Role of Nse1 Subunit of SMC5/6 Complex as a Ubiquitin Ligase
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Expression and Purification of Recombinant Proteins
2.3. In Vitro Ubiquitination Assay
2.4. Yeast Two-Hybrid Assay
2.5. Yeast Strains
2.6. Tandem Affinity Purification of Nse1-HBH
2.7. Liquid Chromatography-Mass Spectrometry Analyses
2.8. Protein Modelling
3. Results
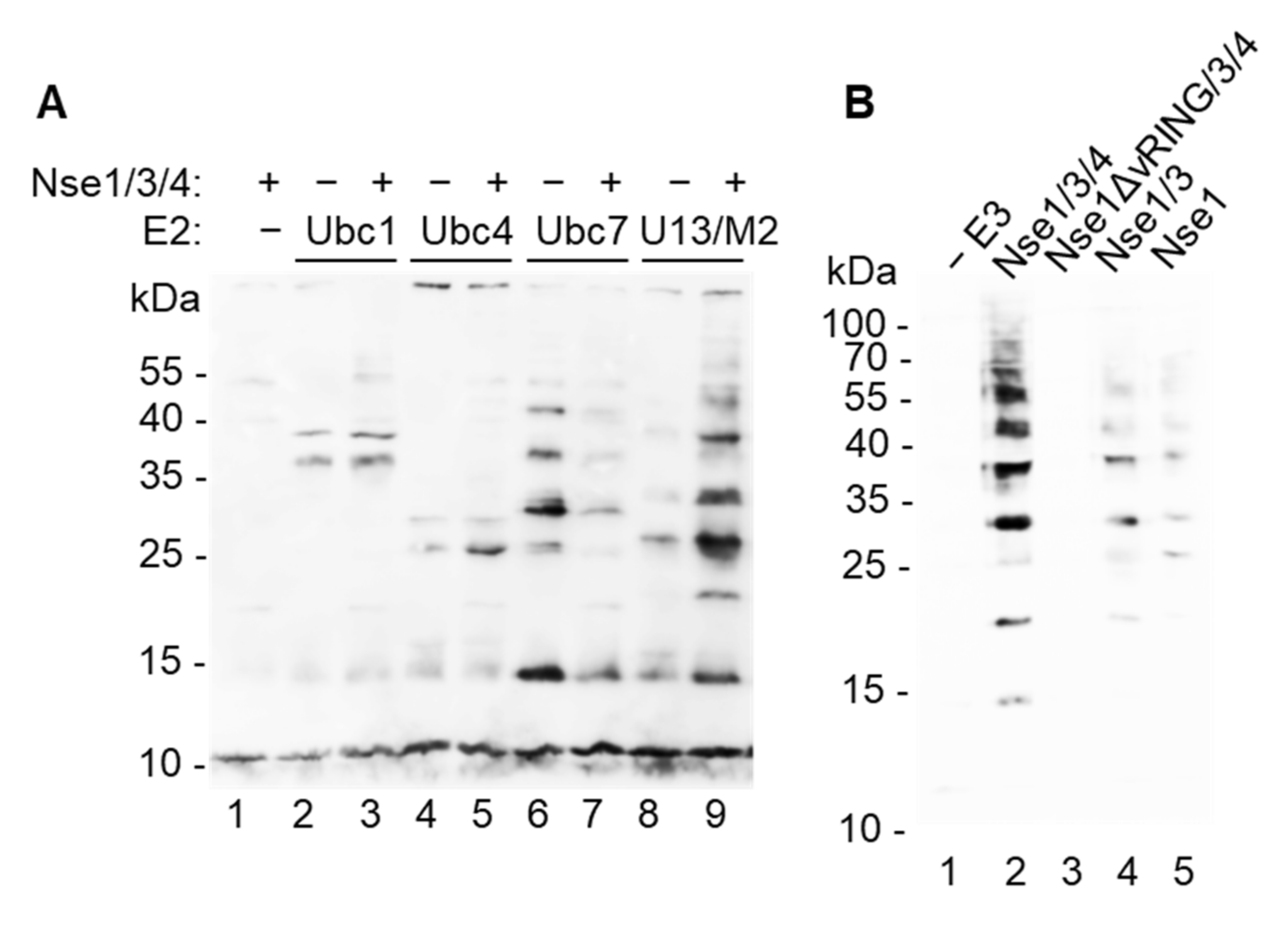
3.1. Nse1 Is a Ubiquitin Ligase
3.2. Nse1 Ligase Activity Is Abolished by the Nse1-R188E Mutation
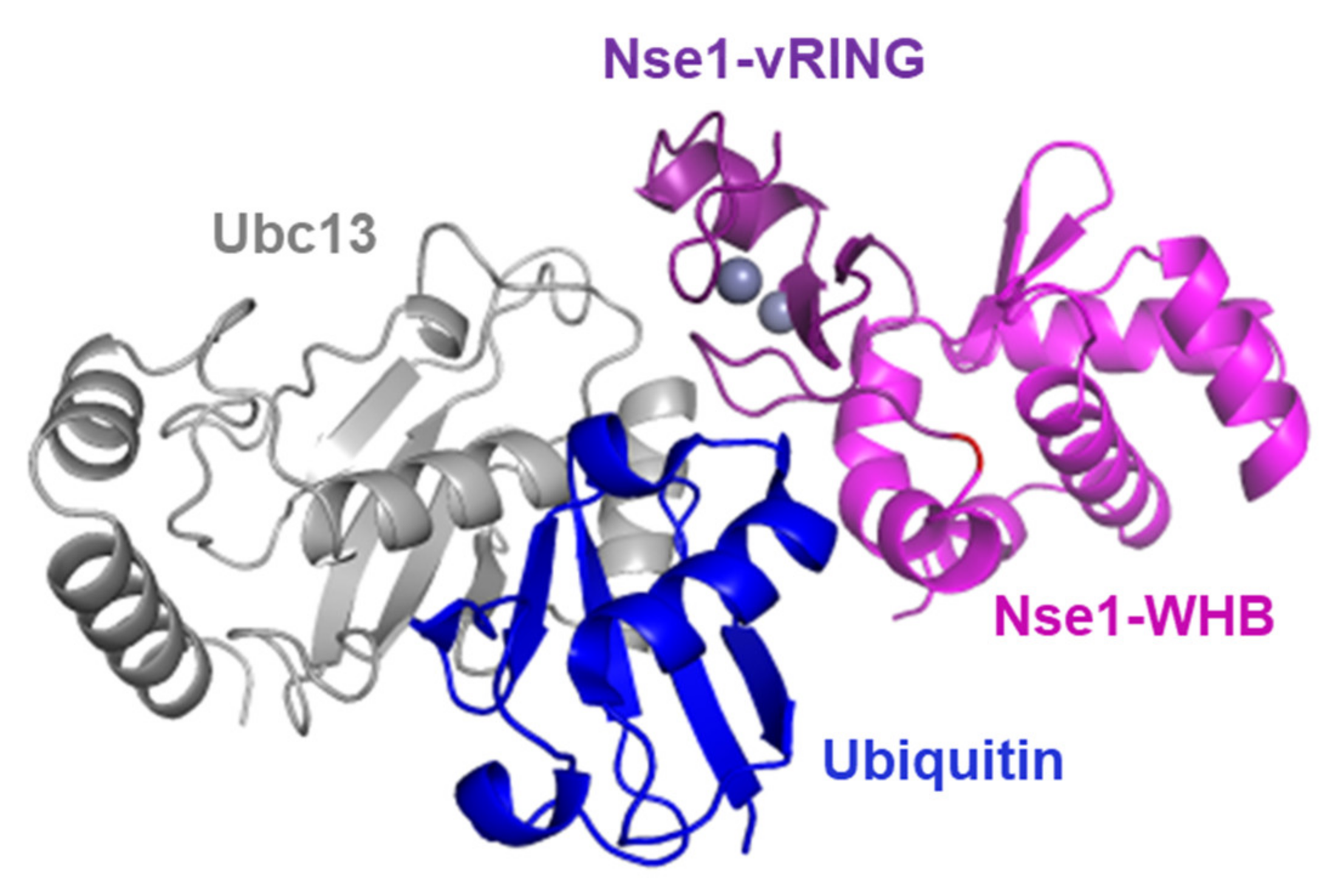
3.3. Nse1 Associates with Ubc13/Mms2 and Ubiquitin
3.4. Nse1 E3 Activity Plays an Important Role during Replication Stress
3.5. Nse4 and Nse3 Are Substrates of Nse1 Ubiquitin Ligase
4. Discussion
4.1. Nse1 Is a Ubiquitin Ligase Working Specifically with Ubc13/Mms2
4.2. Nse1 Mutation R188E Specifically Impairs Its Ubiquitin Ligase Function
4.3. Nse1 Ligase Substrates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uhlmann, F. SMC Complexes: From DNA to Chromosomes. Nat. Rev. Mol. Cell Biol. 2016, 17, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, A.R.; Walicka, M.; Griffiths, D.J.; Murray, J.M.; Watts, F.Z.; McCready, S.; Carr, A.M. The Rad18 Gene of Schizosaccharomyces Pombe Defines a New Subgroup of the SMC Superfamily Involved in DNA Repair. Mol. Cell. Biol. 1995, 15, 7067–7080. [Google Scholar] [CrossRef] [PubMed]
- Irmisch, A.; Ampatzidou, E.; Mizuno, K.; O’Connell, M.J.; Murray, J.M. Smc5/6 Maintains Stalled Replication Forks in a Recombination-Competent Conformation. EMBO J. 2009, 28, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Aragón, L. The Smc5/6 Complex: New and Old Functions of the Enigmatic Long-Distance Relative. Annu. Rev. Genet. 2018, 52, 89–107. [Google Scholar] [CrossRef]
- Palecek, J.J. SMC5/6: Multifunctional Player in Replication. Genes 2018, 10, 7. [Google Scholar] [CrossRef]
- Sergeant, J.; Taylor, E.; Palecek, J.; Fousteri, M.; Andrews, E.A.; Sweeney, S.; Shinagawa, H.; Watts, F.Z.; Lehmann, A.R. Composition and Architecture of the Schizosaccharomyces Pombe Rad18 (Smc5-6) Complex. Mol. Cell. Biol. 2005, 25, 172–184. [Google Scholar] [CrossRef]
- Palecek, J.; Vidot, S.; Feng, M.; Doherty, A.J.; Lehmann, A.R. The Smc5-Smc6 DNA Repair Complex. Bridging of the Smc5-Smc6 Heads by the KLEISIN, Nse4, and Non-Kleisin Subunits. J. Biol. Chem. 2006, 281, 36952–36959. [Google Scholar] [CrossRef]
- Vondrova, L.; Kolesar, P.; Adamus, M.; Nociar, M.; Oliver, A.W.; Palecek, J.J. A Role of the Nse4 Kleisin and Nse1/Nse3 KITE Subunits in the ATPase Cycle of SMC5/6. Sci. Rep. 2020, 10, 9694. [Google Scholar] [CrossRef]
- Palecek, J.J.; Gruber, S. Kite Proteins: A Superfamily of SMC/Kleisin Partners Conserved Across Bacteria, Archaea, and Eukaryotes. Structure 2015, 23, 2183–2190. [Google Scholar] [CrossRef]
- Adamus, M.; Lelkes, E.; Potesil, D.; Ganji, S.R.; Kolesar, P.; Zabrady, K.; Zdrahal, Z.; Palecek, J.J. Molecular Insights into the Architecture of the Human SMC5/6 Complex. J. Mol. Biol. 2020, 432, 3820–3837. [Google Scholar] [CrossRef]
- Guerineau, M.; Kriz, Z.; Kozakova, L.; Bednarova, K.; Janos, P.; Palecek, J. Analysis of the Nse3/MAGE-Binding Domain of the Nse4/EID Family Proteins. PLoS ONE 2012, 7, e35813. [Google Scholar] [CrossRef][Green Version]
- Hudson, J.J.R.; Bednarova, K.; Kozakova, L.; Liao, C.; Guerineau, M.; Colnaghi, R.; Vidot, S.; Marek, J.; Bathula, S.R.; Lehmann, A.R.; et al. Interactions between the Nse3 and Nse4 Components of the SMC5-6 Complex Identify Evolutionarily Conserved Interactions between MAGE and EID Families. PLoS ONE 2011, 6, e17270. [Google Scholar] [CrossRef] [PubMed]
- Jo, A.; Li, S.; Shin, J.W.; Zhao, X.; Cho, Y. Structure Basis for Shaping the Nse4 Protein by the Nse1 and Nse3 Dimer within the Smc5/6 Complex. J. Mol. Biol. 2021, 433, 166910. [Google Scholar] [CrossRef] [PubMed]
- Solé-Soler, R.; Torres-Rosell, J. Smc5/6, an Atypical SMC Complex with Two RING-Type Subunits. Biochem. Soc. Trans. 2020, 48, 2159–2171. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A.P. RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Pebernard, S.; Perry, J.J.P.; Tainer, J.A.; Boddy, M.N. Nse1 RING-like Domain Supports Functions of the Smc5-Smc6 Holocomplex in Genome Stability. Mol. Biol. Cell 2008, 19, 4099–4109. [Google Scholar] [CrossRef]
- Tapia-Alveal, C.; O’Connell, M.J. Nse1-Dependent Recruitment of Smc5/6 to Lesion-Containing Loci Contributes to the Repair Defects of Mutant Complexes. Mol. Biol. Cell 2011, 22, 4669–4682. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.M.; Gao, J.; Wang, J.; Yang, M.; Potts, P.R. MAGE-RING Protein Complexes Comprise a Family of E3 Ubiquitin Ligases. Mol. Cell 2010, 39, 963–974. [Google Scholar] [CrossRef]
- Kozakova, L.; Vondrova, L.; Stejskal, K.; Charalabous, P.; Kolesar, P.; Lehmann, A.R.; Uldrijan, S.; Sanderson, C.M.; Zdrahal, Z.; Palecek, J.J. The Melanoma-Associated Antigen 1 (MAGEA1) Protein Stimulates the E3 Ubiquitin-Ligase Activity of TRIM31 within a TRIM31-MAGEA1-NSE4 Complex. Cell Cycle 2015, 14, 920–930. [Google Scholar] [CrossRef]
- Weon, J.L.; Yang, S.W.; Potts, P.R. Cytosolic Iron-Sulfur Assembly Is Evolutionarily Tuned by a Cancer-Amplified Ubiquitin Ligase. Mol. Cell 2018, 69, 113–125.e6. [Google Scholar] [CrossRef]
- Olsen, S.K.; Lima, C.D. Structure of a Ubiquitin E1-E2 Complex: Insights to E1-E2 Thioester Transfer. Mol. Cell 2013, 49, 884–896. [Google Scholar] [CrossRef]
- Zabrady, K.; Adamus, M.; Vondrova, L.; Liao, C.; Skoupilova, H.; Novakova, M.; Jurcisinova, L.; Alt, A.; Oliver, A.W.; Lehmann, A.R.; et al. Chromatin Association of the SMC5/6 Complex Is Dependent on Binding of Its NSE3 Subunit to DNA. Nucleic Acids Res. 2016, 44, 1064–1079. [Google Scholar] [CrossRef]
- Moreno, S.; Klar, A.; Nurse, P. Molecular Genetic Analysis of Fission Yeast Schizosaccharomyces Pombe. Methods Enzymol. 1991, 194, 795–823. [Google Scholar] [CrossRef]
- Kaiser, P.; Meierhofer, D.; Wang, X.; Huang, L. Tandem Affinity Purification Combined with Mass Spectrometry to Identify Components of Protein Complexes. Methods Mol. Biol. Clifton 2008, 439, 309–326. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kiss, L.; Clift, D.; Renner, N.; Neuhaus, D.; James, L.C. RING Domains Act as Both Substrate and Enzyme in a Catalytic Arrangement to Drive Self-Anchored Ubiquitination. Nat. Commun. 2021, 12, 1220. [Google Scholar] [CrossRef]
- Furmanová, K.; Byška, J.; Gröller, E.M.; Viola, I.; Paleček, J.J.; Kozlíková, B. COZOID: Contact Zone Identifier for Visual Analysis of Protein-Protein Interactions. BMC Bioinformatics 2018, 19, 125. [Google Scholar] [CrossRef]
- Andrews, E.A.; Palecek, J.; Sergeant, J.; Taylor, E.; Lehmann, A.R.; Watts, F.Z. Nse2, a Component of the Smc5-6 Complex, Is a SUMO Ligase Required for the Response to DNA Damage. Mol. Cell. Biol. 2005, 25, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zhang, W.; Xiao, Z.; Gan, H.; Lin, X.; Liao, S.; Han, C. Mining and Characterization of Ubiquitin E3 Ligases Expressed in the Mouse Testis. BMC Genom. 2012, 13, 495. [Google Scholar] [CrossRef] [PubMed]
- Christensen, D.E.; Brzovic, P.S.; Klevit, R.E. E2-BRCA1 RING Interactions Dictate Synthesis of Mono- or Specific Polyubiquitin Chain Linkages. Nat. Struct. Mol. Biol. 2007, 14, 941–948. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.-L.; Pyrowolakis, G.; Jentsch, S. RAD6-Dependent DNA Repair Is Linked to Modification of PCNA by Ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Dou, H.; Buetow, L.; Sibbet, G.J.; Cameron, K.; Huang, D.T. BIRC7-E2 Ubiquitin Conjugate Structure Reveals the Mechanism of Ubiquitin Transfer by a RING Dimer. Nat. Struct. Mol. Biol. 2012, 19, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Plechanovová, A.; Jaffray, E.G.; Tatham, M.H.; Naismith, J.H.; Hay, R.T. Structure of a RING E3 Ligase and Ubiquitin-Loaded E2 Primed for Catalysis. Nature 2012, 489, 115–120. [Google Scholar] [CrossRef]
- Pruneda, J.N.; Littlefield, P.J.; Soss, S.E.; Nordquist, K.A.; Chazin, W.J.; Brzovic, P.S.; Klevit, R.E. Structure of an E3:E2~Ub Complex Reveals an Allosteric Mechanism Shared among RING/U-Box Ligases. Mol. Cell 2012, 47, 933–942. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolesar, P.; Stejskal, K.; Potesil, D.; Murray, J.M.; Palecek, J.J. Role of Nse1 Subunit of SMC5/6 Complex as a Ubiquitin Ligase. Cells 2022, 11, 165. https://doi.org/10.3390/cells11010165
Kolesar P, Stejskal K, Potesil D, Murray JM, Palecek JJ. Role of Nse1 Subunit of SMC5/6 Complex as a Ubiquitin Ligase. Cells. 2022; 11(1):165. https://doi.org/10.3390/cells11010165
Chicago/Turabian StyleKolesar, Peter, Karel Stejskal, David Potesil, Johanne M. Murray, and Jan J. Palecek. 2022. "Role of Nse1 Subunit of SMC5/6 Complex as a Ubiquitin Ligase" Cells 11, no. 1: 165. https://doi.org/10.3390/cells11010165
APA StyleKolesar, P., Stejskal, K., Potesil, D., Murray, J. M., & Palecek, J. J. (2022). Role of Nse1 Subunit of SMC5/6 Complex as a Ubiquitin Ligase. Cells, 11(1), 165. https://doi.org/10.3390/cells11010165





