Chromatin Organization and Function in Drosophila
Abstract
1. Introduction
2. The Drosophila Genome
3. Chromatin Composition and Structure
3.1. Nucleosome Dynamics
3.2. Core Histones and Their Variants
3.3. Linker Histones
4. Covalent Modifications of Chromatin
4.1. DNA Methylation
4.2. Histone Modifications
5. Functional Organization of the Genome
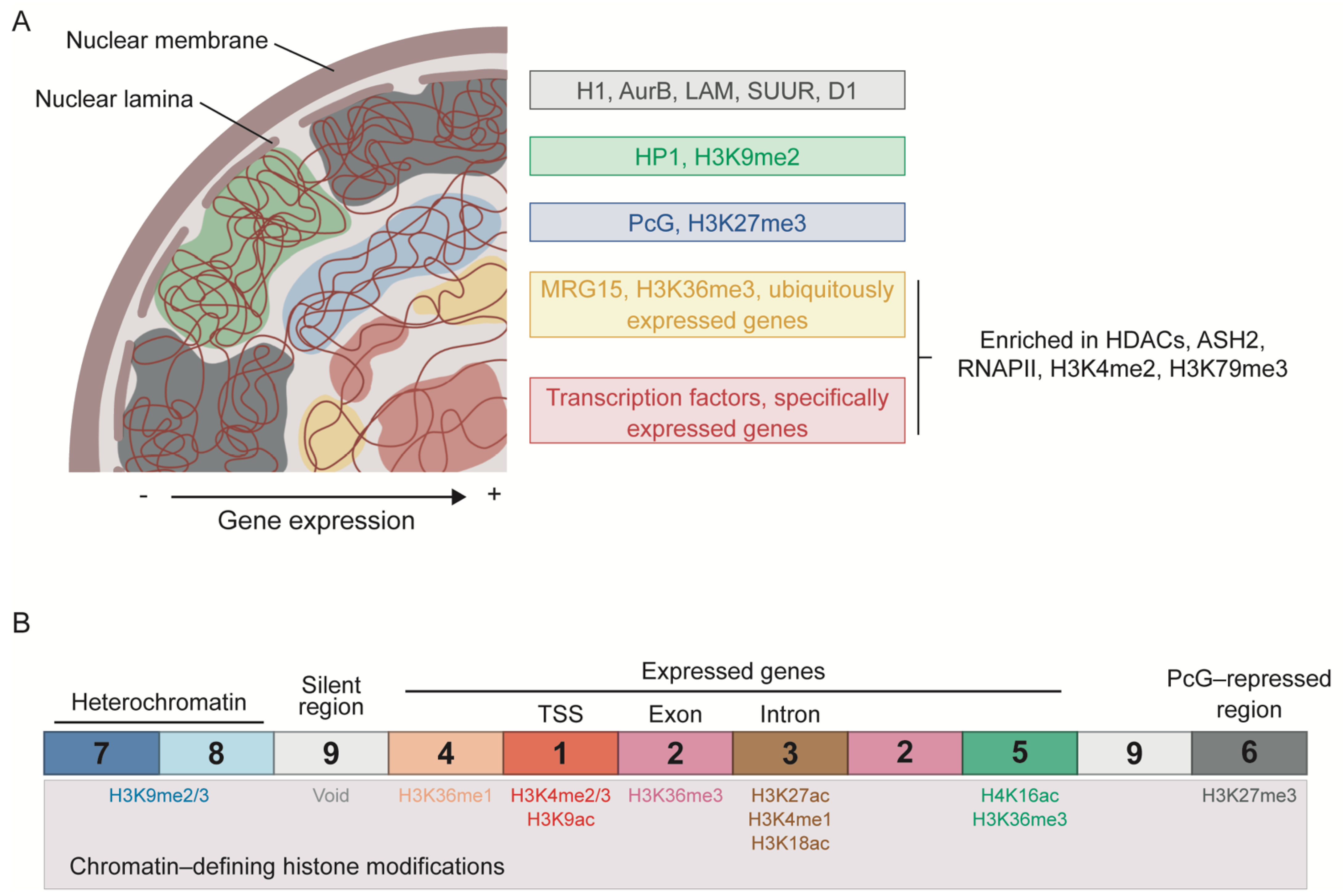
5.1. Segmentation of the Genome into Chromatin States
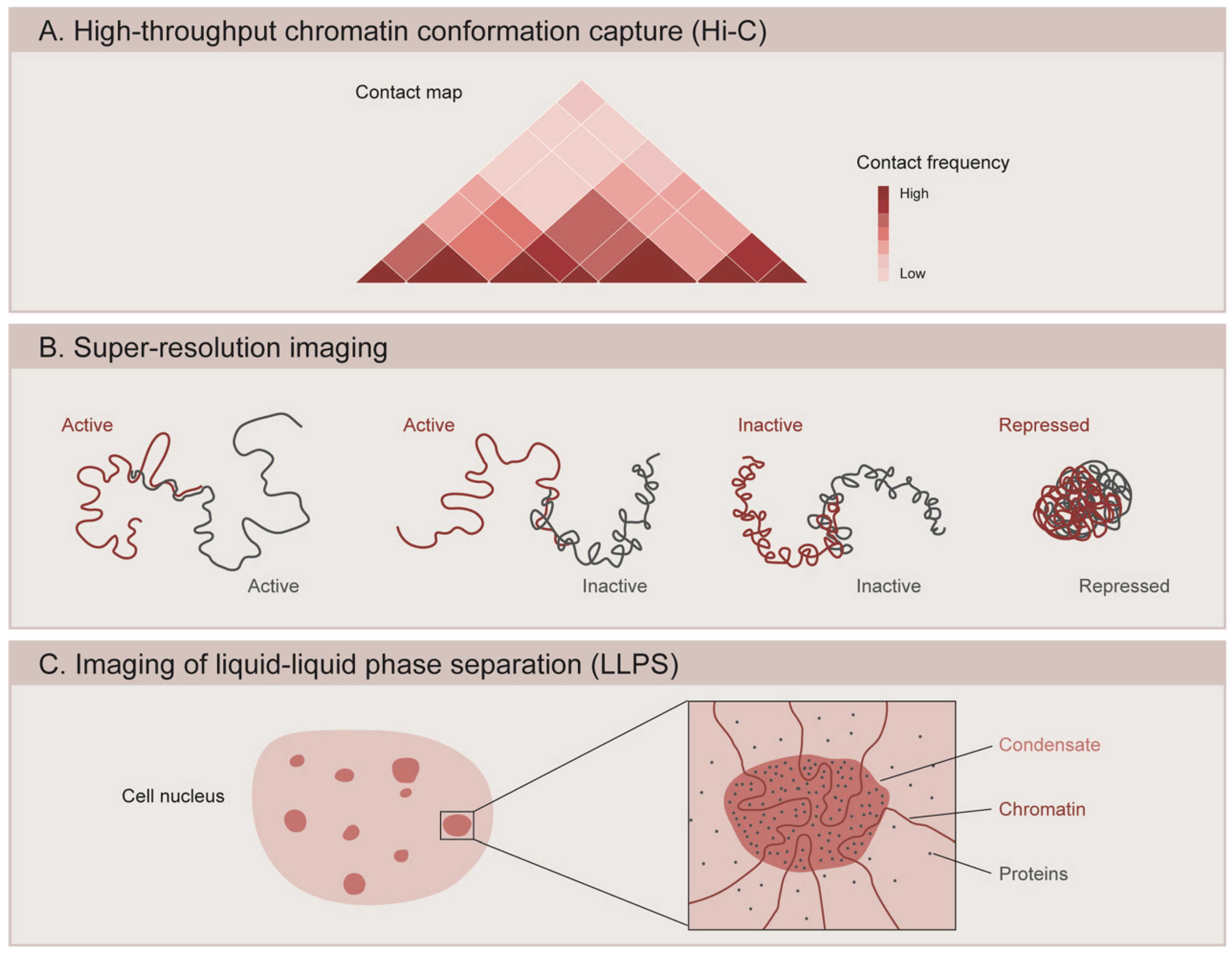
5.2. 3D Organization of the Genome
5.3. Regulation of Gene Expression by Chromatin Organization
5.4. Nuclear Lamina and Pericentromeric Heterochromatin
5.5. Nuclear Pore Complexes
6. Liquid–Liquid Phase Separation
7. Conclusions and Future Perspectives
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The Genome Sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef]
- Myers, E.W. A Whole-Genome Assembly of Drosophila. Science 2000, 287, 2196–2204. [Google Scholar] [CrossRef]
- Hoskins, R.A.; Carlson, J.W.; Wan, K.H.; Park, S.; Mendez, I.; Galle, S.E.; Booth, B.W.; Pfeiffer, B.D.; George, R.A.; Svirskas, R.; et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015, 25, 445–458. [Google Scholar] [CrossRef]
- Zhimulev, I.F.; Belyaeva, E.S.; Vatolina, T.Y.; Demakov, S.A. Banding patterns in Drosophila melanogaster polytene chromosomes correlate with DNA-binding protein occupancy. BioEssays 2012, 34, 498–508. [Google Scholar] [CrossRef]
- Zykova, T.Y.; Levitsky, V.G.; Belyaeva, E.S.; Zhimulev, I.F. Polytene Chromosomes—A Portrait of Functional Organization of the Drosophila Genome. Curr. Genom. 2018, 19, 179–191. [Google Scholar] [CrossRef]
- Grewal, S.I.S.; Elgin, S.C.R. Heterochromatin: New possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002, 12, 178–187. [Google Scholar] [CrossRef]
- Craig, J.M. Heterochromatin? many flavours, common themes. BioEssays 2005, 27, 17–28. [Google Scholar] [CrossRef]
- Pimpinelli, S.; Wakimoto, B.T. Expanding the boundaries of heterochromatin. Genetica 2003, 117, 111–116. [Google Scholar] [CrossRef]
- Pimpinelli, S.; Berloco, M.; Fanti, L.; Dimitri, P.; Bonaccorsi, S.; Marchetti, E.; Caizzi, R.; Caggese, C.; Gatti, M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc. Natl. Acad. Sci. USA 1995, 92, 3804–3808. [Google Scholar] [CrossRef]
- Eissenberg, J.C.; Elgin, S.C.R. HP1a: A structural chromosomal protein regulating transcription. Trends Genet. 2014, 30, 103–110. [Google Scholar] [CrossRef]
- Marsano, R.M.; Giordano, E.; Messina, G.; Dimitri, P. A New Portrait of Constitutive Heterochromatin: Lessons from Drosophila melanogaster. Trends Genet. 2019, 35, 615–631. [Google Scholar] [CrossRef]
- Swenson, J.M.; Colmenares, S.U.; Strom, A.R.; Costes, S.V.; Karpen, G.H. The composition and organization of Drosophila heterochromatin are heterogeneous and dynamic. eLife 2016, 5, e16096. [Google Scholar] [CrossRef]
- Filion, G.J.; van Bemmel, J.G.; Braunschweig, U.; Talhout, W.; Kind, J.; Ward, L.D.; Brugman, W.; de Castro, I.J.; Kerkhoven, R.M.; Bussemaker, H.J.; et al. Systematic Protein Location Mapping Reveals Five Principal Chromatin Types in Drosophila Cells. Cell 2010, 143, 212–224. [Google Scholar] [CrossRef]
- Kornberg, R.D. Chromatin Structure: A Repeating Unit of Histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Iwasaki, W.; Miya, Y.; Horikoshi, N.; Osakabe, A.; Taguchi, H.; Tachiwana, H.; Shibata, T.; Kagawa, W.; Kurumizaka, H. Contribution of histone N-terminal tails to the structure and stability of nucleosomes. FEBS Open Bio 2013, 3, 363–369. [Google Scholar] [CrossRef]
- Cutter, A.R.; Hayes, J.J. A brief review of nucleosome structure. FEBS Lett. 2015, 589, 2914–2922. [Google Scholar] [CrossRef]
- Allan, J.; Hartman, P.G.; Crane-Robinson, C.; Aviles, F.X. The structure of histone H1 and its location in chromatin. Nature 1980, 288, 675–679. [Google Scholar] [CrossRef]
- Thoma, F.; Koller, T. Influence of histone H1 on chromatin structure. Cell 1977, 12, 101–107. [Google Scholar] [CrossRef]
- Thoma, F.; Koller, T.; Klug, A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979, 83, 403–427. [Google Scholar] [CrossRef]
- Simpson, R.T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry 1978, 17, 5524–5531. [Google Scholar] [CrossRef]
- Bednar, J.; Garcia-Saez, I.; Boopathi, R.; Cutter, A.R.; Papai, G.; Reymer, A.; Syed, S.H.; Lone, I.N.; Tonchev, O.; Crucifix, C.; et al. Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Mol. Cell 2017, 66, 384–397.e8. [Google Scholar] [CrossRef]
- Cutter, A.R.; Hayes, J.J. Linker histones: Novel insights into structure-specific recognition of the nucleosome. Biochem. Cell Biol. 2017, 95, 171–178. [Google Scholar] [CrossRef]
- Ricci, M.A.; Manzo, C.; García-Parajo, M.F.; Lakadamyali, M.; Cosma, M.P. Chromatin Fibers Are Formed by Heterogeneous Groups of Nucleosomes In Vivo. Cell 2015, 160, 1145–1158. [Google Scholar] [CrossRef]
- Mavrich, T.N.; Ioshikhes, I.P.; Venters, B.J.; Jiang, C.; Tomsho, L.P.; Qi, J.; Schuster, S.C.; Albert, I.; Pugh, B.F. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008, 18, 1073–1083. [Google Scholar] [CrossRef]
- Kaplan, N.; Moore, I.K.; Fondufe-Mittendorf, Y.; Gossett, A.J.; Tillo, D.; Field, Y.; LeProust, E.M.; Hughes, T.R.; Lieb, J.D.; Widom, J.; et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 2009, 458, 362–366. [Google Scholar] [CrossRef]
- Yuan, G.C.; Liu, Y.-J.; Dion, M.F.; Slack, M.D.; Wu, L.F.; Altschuler, S.J.; Rando, O.J. Genome-Scale Identification of Nucleosome Positions in S. cerevisiae. Science 2005, 309, 626–630. [Google Scholar] [CrossRef]
- Struhl, K.; Segal, E. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013, 20, 267–273. [Google Scholar] [CrossRef]
- Martin, R.L.; Maiorano, J.; Beitel, G.J.; Marko, J.F.; McVicker, G.; Fondufe-Mittendorf, Y.N. A comparison of nucleosome organization in Drosophila cell lines. PLoS ONE 2017, 12, e0178590. [Google Scholar] [CrossRef]
- Li, G.; Levitus, M.; Bustamante, C.; Widom, J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005, 12, 46–53. [Google Scholar] [CrossRef]
- Poirier, M.G.; Oh, E.; Tims, H.S.; Widom, J. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 2009, 16, 938–944. [Google Scholar] [CrossRef]
- Zhou, K.; Gaullier, G.; Luger, K. Nucleosome structure and dynamics are coming of age. Nat. Struct. Mol. Biol. 2019, 26, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.K.M.; Pugh, B.F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 2017, 18, 548–562. [Google Scholar] [CrossRef] [PubMed]
- Horard, B.; Loppin, B. Histone storage and deposition in the early Drosophila embryo. Chromosoma 2015, 124, 163–175. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants at a glance. J. Cell Sci. 2021, 134, jcs244749. [Google Scholar] [CrossRef] [PubMed]
- Baldi, S.; Becker, P.B. The variant histone H2A.V of Drosophila—Three roles, two guises. Chromosoma 2013, 122, 245–258. [Google Scholar] [CrossRef]
- Prozzillo, Y.; Cuticone, S.; Ferreri, D.; Fattorini, G.; Messina, G.; Dimitri, P. In Vivo Silencing of Genes Coding for dTip60 Chromatin Remodeling Complex Subunits Affects Polytene Chromosome Organization and Proper Development in Drosophila melanogaster. Int. J. Mol. Sci. 2021, 22, 4525. [Google Scholar] [CrossRef]
- Messina, G.; Damia, E.; Fanti, L.; Atterrato, M.T.; Celauro, E.; Mariotti, F.R.; Accardo, M.C.; Walther, M.; Vernì, F.; Picchioni, D.; et al. Yeti, a Drosophila melanogaster essential gene, encodes a protein required for chromatin organization. J. Cell Sci. 2014, 127, 2577–2588. [Google Scholar] [CrossRef]
- Weber, C.M.; Henikoff, J.G.; Henikoff, S. H2A.Z nucleosomes enriched over active genes are homotypic. Nat. Struct. Mol. Biol. 2010, 17, 1500–1507. [Google Scholar] [CrossRef]
- Weber, C.M.; Ramachandran, S.; Henikoff, S. Nucleosomes are context-specific, H2A.Z-Modulated barriers to RNA polymerase. Mol. Cell 2014, 53, 819–830. [Google Scholar] [CrossRef]
- Swaminathan, J.; Baxter, E.M.; Corces, V.G. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 2005, 19, 65–76. [Google Scholar] [CrossRef]
- Leach, T.J.; Mazzeo, M.; Chotkowski, H.L.; Madigan, J.P.; Wotring, M.G.; Glaser, R.L. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J. Biol. Chem. 2000, 275, 23267–23272. [Google Scholar] [CrossRef]
- Akhmanova, A.S.; Bindels, P.C.T.; Xu, J.; Miedema, K.; Kremer, H.; Hennig, W.; Xu, J.; Hennig, W. Structure and expression of histone H3.3 genes in Drosophila melanogaster and Drosophila hydei. Genome 1995, 38, 586–600. [Google Scholar] [CrossRef]
- Ahmad, K.; Henikoff, S. The Histone Variant H3.3 Marks Active Chromatin by Replication-Independent Nucleosome Assembly. Mol. Cell 2002, 9, 1191–1200. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.-M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef]
- Mito, Y.; Henikoff, J.G.; Henikoff, S. Histone Replacement Marks the Boundaries of cis-Regulatory Domains. Science 2007, 315, 1408–1411. [Google Scholar] [CrossRef]
- Hödl, M.; Basler, K. Transcription in the Absence of Histone H3.3. Curr. Biol. 2009, 19, 1221–1226. [Google Scholar] [CrossRef]
- Braunschweig, U.; Hogan, G.J.; Pagie, L.; van Steensel, B. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 2009, 28, 3635–3645. [Google Scholar] [CrossRef] [PubMed]
- Nagel, S.; Grossbach, U. Histone H1 Genes and Histone Gene Clusters in the Genus Drosophila. J. Mol. Evol. 2000, 51, 286–298. [Google Scholar] [CrossRef]
- Pérez-Montero, S.; Carbonell, A.; Morán, T.; Vaquero, A.; Azorín, F. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev. Cell 2013, 26, 578–590. [Google Scholar] [CrossRef]
- Nalabothula, N.; McVicker, G.; Maiorano, J.; Martin, R.; Pritchard, J.K.; Fondufe-Mittendorf, Y.N. The chromatin architectural proteins HMGD1 and H1 bind reciprocally and have opposite effects on chromatin structure and gene regulation. BMC Genomics 2014, 15, 92. [Google Scholar] [CrossRef]
- Lu, X.; Wontakal, S.N.; Kavi, H.; Kim, B.J.; Guzzardo, P.M.; Emelyanov, A.V.; Xu, N.; Hannon, G.J.; Zavadil, J.; Fyodorov, D.V.; et al. Drosophila H1 Regulates the Genetic Activity of Heterochromatin by Recruitment of Su(var)3-9. Science 2013, 340, 78–81. [Google Scholar] [CrossRef]
- Vujatovic, O.; Zaragoza, K.; Vaquero, A.; Reina, O.; Bernués, J.; Azorín, F. Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity. Nucleic Acids Res. 2012, 40, 5402–5414. [Google Scholar] [CrossRef]
- Lu, X.; Wontakal, S.N.; Emelyanov, A.V.; Morcillo, P.; Konev, A.Y.; Fyodorov, D.V.; Skoultchi, A.I. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009, 23, 452–465. [Google Scholar] [CrossRef]
- Siriaco, G.; Deuring, R.; Chioda, M.; Becker, P.B.; Tamkun, J.W. Drosophila ISWI Regulates the Association of Histone H1 with Interphase Chromosomes in Vivo. Genetics 2009, 182, 661–669. [Google Scholar] [CrossRef]
- Bayona-Feliu, A.; Casas-Lamesa, A.; Reina, O.; Bernués, J.; Azorín, F. Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat. Commun. 2017, 8, 283. [Google Scholar] [CrossRef]
- Cao, K.; Lailler, N.; Zhang, Y.; Kumar, A.; Uppal, K.; Liu, Z.; Lee, E.K.; Wu, H.; Medrzycki, M.; Pan, C.; et al. High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells. PLoS Genet. 2013, 9, e1003417. [Google Scholar] [CrossRef]
- Izzo, A.; Kamieniarz-Gdula, K.; Ramírez, F.; Noureen, N.; Kind, J.; Manke, T.; van Steensel, B.; Schneider, R. The Genomic Landscape of the Somatic Linker Histone Subtypes H1.1 to H1.5 in Human Cells. Cell Rep. 2013, 3, 2142–2154. [Google Scholar] [CrossRef]
- Mayor, R.; Izquierdo-Bouldstridge, A.; Millán-Ariño, L.; Bustillos, A.; Sampaio, C.; Luque, N.; Jordan, A. Genome Distribution of Replication-independent Histone H1 Variants Shows H1.0 Associated with Nucleolar Domains and H1X Associated with RNA Polymerase II-enriched Regions. J. Biol. Chem. 2015, 290, 7474–7491. [Google Scholar] [CrossRef]
- Millán-Ariño, L.; Islam, A.B.M.M.K.; Izquierdo-Bouldstridge, A.; Mayor, R.; Terme, J.-M.; Luque, N.; Sancho, M.; López-Bigas, N.; Jordan, A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014, 42, 4474–4493. [Google Scholar] [CrossRef]
- Fan, Y.; Nikitina, T.; Zhao, J.; Fleury, T.J.; Bhattacharyya, R.; Bouhassira, E.E.; Stein, A.; Woodcock, C.L.; Skoultchi, A.I. Histone H1 Depletion in Mammals Alters Global Chromatin Structure but Causes Specific Changes in Gene Regulation. Cell 2005, 123, 1199–1212. [Google Scholar] [CrossRef]
- Izquierdo-Bouldstridge, A.; Bustillos, A.; Bonet-Costa, C.; Aribau-Miralbés, P.; García-Gomis, D.; Dabad, M.; Esteve-Codina, A.; Pascual-Reguant, L.; Peiró, S.; Esteller, M.; et al. Histone H1 depletion triggers an interferon response in cancer cells via activation of heterochromatic repeats. Nucleic Acids Res. 2017, 45, 11622–11642. [Google Scholar] [CrossRef]
- Sancho, M.; Diani, E.; Beato, M.; Jordan, A. Depletion of Human Histone H1 Variants Uncovers Specific Roles in Gene Expression and Cell Growth. PLoS Genet. 2008, 4, e1000227. [Google Scholar] [CrossRef]
- Healton, S.E.; Pinto, H.D.; Mishra, L.N.; Hamilton, G.A.; Wheat, J.C.; Swist-Rosowska, K.; Shukeir, N.; Dou, Y.; Steidl, U.; Jenuwein, T.; et al. H1 linker histones silence repetitive elements by promoting both histone H3K9 methylation and chromatin compaction. Proc. Natl. Acad. Sci. USA 2020, 117, 14251–14258. [Google Scholar] [CrossRef]
- Carbonell, A.; Pérez-Montero, S.; Climent-Cantó, P.; Reina, O.; Azorín, F. The Germline Linker Histone dBigH1 and the Translational Regulator Bam Form a Repressor Loop Essential for Male Germ Stem Cell Differentiation. Cell Rep. 2017, 21, 3178–3189. [Google Scholar] [CrossRef]
- Climent-Cantó, P.; Carbonell, A.; Tamirisa, S.; Henn, L.; Pérez-Montero, S.; Boros, I.M.; Azorín, F. The tumour suppressor brain tumour (Brat) regulates linker histone dBigH1 expression in the Drosophila female germline and the early embryo. Open Biol. 2021, 11, 200408. [Google Scholar] [CrossRef]
- Henn, L.; Szabó, A.; Imre, L.; Román, Á.; Ábrahám, A.; Vedelek, B.; Nánási, P.; Boros, I.M. Alternative linker histone permits fast paced nuclear divisions in early Drosophila embryo. Nucleic Acids Res. 2020, 48, 9007–9018. [Google Scholar] [CrossRef]
- Dunwell, T.L.; Gerd, P.P. Drosophila genomic methylation: New evidence and new questions. Epigenomics 2014, 6, 459–461. [Google Scholar] [CrossRef]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.-H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef]
- Yu, G.; Wu, Q.; Gao, Y.; Chen, M.; Yang, M. The Epigenetics of Aging in Invertebrates. Int. J. Mol. Sci. 2019, 20, 4535. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhang, G.; Wang, J.; Gao, Y.; Sun, R.; Cao, Z.; Chen, Z.; Zheng, X.; Yuan, J.; Luo, Y.; et al. 6mA-DNA-binding factor Jumu controls maternal-to-zygotic transition upstream of Zelda. Nat. Commun. 2019, 10, 2219. [Google Scholar] [CrossRef]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef]
- Akhtar, A.; Becker, P.B. Activation of Transcription through Histone H4 Acetylation by MOF, an Acetyltransferase Essential for Dosage Compensation in Drosophila. Mol. Cell 2000, 5, 367–375. [Google Scholar] [CrossRef]
- Shogren-Knaak, M.; Haruhiko, I.; Sun, J.-M.; Pazin, M.; Davie, J.R.; Peterson, C.L. Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef]
- Lu, X.; Simon, M.D.; Chodaparambil, J.V.; Hansen, J.C.; Shokat, K.M.; Luger, K. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat. Struct. Mol. Biol. 2008, 15, 1122–1124. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.-M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef]
- Verdin, E.; Ott, M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015, 16, 258–264. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. The Yin and Yang of Histone Marks in Transcription. Annu. Rev. Genom. Hum. Genet. 2021, 22, 147–170. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Lluch, S.; Blanco, E.; Tilgner, H.; Curado, J.; Ruiz-Romero, M.; Corominas, M.; Guigó, R. Absence of canonical marks of active chromatin in developmentally regulated genes. Nat. Genet. 2015, 47, 1158–1167. [Google Scholar] [CrossRef]
- Hödl, M.; Basler, K. Transcription in the Absence of Histone H3.2 and H3K4 Methylation. Curr. Biol. 2012, 22, 2253–2257. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, Z.; Dong, Q.; Xiong, J.; Zhu, B. Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 2020, 21, 45. [Google Scholar] [CrossRef]
- Li, X.-Y.; Harrison, M.M.; Villalta, J.E.; Kaplan, T.; Eisen, M.B. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 2014, 3, e03737. [Google Scholar] [CrossRef]
- Chen, K.; Johnston, J.; Shao, W.; Meier, S.; Staber, C.; Zeitlinger, J. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife 2013, 2, e00861. [Google Scholar] [CrossRef] [PubMed]
- Regadas, I.; Dahlberg, O.; Vaid, R.; Ho, O.; Belikov, S.; Dixit, G.; Deindl, S.; Wen, J.; Mannervik, M. A unique histone 3 lysine 14 chromatin signature underlies tissue-specific gene regulation. Mol. Cell 2021, 81, 1766–1780.e10. [Google Scholar] [CrossRef]
- Graves, H.K.; Wang, P.; Lagarde, M.; Chen, Z.; Tyler, J.K. Mutations that prevent or mimic persistent post-translational modifications of the histone H3 globular domain cause lethality and growth defects in Drosophila. Epigenet. Chromatin 2016, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Kharchenko, P.V.; Alekseyenko, A.A.; Schwartz, Y.B.; Minoda, A.; Riddle, N.C.; Ernst, J.; Sabo, P.J.; Larschan, E.; Gorchakov, A.A.; Gu, T.; et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 2011, 471, 480–485. [Google Scholar] [CrossRef]
- Bantignies, F.; Roure, V.; Comet, I.; Leblanc, B.; Schuettengruber, B.; Bonnet, J.; Tixier, V.; Mas, A.; Cavalli, G. Polycomb-Dependent Regulatory Contacts between Distant Hox Loci in Drosophila. Cell 2011, 144, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Boettiger, A.N.; Bintu, B.; Moffitt, J.R.; Wang, S.; Beliveau, B.J.; Fudenberg, G.; Imakaev, M.; Mirny, L.A.; Wu, C.; Zhuang, X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 2016, 529, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Kempfer, R.; Pombo, A. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 2020, 21, 207–226. [Google Scholar] [CrossRef]
- Dekker, J.; Marti-Renom, M.A.; Mirny, L.A. Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nat. Rev. Genet. 2013, 14, 390–403. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 2012, 148, 458–472. [Google Scholar] [CrossRef]
- Bonev, B.; Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016, 17, 661–678. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Misteli, T. The Self-Organizing Genome: Principles of Genome Architecture and Function. Cell 2020, 183, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Wutz, G.; Várnai, C.; Nagasaka, K.; Cisneros, D.A.; Stocsits, R.R.; Tang, W.; Schoenfelder, S.; Jessberger, G.; Muhar, M.; Hossain, M.J.; et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017, 36, 3573–3599. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Bauer, B.; Goetz, D.; Tang, W.; Wutz, G.; Peters, J. DNA loop extrusion by human cohesin. Science 2019, 366, 1338–1345. [Google Scholar] [CrossRef]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huang, S.-C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.-R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e24. [Google Scholar] [CrossRef]
- de Wit, E.; Vos, E.S.M.; Holwerda, S.J.B.; Valdes-Quezada, C.; Verstegen, M.J.A.M.; Teunissen, H.; Splinter, E.; Wijchers, P.J.; Krijger, P.H.L.; de Laat, W. CTCF Binding Polarity Determines Chromatin Looping. Mol. Cell 2015, 60, 676–684. [Google Scholar] [CrossRef]
- Hanssen, L.L.P.; Kassouf, M.T.; Oudelaar, A.M.; Biggs, D.; Preece, C.; Downes, D.J.; Gosden, M.; Sharpe, J.A.; Sloane-Stanley, J.A.; Hughes, J.R.; et al. Tissue-specific CTCF–cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 2017, 19, 952–961. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.-L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944. [Google Scholar] [CrossRef]
- Rowley, M.J.; Nichols, M.H.; Lyu, X.; Ando-Kuri, M.; Rivera, I.S.M.; Hermetz, K.; Wang, P.; Ruan, Y.; Corces, V.G. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol. Cell 2017, 67, 837–852. [Google Scholar] [CrossRef]
- Hug, C.B.; Grimaldi, A.G.; Kruse, K.; Vaquerizas, J.M. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 2017, 169, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Bhardwaj, V.; Arrigoni, L.; Lam, K.C.; Grüning, B.A.; Villaveces, J.; Habermann, B.; Akhtar, A.; Manke, T. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 2018, 9, 189. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, A.; Mohana, G.; Dorier, J.; Özdemir, I.; Omer, A.; Cousin, P.; Semenova, A.; Taschner, M.; Dergai, O.; Marzetta, F.; et al. CTCF loss has limited effects on global genome architecture in Drosophila despite critical regulatory functions. Nat. Commun. 2021, 12, 1011. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, M.C.; Furlong, E.E.M. The Insulator Protein CTCF Is Required for Correct Hox Gene Expression, but Not for Embryonic Development in Drosophila. Genetics 2018, 210, 129–136. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Cavalli, G. Three-dimensional genome organization and function in Drosophila. Genetics 2017, 205, 5–24. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, Q.; Czajkowsky, D.M.; Shao, Z. Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nat. Commun. 2018, 9, 188. [Google Scholar] [CrossRef]
- Ogiyama, Y.; Schuettengruber, B.; Papadopoulos, G.L.; Chang, J.-M.; Cavalli, G. Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Mol. Cell 2018, 71, 73–88. [Google Scholar] [CrossRef]
- Szabo, Q.; Donjon, A.; Jerković, I.; Papadopoulos, G.L.; Cheutin, T.; Bonev, B.; Nora, E.P.; Bruneau, B.G.; Bantignies, F.; Cavalli, G. Regulation of single-cell genome organization into TADs and chromatin nanodomains. Nat. Genet. 2020, 52, 1151–1157. [Google Scholar] [CrossRef]
- Szabo, Q.; Jost, D.; Chang, J.-M.; Cattoni, D.I.; Papadopoulos, G.L.; Bonev, B.; Sexton, T.; Gurgo, J.; Jacquier, C.; Nollmann, M.; et al. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci. Adv. 2018, 4, eaar8082. [Google Scholar] [CrossRef]
- Kundu, S.; Ji, F.; Sunwoo, H.; Jain, G.; Lee, J.T.; Sadreyev, R.I.; Dekker, J.; Kingston, R.E. Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Mol. Cell 2017, 65, 432–446. [Google Scholar] [CrossRef]
- Mateo, L.J.; Murphy, S.E.; Hafner, A.; Cinquini, I.S.; Walker, C.A.; Boettiger, A.N. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 2019, 568, 49–54. [Google Scholar] [CrossRef]
- De, S.; Gehred, N.D.; Fujioka, M.; Chan, F.W.; Jaynes, J.B.; Kassis, J.A. Defining the Boundaries of Polycomb Domains in Drosophila. Genetics 2020, 216, 689–700. [Google Scholar] [CrossRef]
- Szabo, Q.; Bantignies, F.; Cavalli, G. Principles of genome folding into topologically associating domains. Sci. Adv. 2019, 5, eaaw1668. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Razin, S.V. The two waves in single-cell 3D genomics. Semin. Cell Dev. Biol. 2021. [Google Scholar] [CrossRef]
- Flyamer, I.M.; Gassler, J.; Imakaev, M.; Brandão, H.B.; Ulianov, S.V.; Abdennur, N.; Razin, S.V.; Mirny, L.A.; Tachibana-Konwalski, K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 2017, 544, 110–114. [Google Scholar] [CrossRef]
- Cattoni, D.I.; Cardozo Gizzi, A.M.; Georgieva, M.; Di Stefano, M.; Valeri, A.; Chamousset, D.; Houbron, C.; Déjardin, S.; Fiche, J.-B.; González, I.; et al. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat. Commun. 2017, 8, 1753. [Google Scholar] [CrossRef] [PubMed]
- Eagen, K.P.; Hartl, T.A.; Kornberg, R.D. Stable Chromosome Condensation Revealed by Chromosome Conformation Capture. Cell 2015, 163, 934–946. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Khrameeva, E.E.; Gavrilov, A.A.; Flyamer, I.M.; Kos, P.; Mikhaleva, E.A.; Penin, A.A.; Logacheva, M.D.; Imakaev, M.V.; Chertovich, A.; et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 2016, 26, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Li, L.; Qin, Z.S.; Corces, V.G. Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Mol. Cell 2012, 48, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Levo, M.; Barinov, L.; Fujioka, M.; Jaynes, J.B.; Gregor, T. Dynamic interplay between enhancer–promoter topology and gene activity. Nat. Genet. 2018, 50, 1296–1303. [Google Scholar] [CrossRef]
- van Steensel, B.; Furlong, E.E.M. The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 2019, 20, 327–337. [Google Scholar] [CrossRef]
- Misteli, T.; Finn, E.H. Chromatin architecture is a flexible foundation for gene expression. Nat. Genet. 2021, 53, 426–427. [Google Scholar] [CrossRef]
- Cheutin, T.; Cavalli, G. Loss of PRC1 induces higher-order opening of Hox loci independently of transcription during Drosophila embryogenesis. Nat. Commun. 2018, 9, 3898. [Google Scholar] [CrossRef]
- Lupiáñez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kayserili, H.; Opitz, J.M.; Laxova, R.; et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef]
- Arzate-Mejía, R.G.; Josué Cerecedo-Castillo, A.; Guerrero, G.; Furlan-Magaril, M.; Recillas-Targa, F. In situ dissection of domain boundaries affect genome topology and gene transcription in Drosophila. Nat. Commun. 2020, 11, 894. [Google Scholar] [CrossRef] [PubMed]
- Cavalheiro, G.R.; Pollex, T.; Furlong, E.E. To loop or not to loop: What is the role of TADs in enhancer function and gene regulation? Curr. Opin. Genet. Dev. 2021, 67, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Ghavi-Helm, Y.; Jankowski, A.; Meiers, S.; Viales, R.R.; Korbel, J.O.; Furlong, E.E.M. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 2019, 51, 1272–1282. [Google Scholar] [CrossRef]
- Ing-Simmons, E.; Vaid, R.; Bing, X.Y.; Levine, M.; Mannervik, M.; Vaquerizas, J.M. Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nat. Genet. 2021, 53, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Espinola, S.M.; Götz, M.; Bellec, M.; Messina, O.; Fiche, J.-B.; Houbron, C.; Dejean, M.; Reim, I.; Cardozo Gizzi, A.M.; Lagha, M.; et al. Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat. Genet. 2021, 53, 477–486. [Google Scholar] [CrossRef]
- Williamson, I.; Kane, L.; Devenney, P.S.; Flyamer, I.M.; Anderson, E.; Kilanowski, F.; Hill, R.E.; Bickmore, W.A.; Lettice, L.A. Developmentally regulated Shh expression is robust to TAD perturbations. Development 2019, 146, dev179523. [Google Scholar] [CrossRef]
- Shevelyov, Y.Y.; Ulianov, S.V. The Nuclear Lamina as an Organizer of Chromosome Architecture. Cells 2019, 8, 136. [Google Scholar] [CrossRef] [PubMed]
- Pickersgill, H.; Kalverda, B.; de Wit, E.; Talhout, W.; Fornerod, M.; van Steensel, B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 2006, 38, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Peric-Hupkes, D.; Meuleman, W.; Pagie, L.; Bruggeman, S.W.M.; Solovei, I.; Brugman, W.; Gräf, S.; Flicek, P.; Kerkhoven, R.M.; van Lohuizen, M.; et al. Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol. Cell 2010, 38, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Nikolakaki, E.; Mylonis, I.; Giannakouros, T. Lamin B Receptor: Interplay between Structure, Function and Localization. Cells 2017, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Pindyurin, A.V.; Ilyin, A.A.; Ivankin, A.V.; Tselebrovsky, M.V.; Nenasheva, V.V.; Mikhaleva, E.A.; Pagie, L.; van Steensel, B.; Shevelyov, Y.Y. The large fraction of heterochromatin in Drosophila neurons is bound by both B-type lamin and HP1a. Epigenet. Chromatin 2018, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and Lamin A/C Sequentially Tether Peripheral Heterochromatin and Inversely Regulate Differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef]
- Zhu, J.; Adli, M.; Zou, J.Y.; Verstappen, G.; Coyne, M.; Zhang, X.; Durham, T.; Miri, M.; Deshpande, V.; De Jager, P.L.; et al. Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues. Cell 2013, 152, 642–654. [Google Scholar] [CrossRef]
- Kind, J.; Pagie, L.; de Vries, S.S.; Nahidiazar, L.; Dey, S.S.; Bienko, M.; Zhan, Y.; Lajoie, B.; de Graaf, C.A.; Amendola, M.; et al. Genome-wide Maps of Nuclear Lamina Interactions in Single Human Cells. Cell 2015, 163, 134–147. [Google Scholar] [CrossRef]
- Briand, N.; Collas, P. Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 2020, 21, 85. [Google Scholar] [CrossRef]
- Somech, R.; Shaklai, S.; Geller, O.; Amariglio, N.; Simon, A.J.; Rechavi, G.; Gal-Yam, E.N. The nuclear-envelope protein and transcriptional repressor LAP2β interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 2005, 118, 4017–4025. [Google Scholar] [CrossRef]
- Demmerle, J.; Koch, A.J.; Holaska, J.M. The Nuclear Envelope Protein Emerin Binds Directly to Histone Deacetylase 3 (HDAC3) and Activates HDAC3 Activity. J. Biol. Chem. 2012, 287, 22080–22088. [Google Scholar] [CrossRef] [PubMed]
- Milon, B.C.; Cheng, H.; Tselebrovsky, M.V.; Lavrov, S.A.; Nenasheva, V.V.; Mikhaleva, E.A.; Shevelyov, Y.Y.; Nurminsky, D.I. Role of Histone Deacetylases in Gene Regulation at Nuclear Lamina. PLoS ONE 2012, 7, e49692. [Google Scholar] [CrossRef] [PubMed]
- Verboon, J.M.; Rincon-Arano, H.; Werwie, T.R.; Delrow, J.J.; Scalzo, D.; Nandakumar, V.; Groudine, M.; Parkhurst, S.M. Wash Interacts with Lamin and Affects Global Nuclear Organization. Curr. Biol. 2015, 25, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Ulianov, S.V.; Doronin, S.A.; Khrameeva, E.E.; Kos, P.I.; Luzhin, A.V.; Starikov, S.S.; Galitsyna, A.A.; Nenasheva, V.V.; Ilyin, A.A.; Flyamer, I.M.; et al. Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 2019, 10, 1176. [Google Scholar] [CrossRef]
- Akhtar, W.; de Jong, J.; Pindyurin, A.V.; Pagie, L.; Meuleman, W.; de Ridder, J.; Berns, A.; Wessels, L.F.A.; van Lohuizen, M.; van Steensel, B. Chromatin Position Effects Assayed by Thousands of Reporters Integrated in Parallel. Cell 2013, 154, 914–927. [Google Scholar] [CrossRef]
- Shevelyov, Y.Y.; Lavrov, S.A.; Mikhaylova, L.M.; Nurminsky, I.D.; Kulathinal, R.J.; Egorova, K.S.; Rozovsky, Y.M.; Nurminsky, D.I. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc. Natl. Acad. Sci. USA 2009, 106, 3282–3287. [Google Scholar] [CrossRef]
- Kind, J.; Pagie, L.; Ortabozkoyun, H.; Boyle, S.; de Vries, S.S.; Janssen, H.; Amendola, M.; Nolen, L.D.; Bickmore, W.A.; van Steensel, B. Single-Cell Dynamics of Genome-Nuclear Lamina Interactions. Cell 2013, 153, 178–192. [Google Scholar] [CrossRef]
- Poleshko, A.; Shah, P.P.; Gupta, M.; Babu, A.; Morley, M.P.; Manderfield, L.J.; Ifkovits, J.L.; Calderon, D.; Aghajanian, H.; Sierra-Pagán, J.E.; et al. Genome-Nuclear Lamina Interactions Regulate Cardiac Stem Cell Lineage Restriction. Cell 2017, 171, 573–587.e14. [Google Scholar] [CrossRef]
- van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef]
- Tang, S.-J. Chromatin Organization by Repetitive Elements (CORE): A Genomic Principle for the Higher-Order Structure of Chromosomes. Genes 2011, 2, 502–515. [Google Scholar] [CrossRef]
- van de Werken, H.J.G.; Haan, J.C.; Feodorova, Y.; Bijos, D.; Weuts, A.; Theunis, K.; Holwerda, S.J.B.; Meuleman, W.; Pagie, L.; Thanisch, K.; et al. Small chromosomal regions position themselves autonomously according to their chromatin class. Genome Res. 2017, 27, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.; Feodorova, Y.; Naumova, N.; Imakaev, M.; Lajoie, B.R.; Leonhardt, H.; Joffe, B.; Dekker, J.; Fudenberg, G.; Solovei, I.; et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 2019, 570, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Riddle, N.C.; Minoda, A.; Kharchenko, P.V.; Alekseyenko, A.A.; Schwartz, Y.B.; Tolstorukov, M.Y.; Gorchakov, A.A.; Jaffe, J.D.; Kennedy, C.; Linder-Basso, D.; et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011, 21, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.D.; Shu, S.; Mungall, C.J.; Karpen, G.H. The Release 5.1 Annotation of Drosophila melanogaster Heterochromatin. Science 2007, 316, 1586–1591. [Google Scholar] [CrossRef]
- Lee, Y.C.G.; Ogiyama, Y.; Martins, N.M.C.; Beliveau, B.J.; Acevedo, D.; Wu, C.-T.; Cavalli, G.; Karpen, G.H. Pericentromeric heterochromatin is hierarchically organized and spatially contacts H3K9me2 islands in euchromatin. PLoS Genet. 2020, 16, e1008673. [Google Scholar] [CrossRef]
- Jagannathan, M.; Cummings, R.; Yamashita, Y.M. The modular mechanism of chromocenter formation in Drosophila. eLife 2019, 8, e43938. [Google Scholar] [CrossRef]
- Zhang, P.; Spradling, A.C. The Drosophila salivary gland chromocenter contains highly polytenized subdomains of mitotic heterochromatin. Genetics 1995, 139, 659–670. [Google Scholar] [CrossRef]
- Jones, K.W. Chromosomal and Nuclear Location of Mouse Satellite DNA in Individual Cells. Nature 1970, 225, 912–915. [Google Scholar] [CrossRef] [PubMed]
- Zenk, F.; Zhan, Y.; Kos, P.; Löser, E.; Atinbayeva, N.; Schächtle, M.; Tiana, G.; Giorgetti, L.; Iovino, N. HP1 drives de novo 3D genome reorganization in early Drosophila embryos. Nature 2021, 593, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Erdel, F.; Rademacher, A.; Vlijm, R.; Tünnermann, J.; Frank, L.; Weinmann, R.; Schweigert, E.; Yserentant, K.; Hummert, J.; Bauer, C.; et al. Mouse Heterochromatin Adopts Digital Compaction States without Showing Hallmarks of HP1-Driven Liquid-Liquid Phase Separation. Mol. Cell 2020, 78, 236–249.e7. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef]
- Vaquerizas, J.M.; Suyama, R.; Kind, J.; Miura, K.; Luscombe, N.M.; Akhtar, A. Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Drosophila Genome. PLoS Genet. 2010, 6, e1000846. [Google Scholar] [CrossRef] [PubMed]
- Gozalo, A.; Duke, A.; Lan, Y.; Pascual-Garcia, P.; Talamas, J.A.; Nguyen, S.C.; Shah, P.P.; Jain, R.; Joyce, E.F.; Capelson, M. Core Components of the Nuclear Pore Bind Distinct States of Chromatin and Contribute to Polycomb Repression. Mol. Cell 2020, 77, 67–81.e7. [Google Scholar] [CrossRef]
- Capelson, M.; Liang, Y.; Schulte, R.; Mair, W.; Wagner, U.; Hetzer, M.W. Chromatin-Bound Nuclear Pore Components Regulate Gene Expression in Higher Eukaryotes. Cell 2010, 140, 372–383. [Google Scholar] [CrossRef]
- Kalverda, B.; Pickersgill, H.; Shloma, V.V.; Fornerod, M. Nucleoporins Directly Stimulate Expression of Developmental and Cell-Cycle Genes Inside the Nucleoplasm. Cell 2010, 140, 360–371. [Google Scholar] [CrossRef]
- Pascual-Garcia, P.; Jeong, J.; Capelson, M. Nucleoporin Nup98 Associates with Trx/MLL and NSL Histone-Modifying Complexes and Regulates Hox Gene Expression. Cell Rep. 2014, 9, 433–442. [Google Scholar] [CrossRef]
- Pascual-Garcia, P.; Debo, B.; Aleman, J.R.; Talamas, J.A.; Lan, Y.; Nguyen, N.H.; Won, K.J.; Capelson, M. Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Mol. Cell 2017, 66, 63–76. [Google Scholar] [CrossRef]
- Ibarra, A.; Benner, C.; Tyagi, S.; Cool, J.; Hetzer, M.W. Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 2016, 30, 2253–2258. [Google Scholar] [CrossRef]
- Tan-Wong, S.M.; Wijayatilake, H.D.; Proudfoot, N.J. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009, 23, 2610–2624. [Google Scholar] [CrossRef]
- Strom, A.R.; Brangwynne, C.P. The liquid nucleome—Phase transitions in the nucleus at a glance. J. Cell Sci. 2019, 132, jcs235093. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- King, J.T.; Shakya, A. Phase separation of DNA: From past to present. Biophys. J. 2021, 120, 1139–1149. [Google Scholar] [CrossRef]
- Gibson, B.A.; Doolittle, L.K.; Schneider, M.W.G.; Jensen, L.E.; Gamarra, N.; Henry, L.; Gerlich, D.W.; Redding, S.; Rosen, M.K. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179, 470–484.e21. [Google Scholar] [CrossRef] [PubMed]
- Plys, A.J.; Davis, C.P.; Kim, J.; Rizki, G.; Keenen, M.M.; Marr, S.K.; Kingston, R.E. Phase separation of polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019, 33, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Lau, M.S.; Schwartz, M.G.; Kundu, S.; Savol, A.J.; Wang, P.I.; Marr, S.K.; Grau, D.J.; Schorderet, P.; Sadreyev, R.I.; Tabin, C.J.; et al. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science 2017, 355, 1081–1084. [Google Scholar] [CrossRef]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Pott, S.; Lieb, J.D. What are super-enhancers? Nat. Genet. 2015, 47, 8–12. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Cho, W.-K.; Spille, J.-H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.; Stadler, M.R.; Ortiz, S.A.; Hannon, C.E.; Harrison, M.M.; Darzacq, X.; Eisen, M.B. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 2018, 7, e40497. [Google Scholar] [CrossRef] [PubMed]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855. [Google Scholar] [CrossRef] [PubMed]
- Cardozo Gizzi, A.M.; Cattoni, D.I.; Fiche, J.-B.; Espinola, S.M.; Gurgo, J.; Messina, O.; Houbron, C.; Ogiyama, Y.; Papadopoulos, G.L.; Cavalli, G.; et al. Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms. Mol. Cell 2019, 74, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Cardozo Gizzi, A.M.; Espinola, S.M.; Gurgo, J.; Houbron, C.; Fiche, J.-B.; Cattoni, D.I.; Nollmann, M. Direct and simultaneous observation of transcription and chromosome architecture in single cells with Hi-M. Nat. Protoc. 2020, 15, 840–876. [Google Scholar] [CrossRef]
- Askjaer, P.; Harr, J.C. Genetic approaches to revealing the principles of nuclear architecture. Curr. Opin. Genet. Dev. 2021, 67, 52–60. [Google Scholar] [CrossRef]
- Jerkovic, I.; Cavalli, G. Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 2021, 22, 511–528. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Chattoraj, S.; Castillo, D.; Nguyen, S.C.; Nir, G.; Lioutas, A.; Hershberg, E.A.; Martins, N.M.C.; Reginato, P.L.; Hannan, M.; et al. 3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing. Nat. Methods 2020, 17, 822–832. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llorens-Giralt, P.; Camilleri-Robles, C.; Corominas, M.; Climent-Cantó, P. Chromatin Organization and Function in Drosophila. Cells 2021, 10, 2362. https://doi.org/10.3390/cells10092362
Llorens-Giralt P, Camilleri-Robles C, Corominas M, Climent-Cantó P. Chromatin Organization and Function in Drosophila. Cells. 2021; 10(9):2362. https://doi.org/10.3390/cells10092362
Chicago/Turabian StyleLlorens-Giralt, Palmira, Carlos Camilleri-Robles, Montserrat Corominas, and Paula Climent-Cantó. 2021. "Chromatin Organization and Function in Drosophila" Cells 10, no. 9: 2362. https://doi.org/10.3390/cells10092362
APA StyleLlorens-Giralt, P., Camilleri-Robles, C., Corominas, M., & Climent-Cantó, P. (2021). Chromatin Organization and Function in Drosophila. Cells, 10(9), 2362. https://doi.org/10.3390/cells10092362






