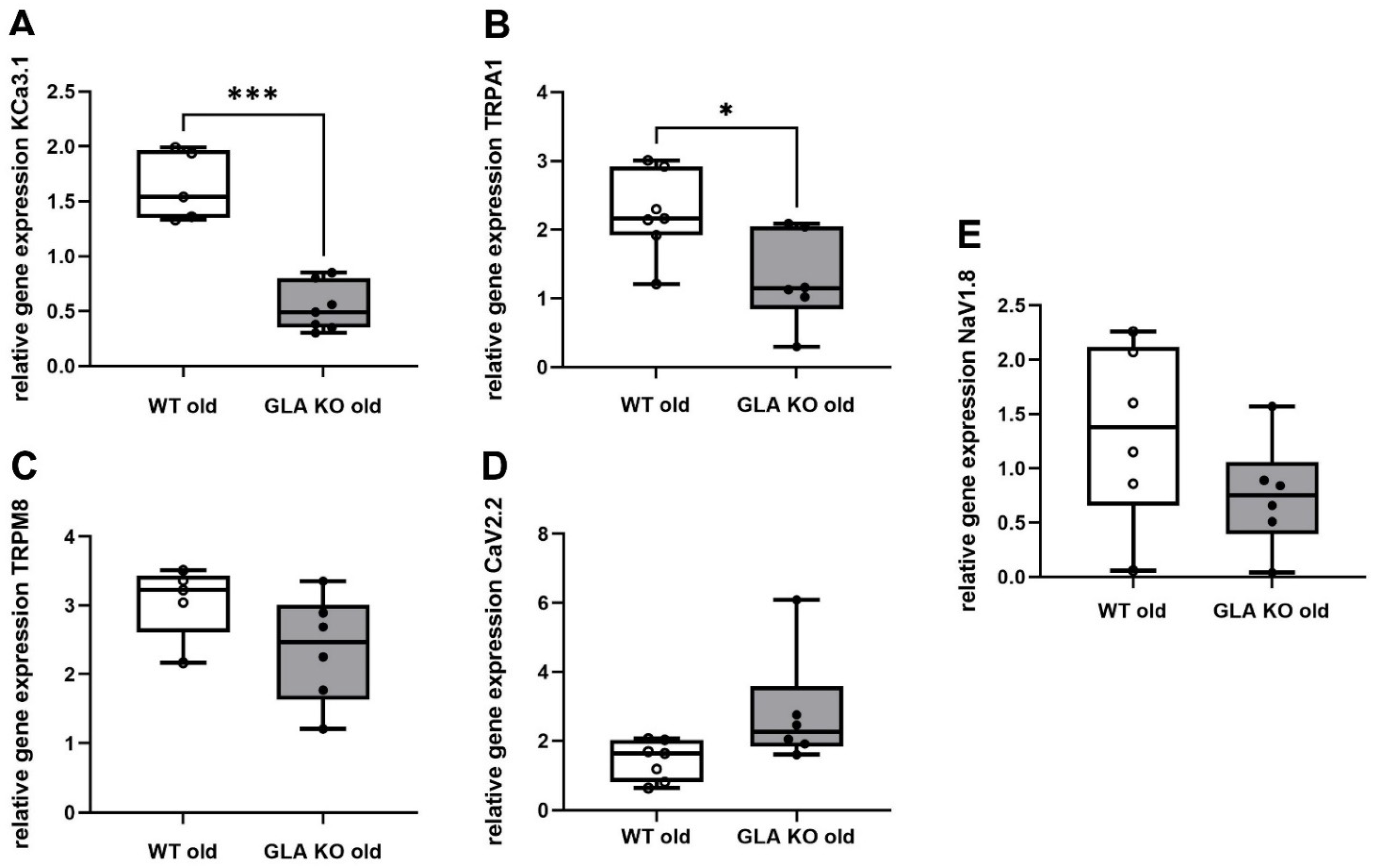
3.5. Lower Ion Channel Gene Expression of KCa3.1 and TRPA1 in DRG of Old GLA KO Compared to Old WT Mice
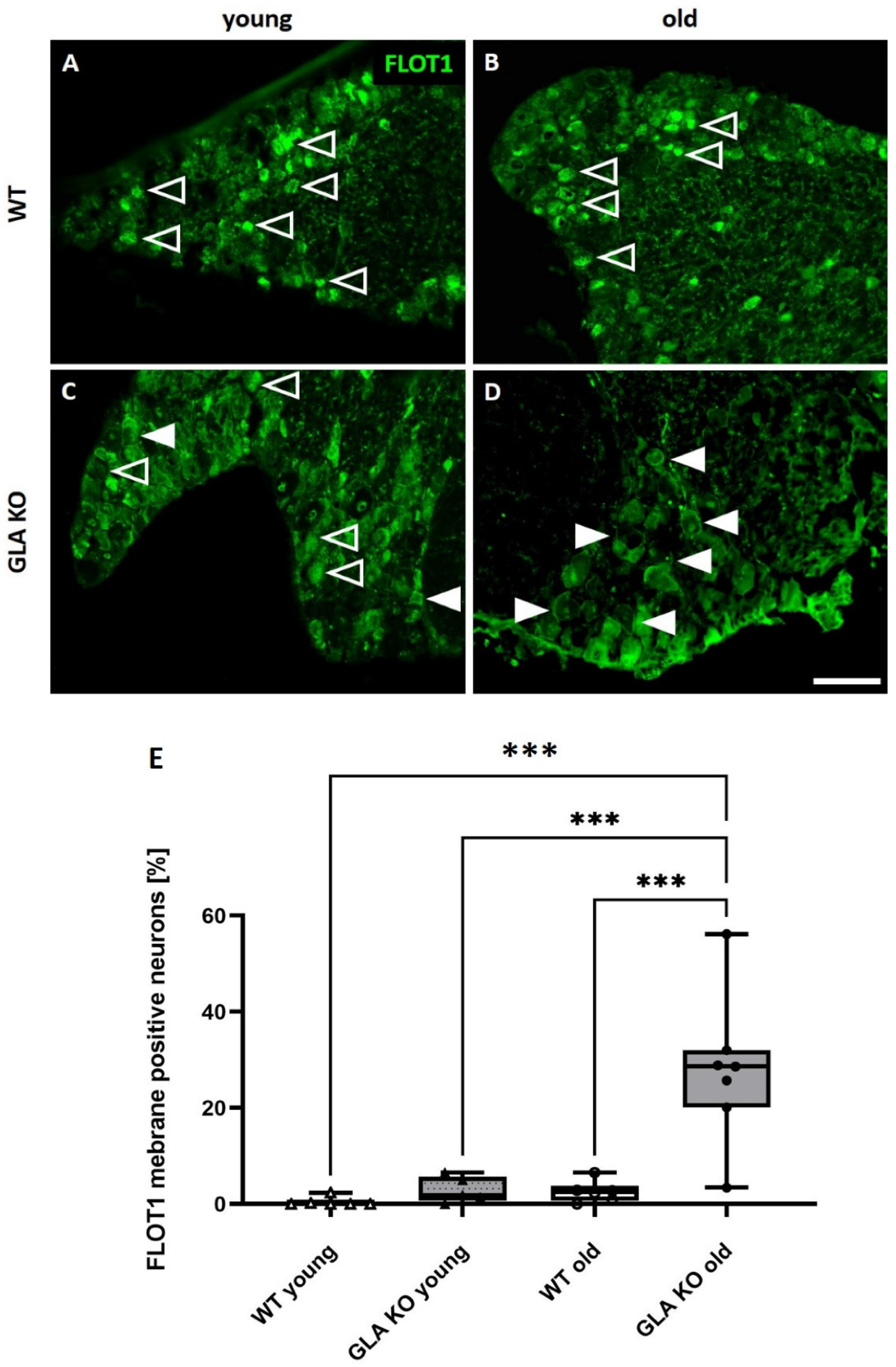
3.6. Old GLA KO Mice Show a Higher Number of FLOT1+ DRG Neurons Displaying a Membranous Distribution Pattern Compared to Young WT and GLA KO and Old WT Mice
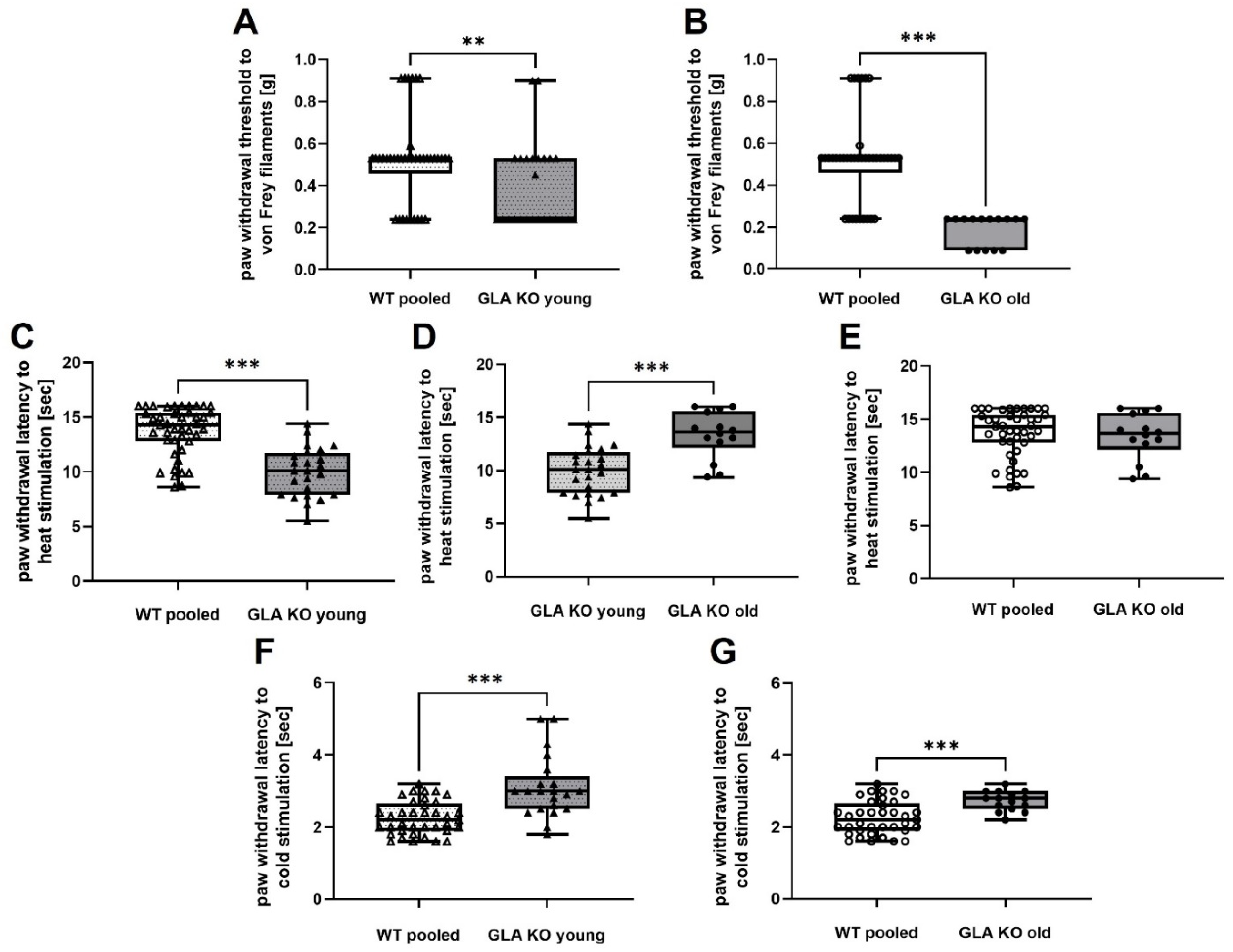
3.7. Old GLA KO Mice Show Mechanical Hypersensitivity and Cold Hyposensitivity Compared to WT Mice, and Age-Dependent Heat Hyposensitivity Compared to Young GLA KO Mice
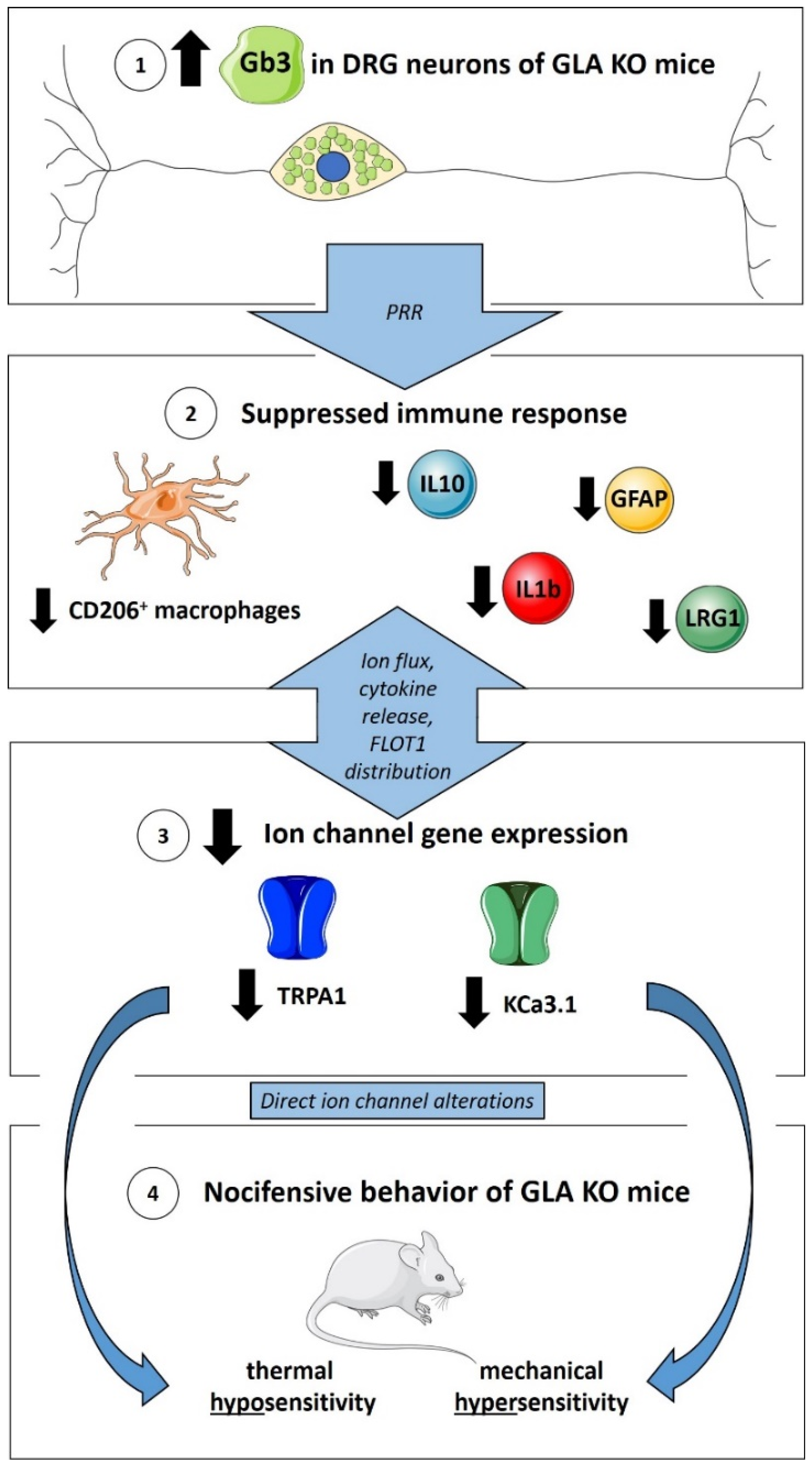
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cairns, T.; Muntze, J.; Gernert, J.; Spingler, L.; Nordbeck, P.; Wanner, C. Hot topics in Fabry disease. Postgrad. Med. J. 2018, 94, 709–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, C.R.; Gahl, W.A. Lysosomal storage diseases. Transl. Sci. Rare Dis. 2017, 2, 1–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bangari, D.S.; Ashe, K.M.; Desnick, R.J.; Maloney, C.; Lydon, J.; Piepenhagen, P.; Budman, E.; Leonard, J.P.; Cheng, S.H.; Marshall, J.; et al. alpha-Galactosidase A knockout mice: Progressive organ pathology resembles the type 2 later-onset phenotype of Fabry disease. Am. J. Pathol. 2015, 185, 651–665. [Google Scholar] [CrossRef] [PubMed]
- Weissmann, C.; Albanese, A.A.; Contreras, N.E.; Gobetto, M.N.; Castellanos, L.C.S.; Uchitel, O.D. Ion channels and pain in Fabry disease. Mol. Pain 2021, 17, 17448069211033172. [Google Scholar] [CrossRef] [PubMed]
- Simonetta, I.; Tuttolomondo, A.; Daidone, M.; Miceli, S.; Pinto, A. Treatment of Anderson-Fabry Disease. Curr. Pharm. Des. 2020, 26, 5089–5099. [Google Scholar] [CrossRef]
- Choi, L.; Vernon, J.; Kopach, O.; Minett, M.S.; Mills, K.; Clayton, P.T.; Meert, T.; Wood, J.N. The Fabry disease-associated lipid Lyso-Gb3 enhances voltage-gated calcium currents in sensory neurons and causes pain. Neurosci. Lett. 2015, 594, 163–168. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, L.; Hose, D.; Griesshammer, A.; Blum, R.; Doring, F.; Dib-Hajj, S.; Waxman, S.; Sommer, C.; Wischmeyer, E.; Üçeyler, N. Characterization of small fiber pathology in a mouse model of Fabry disease. Elife 2018, 7, e39300. [Google Scholar] [CrossRef]
- Kummer, K.K.; Kalpachidou, T.; Kress, M.; Langeslag, M. Signatures of Altered Gene Expression in Dorsal Root Ganglia of a Fabry Disease Mouse Model. Front. Mol. Neurosci. 2017, 10, 449. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.J.; Aoki, K.; Mascari, C.A.; Beltrame, A.K.; Sokumbi, O.; North, P.E.; Tiemeyer, M.; Kriegel, A.J.; Dahms, N.M. alpha-Galactosidase A-deficient rats accumulate glycosphingolipids and develop cardiorenal phenotypes of Fabry disease. FASEB J. 2019, 33, 418–429. [Google Scholar] [CrossRef] [Green Version]
- Godel, T.; Pham, M.; Heiland, S.; Bendszus, M.; Baumer, P. Human dorsal-root-ganglion perfusion measured in-vivo by MRI. Neuroimage 2016, 141, 81–87. [Google Scholar] [CrossRef] [Green Version]
- Üçeyler, N.; Urlaub, D.; Mayer, C.; Uehlein, S.; Held, M.; Sommer, C. Tumor necrosis factor-alpha links heat and inflammation with Fabry pain. Mol. Genet. Metab. 2019, 127, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Godel, T.; Baumer, P.; Pham, M.; Kohn, A.; Muschol, N.; Kronlage, M.; Kollmer, J.; Heiland, S.; Bendszus, M.; Mautner, V.F. Human dorsal root ganglion in vivo morphometry and perfusion in Fabry painful neuropathy. Neurology 2017, 89, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.S.; Chang, C.Y.; Jeon, S.B.; Yoon, H.J.; Ahn, Y.H.; Kim, H.S.; Kim, I.H.; Jeon, S.H.; Johnson, R.S.; Park, E.J. The HIF-1/glial TIM-3 axis controls inflammation-associated brain damage under hypoxia. Nat. Commun. 2015, 6, 6340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohshima, T.; Murray, G.J.; Swaim, W.D.; Longenecker, G.; Quirk, J.M.; Cardarelli, C.O.; Sugimoto, Y.; Pastan, I.; Gottesman, M.M.; Brady, R.O.; et al. alpha-Galactosidase A deficient mice: A model of Fabry disease. Proc. Natl. Acad. Sci. USA 1997, 94, 2540–2544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Üçeyler, N.; Ganendiran, S.; Kramer, D.; Sommer, C. Characterization of pain in fabry disease. Clin. J. Pain 2014, 30, 915–920. [Google Scholar] [CrossRef]
- Üçeyler, N.; He, L.; Schonfeld, D.; Kahn, A.K.; Reiners, K.; Hilz, M.J.; Breunig, F.; Sommer, C. Small fibers in Fabry disease: Baseline and follow-up data under enzyme replacement therapy. J. Peripher. Nerv. Syst. 2011, 16, 304–314. [Google Scholar] [CrossRef]
- Üçeyler, N.; Schroter, N.; Kafke, W.; Kramer, D.; Wanner, C.; Weidemann, F.; Sommer, C. Skin Globotriaosylceramide 3 Load Is Increased in Men with Advanced Fabry Disease. PLoS ONE 2016, 11, e0166484. [Google Scholar] [CrossRef] [Green Version]
- Hakimizadeh, E.; Shamsizadeh, A.; Roohbakhsh, A.; Arababadi, M.K.; Hajizadeh, M.R.; Shariati, M.; Rahmani, M.R.; Allahtavakoli, M. Inhibition of transient receptor potential vanilloid-1 confers neuroprotection, reduces tumor necrosis factor-alpha, and increases IL-10 in a rat stroke model. Fundam. Clin. Pharm. 2017, 31, 420–428. [Google Scholar] [CrossRef]
- Matsui, M.; Kajikuri, J.; Endo, K.; Kito, H.; Ohya, S. KCa3.1 inhibition-induced activation of the JNK/c-Jun signaling pathway enhances IL-10 expression in peripherally-induced regulatory T cells. J. Pharm. Sci. 2022, 148, 1–5. [Google Scholar] [CrossRef]
- Matsui, M.; Kajikuri, J.; Kito, H.; Endo, K.; Hasegawa, Y.; Murate, S.; Ohya, S. Inhibition of Interleukin 10 Transcription through the SMAD2/3 Signaling Pathway by Ca2+-Activated K+ Channel KCa3.1 Activation in Human T-Cell Lymphoma HuT-78 Cells. Mol. Pharm. 2019, 95, 294–302. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.M.; Itson-Zoske, B.; Cai, Y.; Qiu, C.; Pan, B.; Stucky, C.L.; Hogan, Q.H.; Yu, H. Satellite glial cells in sensory ganglia express functional transient receptor potential ankyrin 1 that is sensitized in neuropathic and inflammatory pain. Mol. Pain 2020, 16, 1744806920925425. [Google Scholar] [CrossRef] [PubMed]
- Peiris, M.; Weerts, Z.; Aktar, R.; Masclee, A.A.M.; Blackshaw, A.; Keszthelyi, D. A putative anti-inflammatory role for TRPM8 in irritable bowel syndrome-An exploratory study. Neurogastroenterol. Motil. 2021, 33, e14170. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xie, M.X.; Hu, L.; Wang, X.F.; Mai, J.Z.; Li, Y.Y.; Wu, N.; Zhang, C.; Li, J.; Pang, R.P.; et al. Upregulation of N-type calcium channels in the soma of uninjured dorsal root ganglion neurons contributes to neuropathic pain by increasing neuronal excitability following peripheral nerve injury. Brain Behav. Immun. 2018, 71, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.F.; Zhu, H.Q.; Wei, X.H.; Wang, J.; Li, Y.Y.; Pang, R.P.; Liu, X.G. Interleukin-10 down-regulates voltage gated sodium channels in rat dorsal root ganglion neurons. Exp. Neurol. 2013, 247, 466–475. [Google Scholar] [CrossRef]
- Kwiatkowska, K.; Matveichuk, O.V.; Fronk, J.; Ciesielska, A. Flotillins: At the Intersection of Protein S-Palmitoylation and Lipid-Mediated Signaling. Int. J. Mol. Sci. 2020, 21, 2283. [Google Scholar] [CrossRef] [Green Version]
- Riento, K.; Zhang, Q.; Clark, J.; Begum, F.; Stephens, E.; Wakelam, M.J.; Nichols, B.J. Flotillin proteins recruit sphingosine to membranes and maintain cellular sphingosine-1-phosphate levels. PLoS ONE 2018, 13, e0197401. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Chaplan, S.R.; Bach, F.W.; Pogrel, J.W.; Chung, J.M.; Yaksh, T.L. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 1994, 53, 55–63. [Google Scholar] [CrossRef]
- Hargreaves, K.; Dubner, R.; Brown, F.; Flores, C.; Joris, J. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain 1988, 32, 77–88. [Google Scholar] [CrossRef]
- Brenner, D.S.; Golden, J.P.; Gereau, R.W., IV. A novel behavioral assay for measuring cold sensation in mice. PLoS ONE 2012, 7, e39765. [Google Scholar] [CrossRef] [Green Version]
- Üçeyler, N.; Biko, L.; Hose, D.; Hofmann, L.; Sommer, C. Comprehensive and differential long-term characterization of the alpha-galactosidase A deficient mouse model of Fabry disease focusing on the sensory system and pain development. Mol. Pain 2016, 12, 1744806916646379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozenfeld, P.; Feriozzi, S. Contribution of inflammatory pathways to Fabry disease pathogenesis. Mol. Genet. Metab. 2017, 122, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Lakoma, J.; Rimondini, R.; Ferrer Montiel, A.; Donadio, V.; Liguori, R.; Caprini, M. Increased expression of Trpv1 in peripheral terminals mediates thermal nociception in Fabry disease mouse model. Mol. Pain 2016, 12, 1744806916663729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, S.; Mardell, C.; Wheatland, L.; Zola, H.; Macardle, P.J. A comparison of Verotoxin B-subunit (Stx1B) and CD77 antibody to define germinal centre populations. Cell. Immunol. 2005, 236, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Melton-Celsa, A.R. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol. Spectr. 2014, 2, 2–4. [Google Scholar] [CrossRef] [Green Version]
- Jeannin, P.; Jaillon, S.; Delneste, Y. Pattern recognition receptors in the immune response against dying cells. Curr. Opin. Immunol. 2008, 20, 530–537. [Google Scholar] [CrossRef]
- DeGraba, T.; Azhar, S.; Dignat-George, F.; Brown, E.; Boutiere, B.; Altarescu, G.; McCarron, R.; Schiffmann, R. Profile of endothelial and leukocyte activation in Fabry patients. Ann. Neurol. 2000, 47, 229–233. [Google Scholar] [CrossRef]
- Maruyama, H.; Taguchi, A.; Mikame, M.; Lu, H.; Tada, N.; Ishijima, M.; Kaneko, H.; Kawai, M.; Goto, S.; Saito, A.; et al. Low bone mineral density due to secondary hyperparathyroidism in the Gla(tm)Tg(CAG-A4GALT) mouse model of Fabry disease. FASEB Bioadv. 2020, 2, 365–381. [Google Scholar] [CrossRef]
- Maruyama, H.; Taguchi, A.; Nishikawa, Y.; Guili, C.; Mikame, M.; Nameta, M.; Yamaguchi, Y.; Ueno, M.; Imai, N.; Ito, Y.; et al. Medullary thick ascending limb impairment in the Gla(tm)Tg(CAG-A4GALT) Fabry model mice. FASEB J. 2018, 32, 4544–4559. [Google Scholar] [CrossRef] [Green Version]
- Zigmond, R.E.; Echevarria, F.D. Macrophage biology in the peripheral nervous system after injury. Prog. Neurobiol. 2019, 173, 102–121. [Google Scholar] [CrossRef]
- Makita, N.; Hizukuri, Y.; Yamashiro, K.; Murakawa, M.; Hayashi, Y. IL-10 enhances the phenotype of M2 macrophages induced by IL-4 and confers the ability to increase eosinophil migration. Int. Immunol. 2015, 27, 131–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, S.; Wang, Y.; Yao, Y.; Cao, Z.; Yin, J.; Zi, L.; Chen, H.; Fu, Y.; Wang, X.; Zhao, Q. Inhibition of KCa3.1 Channels Suppresses Atrial Fibrillation via the Attenuation of Macrophage Pro-inflammatory Polarization in a Canine Model with Prolonged Rapid Atrial Pacing. Front. Cardiovasc. Med. 2021, 8, 656631. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Wang, X.; Shao, L.; Jiang, G.T.; Min, J.W.; Mei, X.Y.; He, X.H.; Liu, W.H.; Huang, W.X.; Peng, B.W. TRPV1 mediates astrocyte activation and interleukin-1beta release induced by hypoxic ischemia (HI). J. Neuroinflamm. 2019, 16, 114. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, T.; Hagman, S.; Ristola, M.; Sukki, L.; Veijula, K.; Kreutzer, J.; Kallio, P.; Narkilahti, S. Co-stimulation with IL-1beta and TNF-alpha induces an inflammatory reactive astrocyte phenotype with neurosupportive characteristics in a human pluripotent stem cell model system. Sci. Rep. 2019, 9, 16944. [Google Scholar] [CrossRef] [PubMed]
- Berta, T.; Qadri, Y.; Tan, P.H.; Ji, R.R. Targeting dorsal root ganglia and primary sensory neurons for the treatment of chronic pain. Expert Opin. Ther. Targets 2017, 21, 695–703. [Google Scholar] [CrossRef]
- Lontra, M.B.; Savaris, R.F.; Cavazzola, L.T.; Maissiat, J. Comparison of leucine-rich alpha-2-glycoprotein-1 (LRG-1) plasma levels between patients with and without appendicitis, a case-controlled study. Sci. Rep. 2021, 11, 5574. [Google Scholar] [CrossRef]
- Yasutomi, E.; Inokuchi, T.; Hiraoka, S.; Takei, K.; Igawa, S.; Yamamoto, S.; Ohmori, M.; Oka, S.; Yamasaki, Y.; Kinugasa, H.; et al. Leucine-rich alpha-2 glycoprotein as a marker of mucosal healing in inflammatory bowel disease. Sci. Rep. 2021, 11, 11086. [Google Scholar] [CrossRef]
- Hong, Q.; Zhang, L.; Fu, J.; Verghese, D.A.; Chauhan, K.; Nadkarni, G.N.; Li, Z.; Ju, W.; Kretzler, M.; Cai, G.Y.; et al. LRG1 Promotes Diabetic Kidney Disease Progression by Enhancing TGF-beta-Induced Angiogenesis. J. Am. Soc. Nephrol. 2019, 30, 546–562. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Sun, H.; Liu, D.; Wang, H.; Liu, Q.; Chen, H.; Zhong, D.; Li, G. LRG1 Promotes Apoptosis and Autophagy through the TGFbeta-smad1/5 Signaling Pathway to Exacerbate Ischemia/Reperfusion Injury. Neuroscience 2019, 413, 123–134. [Google Scholar] [CrossRef]
- Lu, R.; Flauaus, C.; Kennel, L.; Petersen, J.; Drees, O.; Kallenborn-Gerhardt, W.; Ruth, P.; Lukowski, R.; Schmidtko, A. KCa3.1 channels modulate the processing of noxious chemical stimuli in mice. Neuropharmacology 2017, 125, 386–395. [Google Scholar] [CrossRef]
- Shang, S.; Zhu, F.; Liu, B.; Chai, Z.; Wu, Q.; Hu, M.; Wang, Y.; Huang, R.; Zhang, X.; Wu, X.; et al. Intracellular TRPA1 mediates Ca2+ release from lysosomes in dorsal root ganglion neurons. J. Cell Biol. 2016, 215, 369–381. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Li, C.; Wu, Y.; Shen, L.; Ma, J.; Qian, J.; Ge, J. Role of KCa3.1 Channels in Macrophage Polarization and Its Relevance in Atherosclerotic Plaque Instability. Arter. Thromb. Vasc. Biol. 2017, 37, 226–236. [Google Scholar] [CrossRef] [Green Version]
- Qiu, M.R.; Campbell, T.J.; Breit, S.N. A potassium ion channel is involved in cytokine production by activated human macrophages. Clin. Exp. Immunol. 2002, 130, 67–74. [Google Scholar] [CrossRef]
- Ohya, S.; Matsui, M.; Kajikuri, J.; Endo, K.; Kito, H. Increased Interleukin-10 Expression by the Inhibition of Ca2+-Activated K+ Channel KCa3.1 in CD4+CD25+ Regulatory T Cells in the Recovery Phase in an Inflammatory Bowel Disease Mouse Model. J. Pharmacol. Exp. Ther. 2021, 377, 75–85. [Google Scholar] [CrossRef]
- Wei, T.; Wang, Y.; Xu, W.; Liu, Y.; Chen, H.; Yu, Z. KCa3.1 deficiency attenuates neuroinflammation by regulating an astrocyte phenotype switch involving the PI3K/AKT/GSK3beta pathway. Neurobiol. Dis. 2019, 132, 104588. [Google Scholar] [CrossRef]
- Kwan, K.Y.; Allchorne, A.J.; Vollrath, M.A.; Christensen, A.P.; Zhang, D.S.; Woolf, C.J.; Corey, D.P. TRPA1 contributes to cold, mechanical, and chemical nociception but is not essential for hair-cell transduction. Neuron 2006, 50, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Miyake, T.; Nakamura, S.; Zhao, M.; So, K.; Inoue, K.; Numata, T.; Takahashi, N.; Shirakawa, H.; Mori, Y.; Nakagawa, T.; et al. Cold sensitivity of TRPA1 is unveiled by the prolyl hydroxylation blockade-induced sensitization to ROS. Nat. Commun. 2016, 7, 12840. [Google Scholar] [CrossRef] [Green Version]
- Brierley, S.M.; Castro, J.; Harrington, A.M.; Hughes, P.A.; Page, A.J.; Rychkov, G.Y.; Blackshaw, L.A. TRPA1 contributes to specific mechanically activated currents and sensory neuron mechanical hypersensitivity. J. Physiol. 2011, 589, 3575–3593. [Google Scholar] [CrossRef]
- Lehto, S.G.; Weyer, A.D.; Youngblood, B.D.; Zhang, M.; Yin, R.; Wang, W.; Teffera, Y.; Cooke, M.; Stucky, C.L.; Schenkel, L.; et al. Selective antagonism of TRPA1 produces limited efficacy in models of inflammatory- and neuropathic-induced mechanical hypersensitivity in rats. Mol. Pain 2016, 12, 1744806916677761. [Google Scholar] [CrossRef] [Green Version]
- Lennertz, R.C.; Kossyreva, E.A.; Smith, A.K.; Stucky, C.L. TRPA1 mediates mechanical sensitization in nociceptors during inflammation. PLoS ONE 2012, 7, e43597. [Google Scholar] [CrossRef]
- Staal, R.G.W.; Khayrullina, T.; Zhang, H.; Davis, S.; Fallon, S.M.; Cajina, M.; Nattini, M.E.; Hu, A.; Zhou, H.; Poda, S.B.; et al. Inhibition of the potassium channel KCa3.1 by senicapoc reverses tactile allodynia in rats with peripheral nerve injury. Eur. J. Pharm. 2017, 795, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.F.; Chandra, D.; McMahon, T.; Wang, D.; Dadgar, J.; Kharazia, V.N.; Liang, Y.J.; Waxman, S.G.; Dib-Hajj, S.D.; Messing, R.O. PKCepsilon phosphorylation of the sodium channel NaV1.8 increases channel function and produces mechanical hyperalgesia in mice. J. Clin. Investig. 2012, 122, 1306–1315. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Lin, X.J.; Ling, Z.H.; Cai, Y.Z. Effect of down-regulation of voltage-gated sodium channel Nav1.7 on activation of astrocytes and microglia in DRG in rats with cancer pain. Asian Pac. J. Trop. Med. 2015, 8, 405–411. [Google Scholar] [CrossRef] [Green Version]
- Chidiac, C.; Xue, Y.; Muniz Moreno, M.D.M.; Bakr Rasheed, A.A.; Lorentz, R.; Birling, M.C.; Gaveriaux-Ruff, C.; Herault, Y. The Human SCN10A(G1662S) Point Mutation Established in Mice Impacts on Mechanical, Heat, and Cool Sensitivity. Front. Pharm. 2021, 12, 780132. [Google Scholar] [CrossRef]
- Redpath, G.M.I.; Ecker, M.; Kapoor-Kaushik, N.; Vartoukian, H.; Carnell, M.; Kempe, D.; Biro, M.; Ariotti, N.; Rossy, J. Flotillins promote T cell receptor sorting through a fast Rab5-Rab11 endocytic recycling axis. Nat. Commun. 2019, 10, 4392. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Gao, Y.; Huang, Q.; Wang, Y.; Mo, X.; Wang, P.; Zhang, Y.; Xie, C.; Li, D.; Yao, J. Flotillin-1 Interacts with and Sustains the Surface Levels of TRPV2 Channel. Front. Cell Dev. Biol. 2021, 9, 634160. [Google Scholar] [CrossRef]


| Target Genes | Target Proteins | Assay ID |
|---|---|---|
| Bcl2 | Apoptosis regulator Bcl 2 | Mm00477631_m1 |
| C3 | Complement 3 | Mm01232779_m1 |
| CASP3 | Caspase 3 | Mm01195085_m1 |
| CaV2.2 | Voltage-gated calcium channel 2.2 | Mm01333678_m1 |
| CCL2 | C-C motif chemokine 2 | Mm00441242_m1 |
| CCL5 | C-C motif chemokine 5 | Mm01302427_m1 |
| CD28 | Cluster of differentiation 28 | Mm01253994_m1 |
| CD4 | Cluster of Differentiation 4 | Mm00442754_m1 |
| CD40 | Cluster of Differentiation 40 | Mm00441891_m1 |
| CD40lg | Cluster of Differentiation 40 ligand | Mm00441911_m1 |
| CD68 | Cluster of Differentiation 68 | Mm03047343_m1 |
| CD80 | Cluster of Differentiation 80 | Mm00711660_m1 |
| GFAP | Glial fibrillary acidic protein | Mm01253033_m1 |
| HMOX1 | Heme oxygenase 1 | Mm00516005_m1 |
| ICAM1 | Intercellular adhesion molecule 1 | Mm00516023_m1 |
| IKBKB | Inhibitor of nuclear factor kappa-B kinase subunit beta | Mm01222247_m1 |
| IL1b | Interleukin 1b | Mm00434228_m1 |
| IL4 | Interleukin 4 | Mm00445259_m1 |
| IL6 | Interleukin 6 | Mm00446190_m1 |
| IL10 | Interleukin 10 | Mm01288386_m1 |
| KCa3.1 | Calcium-activated potassium channel 3.1 | Mm00464686_m1 |
| LRG1 | Leucine-rich alpha-2-glycoprotein 1 | Mm01278767_m1 |
| NaV1.8 | Voltage-gated sodium channel 1.8 | Mm00501467_m1 |
| NFATC3 | Nuclear factor of activated T-cells, cytoplasmic 3 | Mm01249200_m1 |
| NLRP3 | NACHT, LRR, and PYD domain-containing protein 3 | Mm00840904_m1 |
| STAT3 | Signal transducer and activator of transcription 3 | Mm01219775_m1 |
| TGF1b | Transforming growth factor 1 beta | Mm01178820_m1 |
| TNFa | Tumor necrosis factor alpha | Mm00443258_m1 |
| TRPA1 | Transient receptor potential ankyrin 1 | Mm01227437_m1 |
| TRPM8 | Transient receptor potential melastatin 8 | Mm01299593_m1 |
| VEGFa | Vascular endothelial growth factor A | Mm00437306_m1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spitzel, M.; Wagner, E.; Breyer, M.; Henniger, D.; Bayin, M.; Hofmann, L.; Mauceri, D.; Sommer, C.; Üçeyler, N. Dysregulation of Immune Response Mediators and Pain-Related Ion Channels Is Associated with Pain-like Behavior in the GLA KO Mouse Model of Fabry Disease. Cells 2022, 11, 1730. https://doi.org/10.3390/cells11111730
Spitzel M, Wagner E, Breyer M, Henniger D, Bayin M, Hofmann L, Mauceri D, Sommer C, Üçeyler N. Dysregulation of Immune Response Mediators and Pain-Related Ion Channels Is Associated with Pain-like Behavior in the GLA KO Mouse Model of Fabry Disease. Cells. 2022; 11(11):1730. https://doi.org/10.3390/cells11111730
Chicago/Turabian StyleSpitzel, Marlene, Elise Wagner, Maximilian Breyer, Dorothea Henniger, Mehtap Bayin, Lukas Hofmann, Daniela Mauceri, Claudia Sommer, and Nurcan Üçeyler. 2022. "Dysregulation of Immune Response Mediators and Pain-Related Ion Channels Is Associated with Pain-like Behavior in the GLA KO Mouse Model of Fabry Disease" Cells 11, no. 11: 1730. https://doi.org/10.3390/cells11111730
APA StyleSpitzel, M., Wagner, E., Breyer, M., Henniger, D., Bayin, M., Hofmann, L., Mauceri, D., Sommer, C., & Üçeyler, N. (2022). Dysregulation of Immune Response Mediators and Pain-Related Ion Channels Is Associated with Pain-like Behavior in the GLA KO Mouse Model of Fabry Disease. Cells, 11(11), 1730. https://doi.org/10.3390/cells11111730







