PD-L1 Expression Fluctuates Concurrently with Cyclin D in Glioblastoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Transfection
2.2. Cell Counting
2.3. Flow Cytometry
2.4. Western Blot
2.5. Sub-Cellular Fractionation
2.6. Quantitative PCR (qPCR)
2.7. Fluorescence Microscopy
2.8. Chromatin Immunoprecipitation
2.9. Statistical Analysis
3. Results
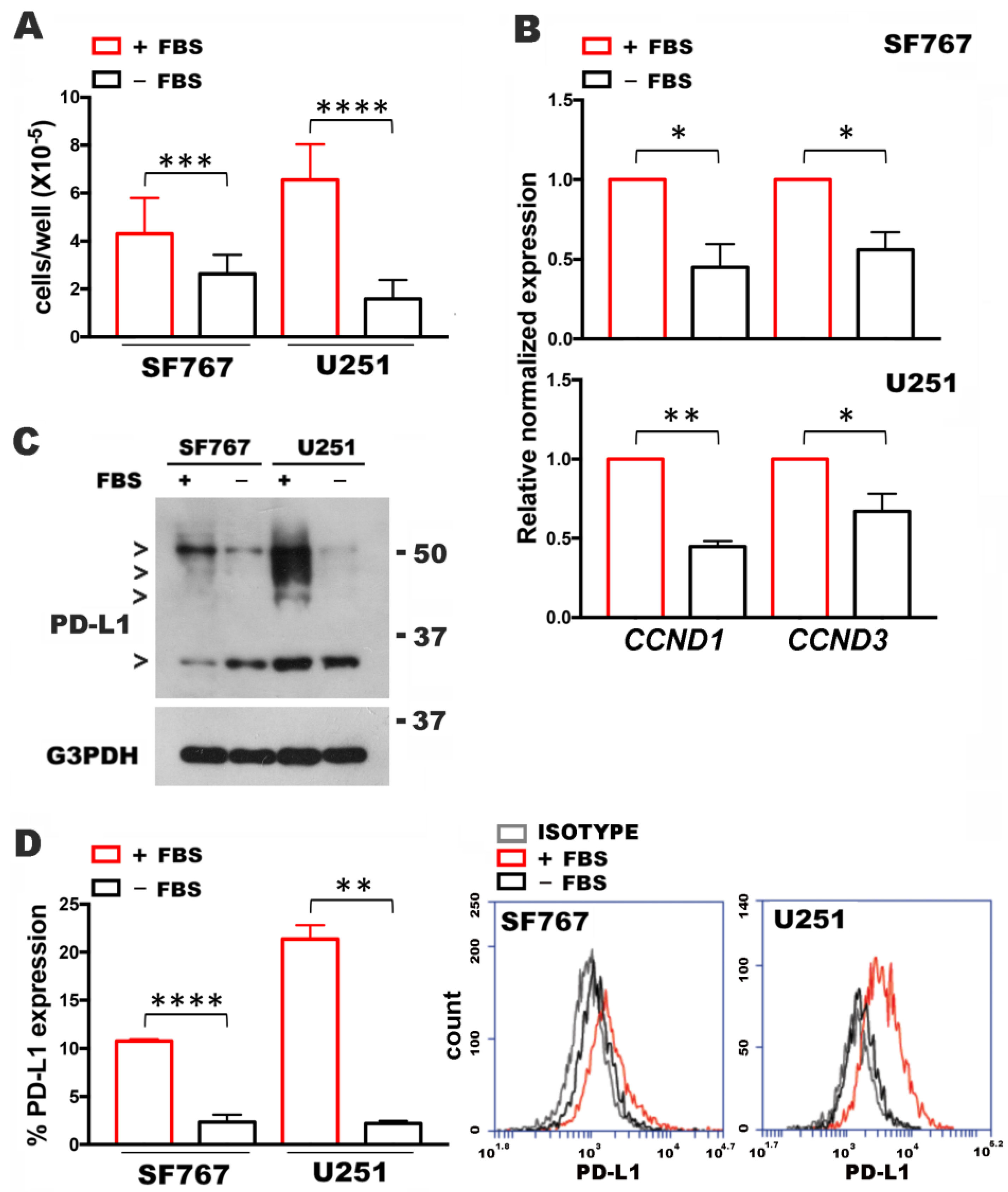
3.1. Cell Growth Affects PD-L1 Expression
3.2. PD-L1 Levels Correlate with Cyclin D Levels
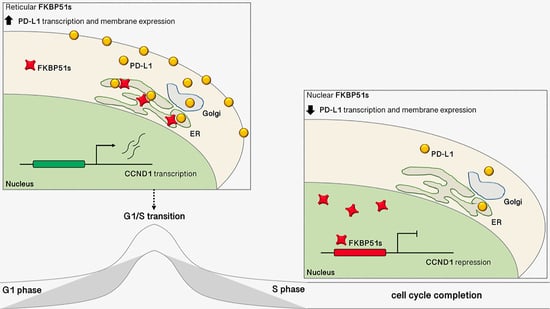
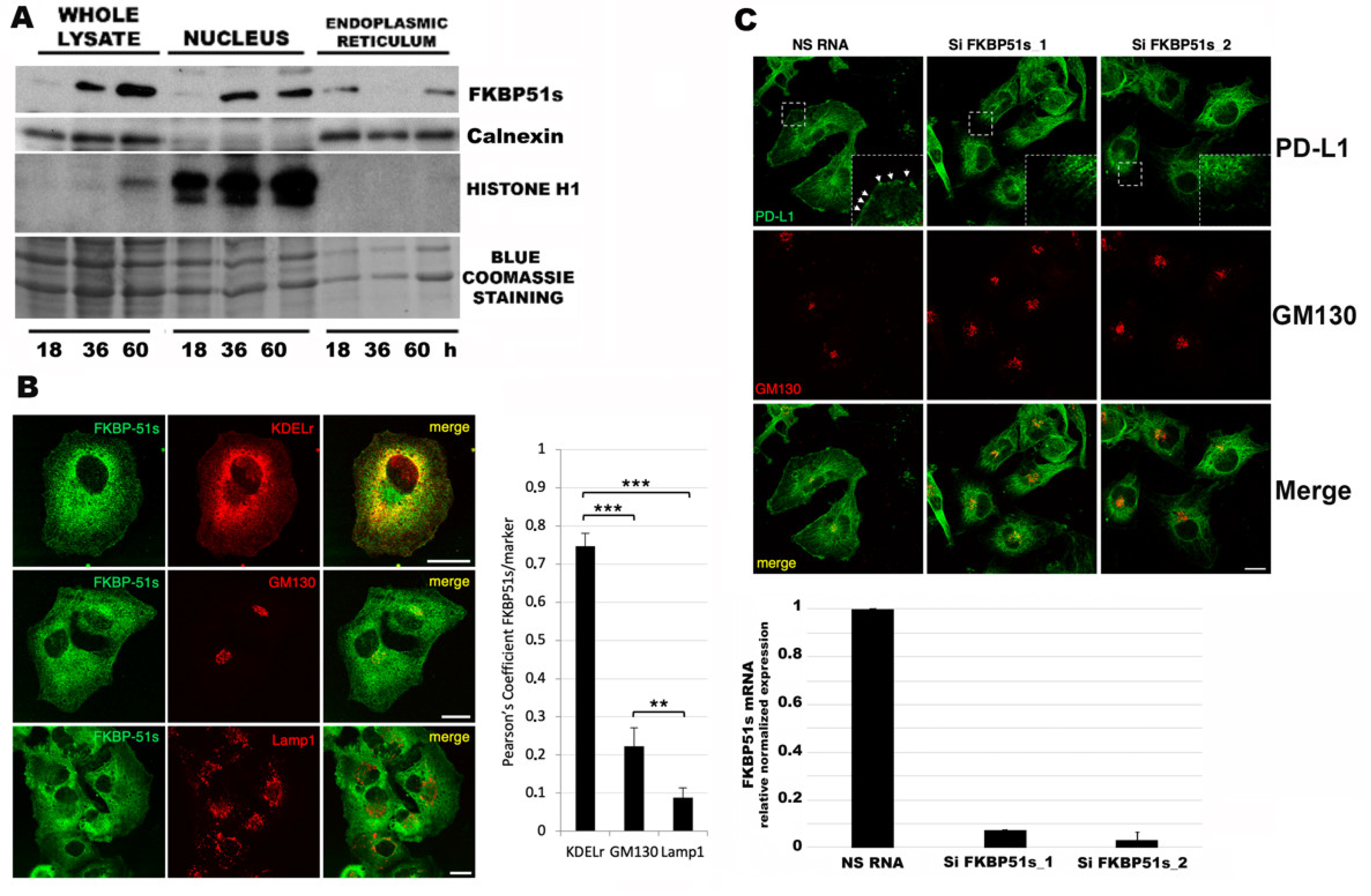
3.3. FKBP51s Localizes in ER When CCND1 Peaks but in the Nucleus during Cell Division
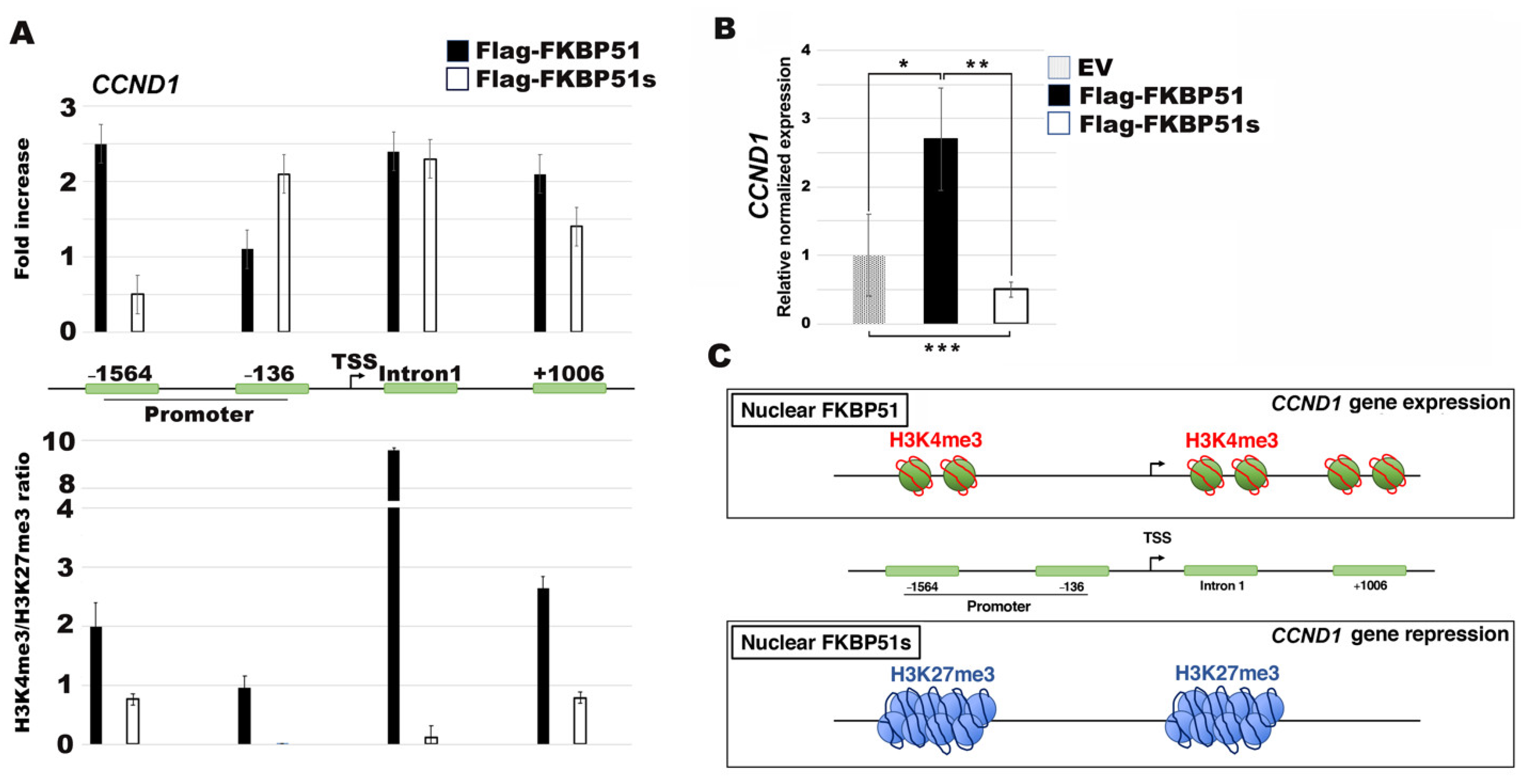
3.4. FKBP51s Interacts with CCND1 Promoter in a Closed Chromatin State
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-Associated B7-H1 Promotes T-Cell Apoptosis: A Potential Mechanism of Immune Evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef]
- Romano, S.; Tufano, M.; D’Arrigo, P.; Vigorito, V.; Russo, S.; Romano, M.F. Cell Stemness, Epithelial-to-Mesenchymal Transition, and Immunoevasion: Intertwined Aspects in Cancer Metastasis. Semin. Cancer Biol. 2020, 60, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Diao, L.; Cuentas, E.R.P.; Denning, W.L.; Chen, L.; Fan, Y.H.; Byers, L.A.; Wang, J.; Papadimitrakopoulou, V.A.; Behrens, C.; et al. Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clin. Cancer Res. 2016, 22, 3630–3642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Arrigo, P.; Russo, M.; Rea, A.; Tufano, M.; Guadagno, E.; Del Basso De Caro, M.L.; Pacelli, R.; Hausch, F.; Staibano, S.; Ilardi, G.; et al. A Regulatory Role for the Co-Chaperone FKBP51s in PD-L1 Expression in Glioma. Oncotarget 2017, 8, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Cazacu, S.; Xiang, C.; Zenklusen, J.C.; Fine, H.A.; Berens, M.; Armstrong, B.; Brodie, C.; Mikkelsen, T. FK506 Binding Protein Mediates Glioma Cell Growth and Sensitivity to Rapamycin Treatment by Regulating NF-ΚB Signaling Pathway. Neoplasia 2008, 10, 235–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotoli, D.; Morales, M.; Maeso, M.D.; Ávila, J.; Pérez-Rodríguez, N.D.; Mobasheri, A.; van Noorden, C.J.F.; Martín-Vasallo, P. IQGAP1, AmotL2, and FKBP51 Scaffoldins in the Glioblastoma Microenvironment. J. Histochem. Cytochem. 2019, 67, 481–494. [Google Scholar] [CrossRef]
- Romano, S.; Sorrentino, A.; Di Pace, A.L.; Nappo, G.; Mercogliano, C.; Romano, M.F. The Emerging Role of Large Immunophilin FK506 Binding Protein 51 in Cancer. Curr. Med. Chem. 2011, 18, 5424–5429. [Google Scholar] [CrossRef] [Green Version]
- Romano, S.; D’Angelillo, A.; Staibano, S.; Simeone, E.; D’Arrigo, P.; Ascierto, P.A.; Scalvenzi, M.; Mascolo, M.; Ilardi, G.; Merolla, F.; et al. Immunomodulatory Pathways Regulate Expression of a Spliced FKBP51 Isoform in Lymphocytes of Melanoma Patients. Pigment. Cell Melanoma Res. 2015, 28, 442–452. [Google Scholar] [CrossRef] [Green Version]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Nduom, E.K.; Wei, J.; Yaghi, N.K.; Huang, N.; Kong, L.Y.; Gabrusiewicz, K.; Ling, X.; Zhou, S.; Ivan, C.; Chen, J.Q.; et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016, 18, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Johanns, T.M.; Miller, C.A.; Dorward, I.G.; Tsien, C.; Chang, E.; Perry, A.; Uppaluri, R.; Ferguson, C.; Schmidt, R.E.; Dahiya, S.; et al. Immunogenomics of Hypermutated Glioblastoma: A patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 2016, 6, 1230–1236. [Google Scholar] [CrossRef] [Green Version]
- Reardon, D.A.; Lassman, A.B.; van den Bent, M.; Kumthekar, P.; Merrell, R.; Scott, A.M.; Fichtel, L.; Sulman, E.P.; Gomez, E.; Fischer, J.; et al. Efficacy and safety results of ABT-414 in combination with radiation and temozolomide in newly diagnosed glioblastoma. Neuro Oncol. 2017, 19, 965–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, K.I.; Peterson, B.; Serritella, A.; Thomas, J.; Reizine, N.; Moya, S.; Tan, C.; Wang, Y.; Catenacci, D.V.T. Spatial and Temporal Heterogeneity of PD-L1 Expression and Tumor Mutational Burden in Gastroesophageal Adenocarcinoma at Baseline Diagnosis and after Chemotherapy. Clin. Cancer Res. 2020, 26, 6453–6463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bu, X.; Wang, H.; Zhu, Y.; Geng, Y.; Nihira, N.T.; Tan, Y.; Ci, Y.; Wu, F.; Dai, X.; et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 2019, 571, E10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzamali, E.; Tzedakis, G.; Sakkalis, V. Modeling How Heterogeneity in Cell Cycle Length Affects Cancer Cell Growth Dynamics in Response to Treatment. Front. Oncol. 2020, 10, 1552. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Xiao, Y.; Nakaya, M.; D’Angelillo, A.; Chang, M.; Jin, J.; Hausch, F.; Masullo, M.; Feng, X.; Romano, M.F.; et al. FKBP51 Employs Both Scaffold and Isomerase Functions to Promote NF-ΚB Activation in Melanoma. Nucleic Acids Res. 2015, 43, 6983–6993. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Caiazza, C.; D’Agostino, M.; Passaro, F.; Faicchia, D.; Mallardo, M.; Paladino, S.; Pierantoni, G.M.; Tramontano, D. Effects of Long-Term Citrate Treatment in the PC3 Prostate Cancer Cell Line. Int. J. Mol. Sci. 2019, 20, 2613. [Google Scholar] [CrossRef] [Green Version]
- Testa, G.; Russo, M.; di Benedetto, G.; Barbato, M.; Parisi, S.; Pirozzi, F.; Tocchetti, C.G.; Abete, P.; Bonaduce, D.; Russo, T.; et al. Bmi1 inhibitor PTC-209 promotes Chemically-induced Direct Cardiac Reprogramming of cardiac fibroblasts into cardiomyocytes. Sci. Rep. 2020, 10, 7129. [Google Scholar] [CrossRef]
- Yang, K.; Hitomi, M.; Stacey, D.W. Variations in cyclin D1 levels through the cell cycle determine the proliferative fate of a cell. Cell Div. 2006, 1, 32. [Google Scholar] [CrossRef] [Green Version]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in Cancer: A Molecular Connection for Cell Cycle Control, Adhesion and Invasion in Tumor and Stroma. Cells 2020, 9, 2648. [Google Scholar] [CrossRef]
- Ben Dori, S.; Aizic, A.; Sabo, E.; Hershkovitz, D. Spatial Heterogeneity of PD-L1 Expression and the Risk for Misclassification of PD-L1 Immunohistochemistry in Non-Small Cell Lung Cancer. Lung Cancer 2020, 147, 91–98. [Google Scholar] [CrossRef]
- Haragan, A.; Field, J.K.; Davies, M.P.A.; Escriu, C.; Gruver, A.; Gosney, J.R. Heterogeneity of PD-L1 Expression in Non-Small Cell Lung Cancer: Implications for Specimen Sampling in Predicting Treatment Response. Lung Cancer 2019, 134, 79–84. [Google Scholar] [CrossRef] [Green Version]
- Hao, C.; Chen, G.; Zhao, H.; Li, Y.; Chen, J.; Zhang, H.; Li, S.; Zhao, Y.; Chen, F.; Li, W.; et al. PD-L1 Expression in Glioblastoma, the Clinical and Prognostic Significance: A Systematic Literature Review and Meta-Analysis. Front. Oncol. 2020, 10, 1015. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.Q.; Liu, F.; Qiu, X.Y.; Chen, X.Q. The Prognostic and Therapeutic Value of PD-L1 in Glioma. Front. Pharmacol. 2019, 9, 1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, S.; Hu, M.; Li, P.; Ma, J.; Xie, L.; Teng, F.; Zhu, Y.; Fan, B.; Mu, D.; Yu, J. Relationship between expression of PD-L1 and tumor angiogenesis, proliferation, and invasion in glioma. Oncotarget 2017, 8, 49702–49712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.L.; Li, G.Z.; Wang, Q.W.; Bao, Z.S.; Wang, Z.; Zhang, C.B.; Jiang, T. PD-L2 expression is correlated with the molecular and clinical features of glioma, and acts as an unfavorable prognostic factor. Oncoimmunology 2018, 8, e1541535. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, C.J.; Kobayashi, D.K.; Johanns, T.M.; Bowman-Kirigin, J.A.; Schaettler, M.O.; Mao, D.D.; Bender, D.; Kelley, D.G.; Uppaluri, R.; et al. GATA2 Regulates Constitutive PD-L1 and PD-L2 Expression in Brain Tumors. Sci. Rep. 2020, 10, 9027. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tufano, M.; D’Arrigo, P.; D’Agostino, M.; Giordano, C.; Marrone, L.; Cesaro, E.; Romano, M.F.; Romano, S. PD-L1 Expression Fluctuates Concurrently with Cyclin D in Glioblastoma Cells. Cells 2021, 10, 2366. https://doi.org/10.3390/cells10092366
Tufano M, D’Arrigo P, D’Agostino M, Giordano C, Marrone L, Cesaro E, Romano MF, Romano S. PD-L1 Expression Fluctuates Concurrently with Cyclin D in Glioblastoma Cells. Cells. 2021; 10(9):2366. https://doi.org/10.3390/cells10092366
Chicago/Turabian StyleTufano, Martina, Paolo D’Arrigo, Massimo D’Agostino, Carolina Giordano, Laura Marrone, Elena Cesaro, Maria Fiammetta Romano, and Simona Romano. 2021. "PD-L1 Expression Fluctuates Concurrently with Cyclin D in Glioblastoma Cells" Cells 10, no. 9: 2366. https://doi.org/10.3390/cells10092366
APA StyleTufano, M., D’Arrigo, P., D’Agostino, M., Giordano, C., Marrone, L., Cesaro, E., Romano, M. F., & Romano, S. (2021). PD-L1 Expression Fluctuates Concurrently with Cyclin D in Glioblastoma Cells. Cells, 10(9), 2366. https://doi.org/10.3390/cells10092366