Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases in Echinoderms: Structure and Possible Functions
Abstract
:1. Introduction
1.1. Echinoderms
1.2. Components of Connective Tissue of Echinoderms
1.3. Proteins Modifying Connective Tissue
2. Matrix Metalloproteinases of Echinoderms
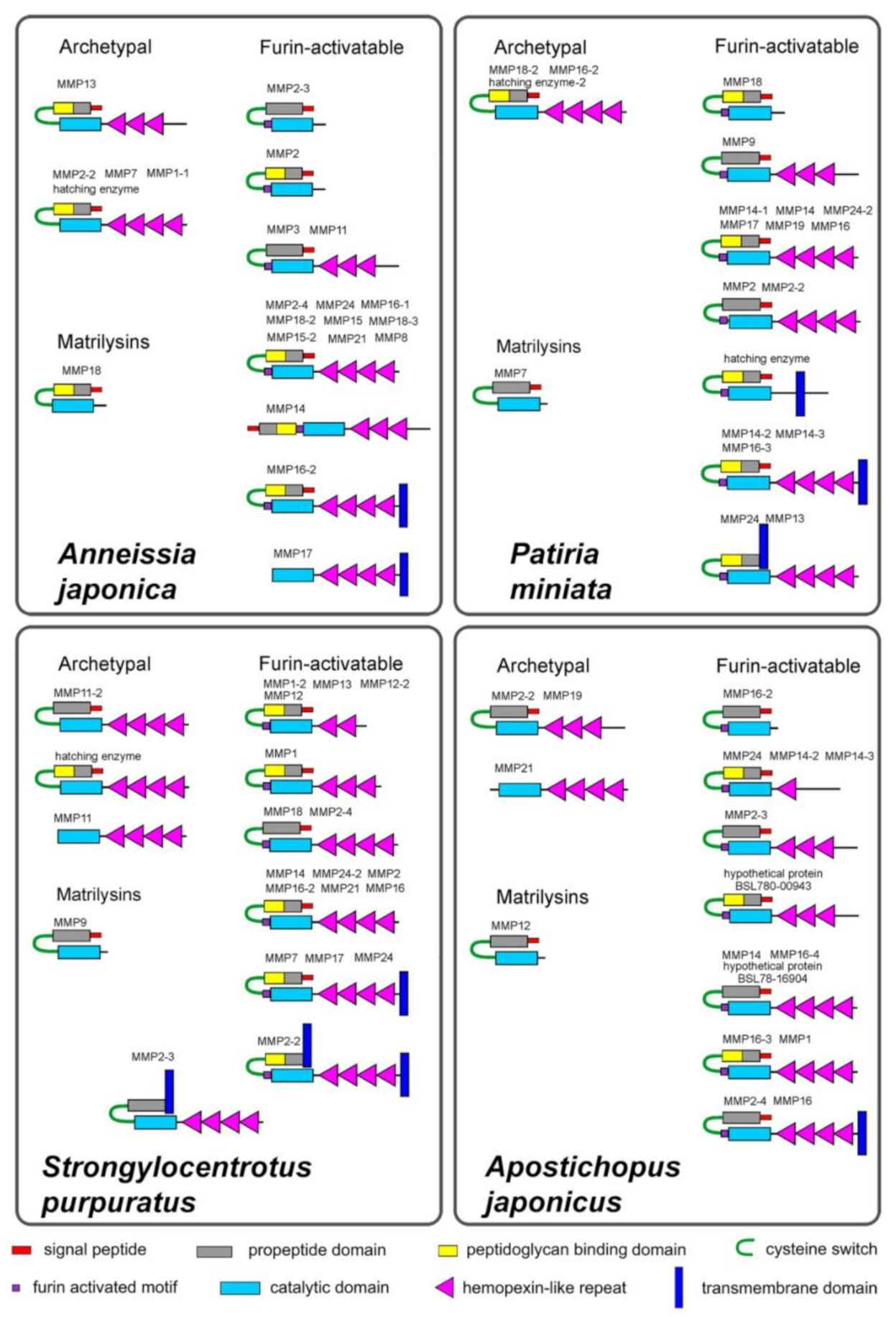
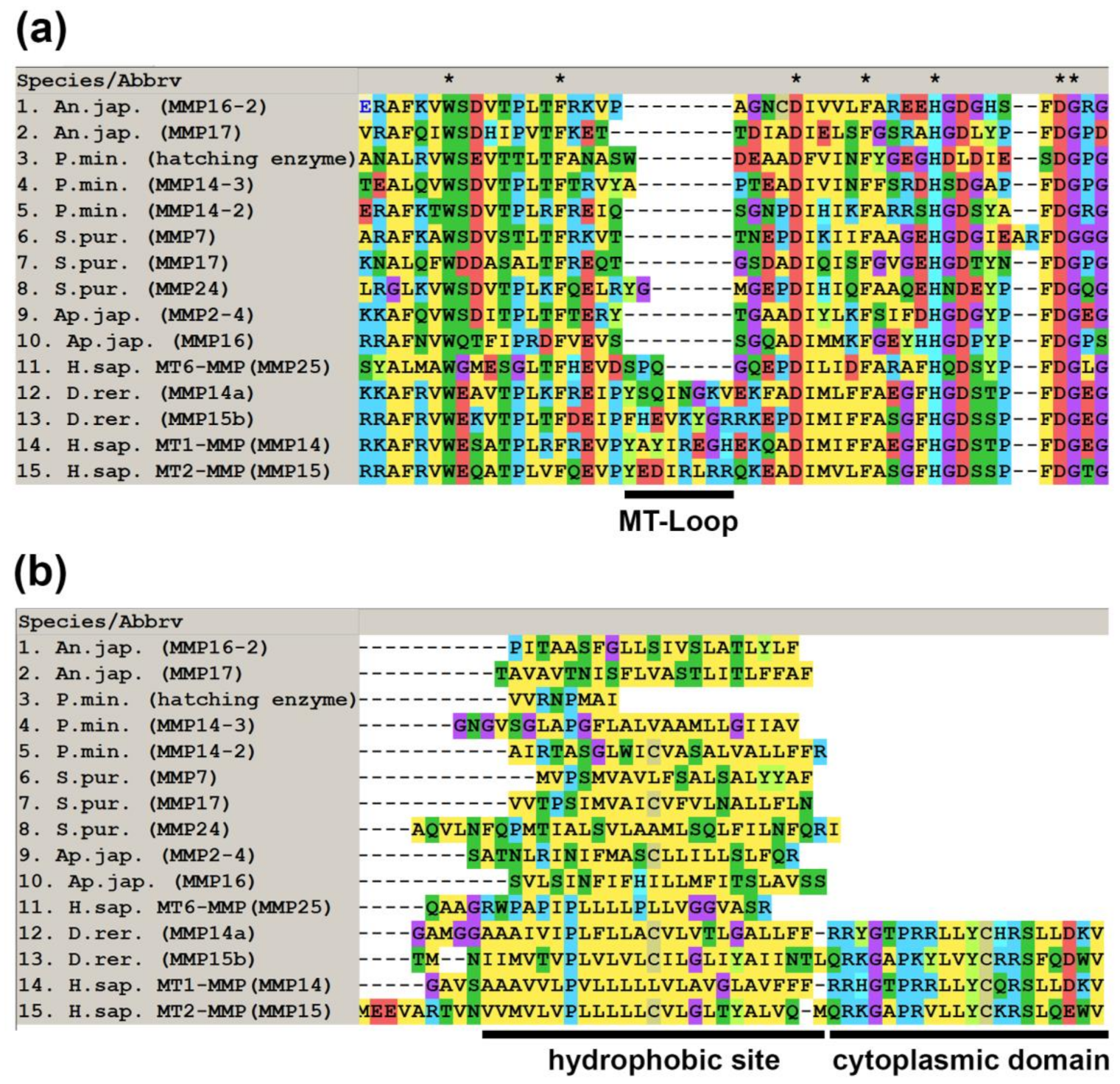
2.1. Domain Structure
2.1.1. Archetypal MMPs
2.1.2. Matrilysins
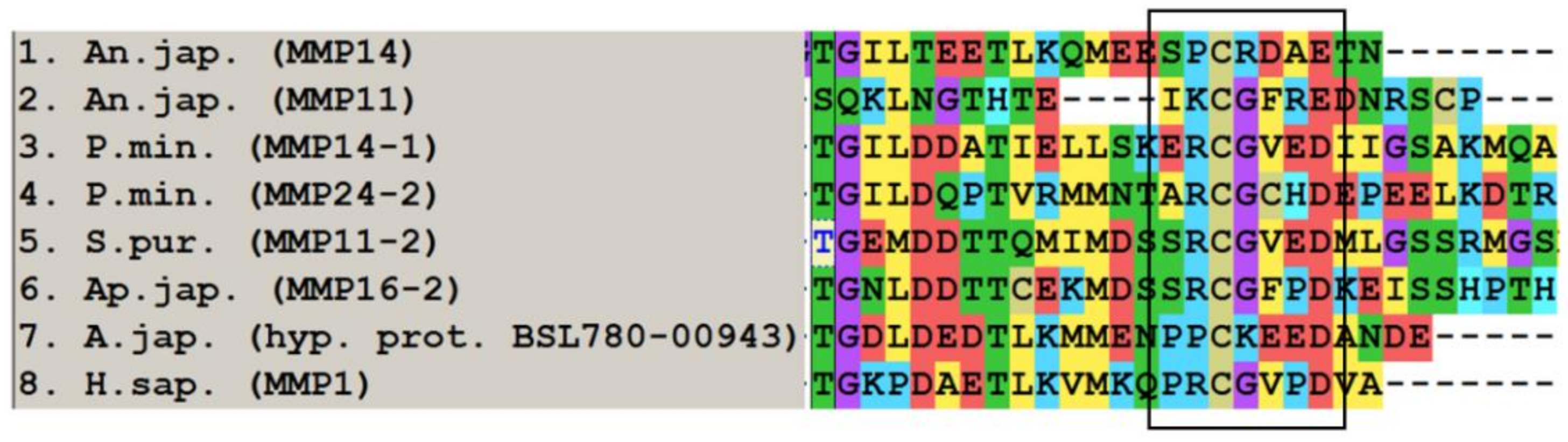
2.1.3. Furin-Activatable MMPs
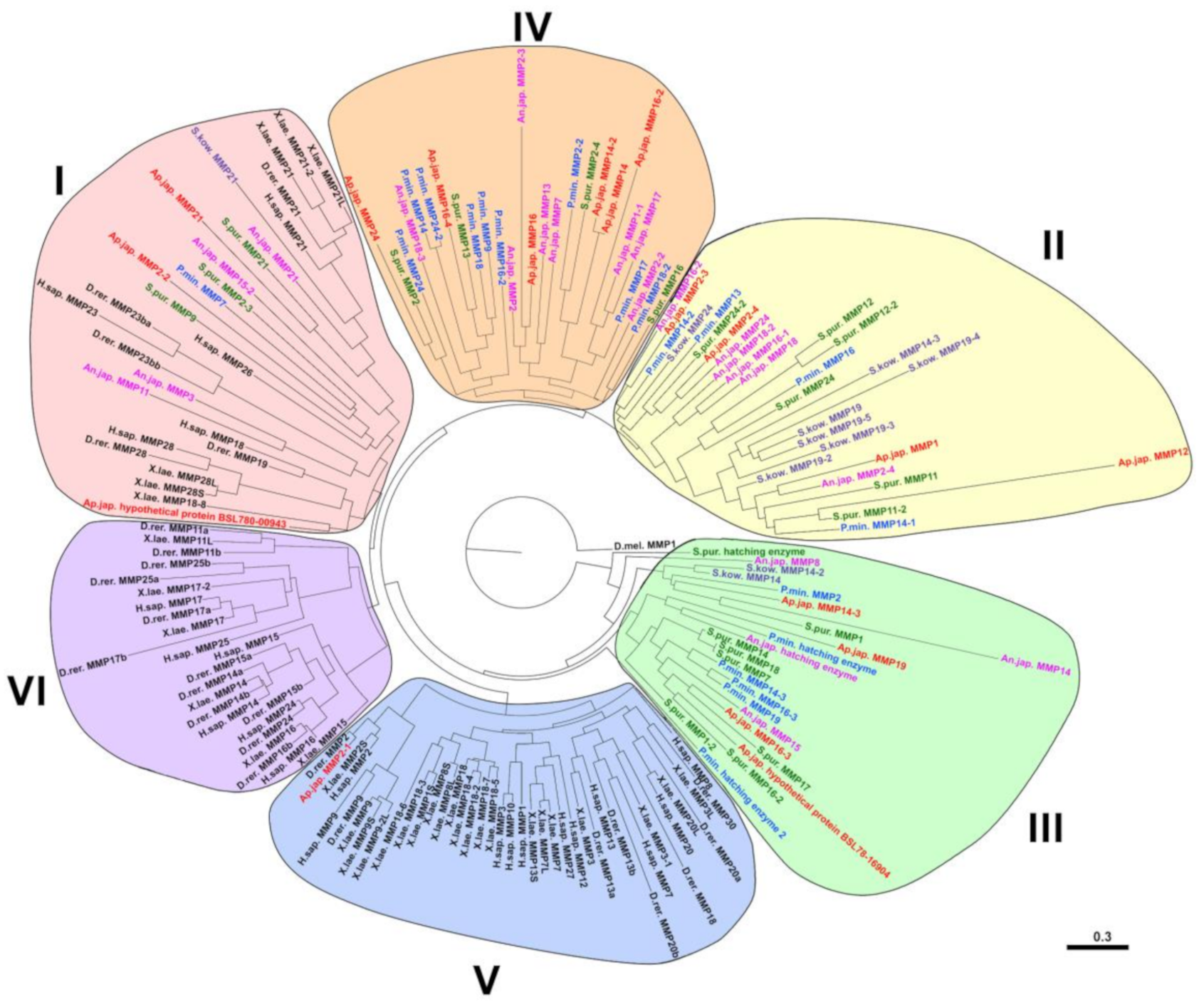
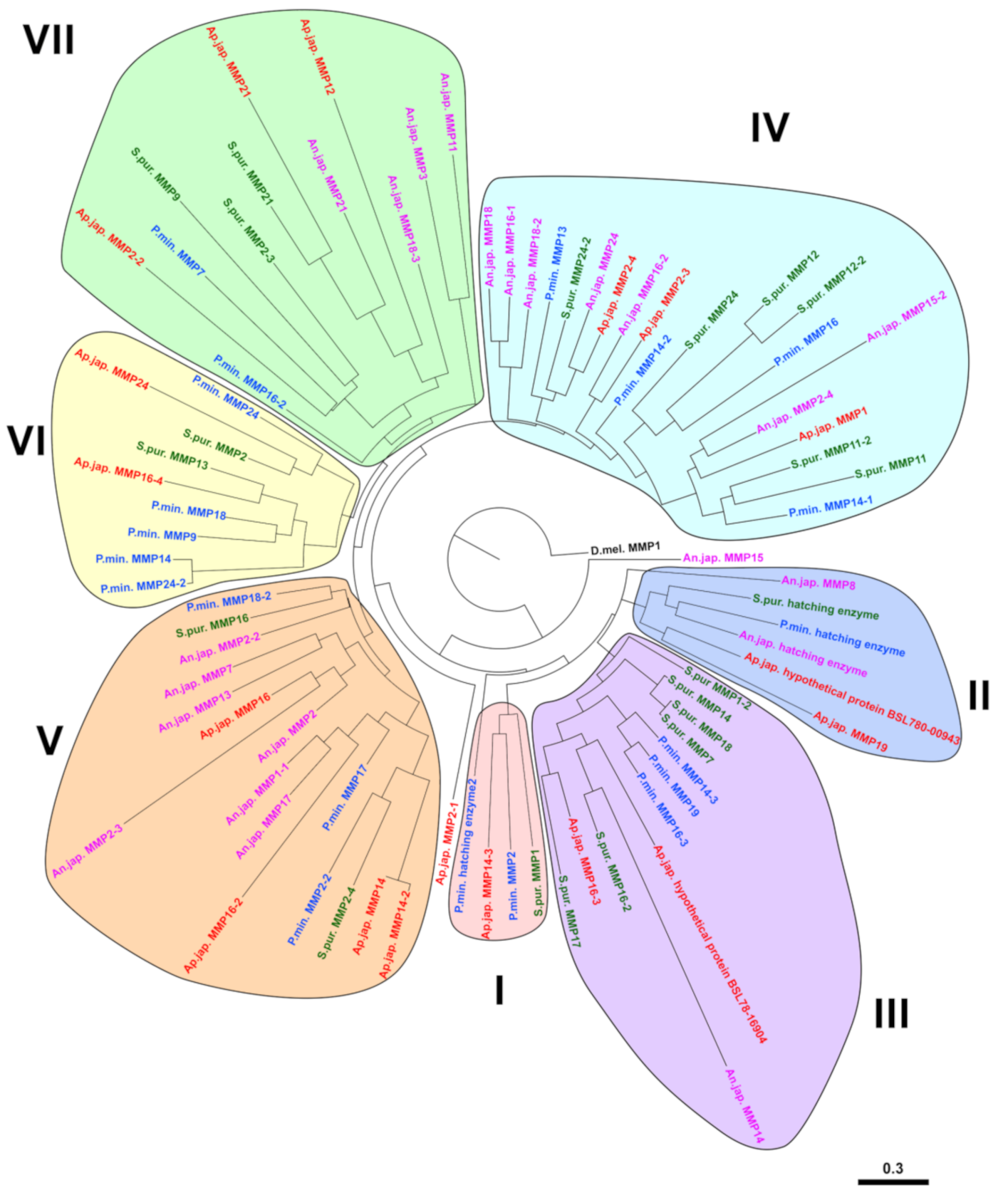
2.2. Evolution of MMPs of Echinoderms
2.3. Substrate Specificity and Function
3. Tissue Inhibitors of Metalloproteinases
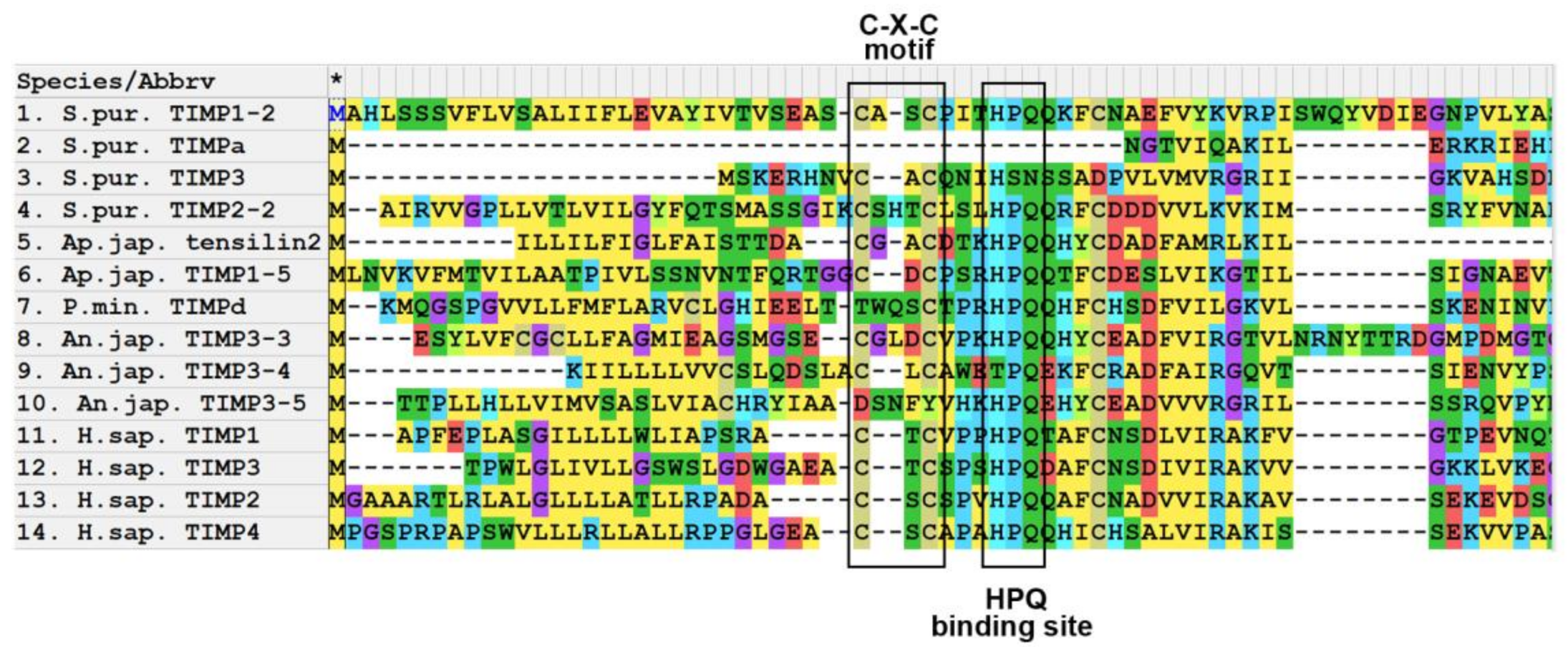
3.1. Domain Structure
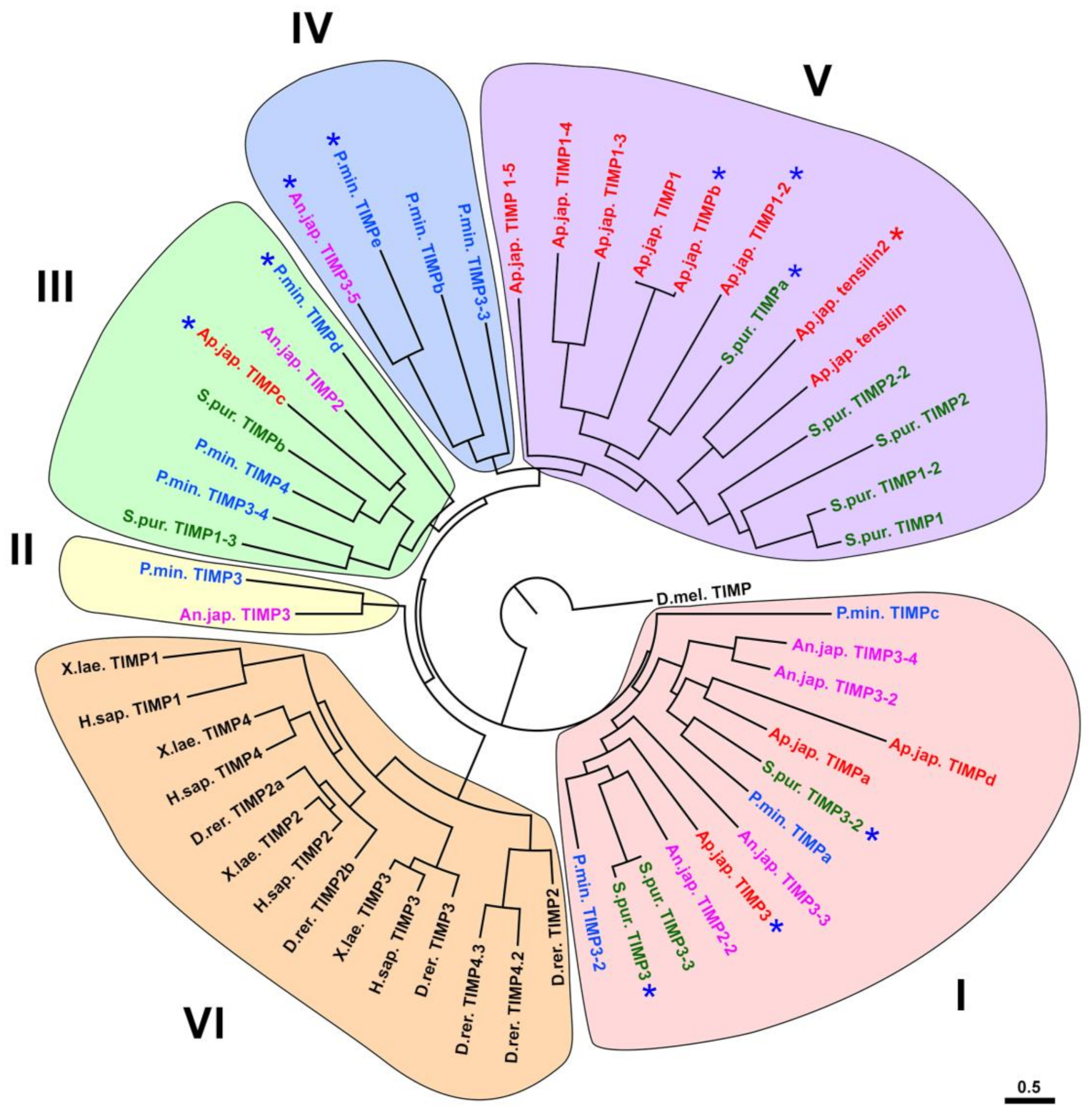
3.2. Evolution of TIMP of Echinoderms
3.3. Functions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Ausich, W.I. Origin of the Crinoidea. In Echinoderms: San Francisco; Mooi, R., Telford, M., Eds.; A.A Balkema: Rotterdam, The Netherlands, 1998; pp. 127–132. [Google Scholar]
- Ausich, W.I.; Baumiller, T.K. Column regeneration in an ordovician crinoid (Echinodermata): Paleobiologic implications. J. Paleontol. 1993, 67, 1068–1070. [Google Scholar] [CrossRef]
- Swalla, B.J.; Smith, A.B. Deciphering deuterostome phylogeny: Molecular, morphological and palaentological perspectives. Philos. Trans. Roy. Soc. B 2008, 363, 1557–1568. [Google Scholar] [CrossRef] [Green Version]
- Bluhm, H.; Gebruk, A. Holothuroidea (Echinodermata) of the Peru Basin—Ecological and taxonomic remarks based on underwater images. Mar. Ecol. 1999, 20, 167–195. [Google Scholar] [CrossRef]
- Solan, M.; Germano, J.D.; Rhoads, D.C.; Smith, C.; Michaud, E.; Parry, D.; Wenzhofer, F.; Kennedy, B.; Henriques, C.; Battle, E.; et al. Towards a greater understanding of pattern, scale and process in marine benthic systems: A picture is worth a thousand worms. J. Exp. Mar. Biol. Ecol. 2003, 285, 313–338. [Google Scholar] [CrossRef]
- Ohta, S. Photographic observations of the deep sea pelagothuriid holothurian Enypniastes (Elasipoda, Holothurioidea). J. Oceanogr. Soc. Jpn. 1985, 41, 121–133. [Google Scholar] [CrossRef]
- Brusca, R.C.; Brusca, G.J. Invertebrates, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2003; p. 936. [Google Scholar]
- Wilkie, I.C. Autotomy as a prelude to regeneration in echinoderms. Microsc. Res. Tech. 2001, 55, 369–396. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, P.V.; Burke, R.D. Echinodermata: Asexual propagation. In Reproductive Biology of Invertebrates, Volume 6, Part B, Asexual Propagation and Reproductive Strategies; Adiyodi, K.G., Adiyodi, R.G., Eds.; Oxford and IBH Publishing Co. PVT. Ltd.: New Delhi, India; Bombay, India; Calcutta, India, 1994; pp. 339–383. [Google Scholar]
- Motokawa, T.; Sato, E.; Umeyama, K. Energy expenditure associated with softening and stiffening of echinoderm connective tissue. Biol. Bull. 2012, 222, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Motokawa, T.; Tsuchi, A. Dynamic mechanical properties of body-wall dermis in various mechanical states and their implications for the behavior of sea cucumbers. Biol. Bull. 2003, 205, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Uthicke, S. Influence of asexual reproduction on the structure and dynamics of Holothuria (Halodeima) atra and Stichopus chloronotus populations of the Great Barrier Reef. Mar. Freshw. Res. 2001, 52, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Wilkie, I.C. Variable tensility in echinoderm collagenous tissues: A review. Mar. Behav. Physiol. 1984, 11, 1–34. [Google Scholar] [CrossRef]
- Motokawa, T. Connective tissue catch in echinoderms. Biol. Rev. 1984, 59, 255–270. [Google Scholar] [CrossRef]
- Szulgit, G. The echinoderm collagen fibril: A hero in the connective tissue research of the 1990s. BioEssays 2007, 29, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Candia Carnevali, M.D. Regeneration in echinoderms: Repair, regrowth, cloning. Invertebr. Surv. J. 2006, 3, 64–76. [Google Scholar]
- Dolmatov, I.Y. Regeneration in echinoderms. Russ. J. Mar. Biol. 1999, 25, 225–233. [Google Scholar]
- Dolmatov, I.Y. Variability of regeneration mechanisms in echinoderms. Russ. J. Mar. Biol. 2020, 46, 391–404. [Google Scholar] [CrossRef]
- Dolmatov, I.Y. Molecular aspects of regeneration mechanisms in holothurians. Genes 2021, 12, 250. [Google Scholar] [CrossRef] [PubMed]
- Dolmatov, I.Y. Asexual reproduction in holothurians. ScientificWorldJournal 2014, 2014, 13. [Google Scholar] [CrossRef]
- Dolmatov, I.Y. Regeneration potential and its changes during ontogenesis of holothurians. Russ. J. Dev. Biol. 1994, 25, 31–37. [Google Scholar]
- Dolmatov, I.Y. New data on asexual reproduction, autotomy, and regeneration in holothurians of the order Dendrochirotida. Russ. J. Mar. Biol. 2014, 40, 228–232. [Google Scholar] [CrossRef]
- Dolmatov, I.Y.; An Khang, N.; Kamenev, Y.O. Asexual reproduction, evisceration, and regeneration in holothurians (Holothuroidea) from Nha Trang Bay of the South China Sea. Russ. J. Mar. Biol. 2012, 38, 243–252. [Google Scholar] [CrossRef]
- Kille, F.R. Regeneration of the reproductive system following binary fission in the sea cucumber Holothuria parvula. Biol. Bull. 1942, 83, 55–66. [Google Scholar] [CrossRef]
- Monticelli, F.S. Sull’ autotomia delle Cucumaria planci (Br.). Atti Accad. Naz. Lincei Cl. Sci. Fis. Mat. Nat. Rendi. 1896, 5, 231–239. [Google Scholar]
- Reichenbach, N.; Holloway, S. Potential for asexual propagation of several commercially important species of tropical sea cucumbers (Echinodermata). J. World Aquac. Soc. 1995, 26, 272–278. [Google Scholar] [CrossRef]
- Reichenbach, N.; Nishar, Y.; Saeed, A. Species and size-related trends in asexual propagation of commercially important species of tropical sea cucumbers (Holothuroidea). J. World Aquac. Soc. 1996, 27, 475–482. [Google Scholar] [CrossRef]
- Torelle, E. Regeneration in holothuria. Zool. Anz. 1910, 35, 15–22. [Google Scholar]
- Kamenev, Y.O.; Dolmatov, I.Y. Posterior regeneration following fission in the holothurian Cladolabes schmeltzii (Dendrochirotida: Holothuroidea). Microsc. Res. Tech. 2015, 78, 540–552. [Google Scholar] [CrossRef]
- Kamenev, Y.O.; Dolmatov, I.Y. Anterior regeneration after fission in the holothurian Cladolabes schmeltzii (Dendrochirotida: Holothuroidea). Microsc. Res. Tech. 2017, 80, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Dolmatov, I.Y.; Ginanova, T.T. Muscle regeneration in holothurians. Microsc. Res. Tech. 2001, 55, 452–463. [Google Scholar] [CrossRef] [PubMed]
- García-Arrarás, J.E.; Estrada-Rodgers, L.; Santiago, R.; Torres, I.I.; Díaz-Miranda, L.; Torres-Avillán, I. Cellular mechanisms in the regeneration of the intestine of the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea: Echinodermata). J. Exp. Zool. 1998, 281, 288–304. [Google Scholar] [CrossRef]
- Mashanov, V.S.; Dolmatov, I.Y.; Heinzeller, T. Transdifferentiation in holothurian gut regeneration. Biol. Bull. 2005, 209, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, C.A.; Bergeron, K.-F.; Whittle, J.; Brandhorst, B.P.; Burke, R.D.; Hynes, R.O. The echinoderm adhesome. Dev. Biol. 2006, 300, 252–266. [Google Scholar] [CrossRef] [Green Version]
- Dolmatov, I.Y.; Afanasyev, S.V.; Boyko, A.V. Molecular mechanisms of fission in echinoderms: Transcriptome analysis. PLoS ONE 2018, 13, e0195836. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Pineda, P.A.; Ramírez-Gómez, F.; Pérez-Ortiz, J.; González-Díaz, S.; Jesús, F.S.; Hernández-Pasos, J.; Valle-Avila, C.D.; Rojas-Cartagena, C.; Suárez-Castillo, E.C.; Tossas, K.; et al. Gene expression profiling of intestinal regeneration in the sea cucumber. BMC Genom. 2009, 10, 262. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Cartagena, C.; Ortiz-Pineda, P.; Ramirez-Gomez, F.; Suarez-Castillo, E.C.; Matos-Cruz, V.; Rodriguez, C.; Ortiz-Zuazaga, H.; Garcia-Arraras, J.E. Distinct profiles of expressed sequence tags during intestinal regeneration in the sea cucumber Holothuria glaberrima. Physiol. Genom. 2007, 31, 203–215. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Chen, M.; Yang, H.; Wang, T.; Liu, B.; Shu, C.; Gardiner, D.M. Large scale gene expression profiling during intestine and body wall regeneration in the sea cucumber Apostichopus japonicus. Comp. Biochem. Physiol. Part D Genom. Proteom. 2011, 6, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, H.; Chen, M.; Ma, D.; Lin, C. RNA-Seq reveals dynamic changes of gene expression in key stages of intestine regeneration in the sea cucumber Apostichopus japonicus. [corrected]. PLoS ONE 2013, 8, e69441. [Google Scholar]
- Naba, A.; Clauser, K.R.; Ding, H.; Whittaker, C.A.; Carr, S.A.; Hynes, R.O. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016, 49, 10–24. [Google Scholar] [CrossRef]
- Hynes, R.O.; Naba, A. Overview of the matrisome—An inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 2012, 4, a004903. [Google Scholar] [CrossRef] [Green Version]
- Boyko, A.V.; Girich, A.S.; Eliseikina, M.G.; Maslennikov, S.I.; Dolmatov, I.Y. Reference assembly and gene expression analysis of Apostichopus japonicus larval development. Sci. Rep. 2019, 9, 1131. [Google Scholar] [CrossRef]
- Boyko, A.V.; Girich, A.S.; Tkacheva, E.S.; Dolmatov, I.Y. The Eupentacta fraudatrix transcriptome provides insights into regulation of cell transdifferentiation. Sci. Rep. 2020, 10, 1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tucker, R.P.; Chiquet-Ehrismann, R. Evidence for the evolution of tenascin and fibronectin early in the chordate lineage. Int. J. Biochem. Cell Biol. 2009, 41, 424–434. [Google Scholar] [CrossRef]
- Chiquet-Ehrismann, R.; Tucker, R.P. Tenascins and the importance of adhesion modulation. Cold Spring Harb. Perspect. Biol. 2011, 3, a004960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzbauer, J.E.; DeSimone, D.W. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb. Perspect. Biol. 2011, 3, a005041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ba, H.; Yao, F.; Yang, L.; Qin, T.; Luan, H.; Li, Z.; Zou, X.; Hou, L. Identification and expression patterns of extracellular matrix-associated genes fibropellin-ia and tenascin involved in regeneration of sea cucumber Apostichopus japonicus. Gene 2015, 565, 96–105. [Google Scholar] [CrossRef]
- Hynes, R.O. The evolution of metazoan extracellular matrix. J. Cell Biol. 2012, 196, 671–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cattaruzza, S.; Perris, R. Proteoglycan control of cell movement during wound healing and cancer spreading. Matrix Biol. 2005, 24, 400–417. [Google Scholar] [CrossRef]
- Hacker, U.; Nybakken, K.; Perrimon, N. Heparan sulphate proteoglycans: The sweet side of development. Nat. Rev. Mol. Cell Biol. 2005, 6, 530–541. [Google Scholar] [CrossRef]
- Kramer, K.L.; Yost, H.J. Heparan sulfate core proteins in cell-cell signaling. Annu. Rev. Genet. 2003, 37, 461–484. [Google Scholar] [CrossRef]
- Thurmond, F.A.; Koob, T.J.; Bowness, J.M.; Trotter, J.A. Partial biochemical and immunologic characterization of fibrillin microfibrils from sea cucumber dermis. Connect. Tissue Res. 1997, 36, 211–222. [Google Scholar] [CrossRef]
- Ribeiro, A.R.; Barbaglio, A.; Oliveira, M.J.; Santos, R.; Coelho, A.V.; Ribeiro, C.C.; Wilkie, I.C.; Carnevali, M.D.C.; Barbosa, M.A. Correlations between the biochemistry and mechanical states of a sea-urchin ligament: A mutable collagenous structure. Biointerphases 2012, 7, 38. [Google Scholar] [CrossRef] [Green Version]
- Vázquez-Vélez, G.E.; Rodríguez-Molina, J.F.; Quiñones-Frías, M.C.; Pagán, M. A proteoglycan-like molecule offers insights into ground substance changes during holothurian intestinal regeneration. J. Histochem. Cytochem. 2016, 64, 381–393. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.C.; Wei, M.; Yue, Y.; Wu, N.; Wang, J.; Zhang, Q. Structural characterization and immunostimulatory activity in vitro of a glycogen from sea urchin-Strongylocentyotus internedius. Carbohydr. Polym. 2021, 258, 117701. [Google Scholar] [CrossRef] [PubMed]
- Eckmair, B.; Jin, C.; Karlsson, N.G.; Abed-Navandi, D.; Wilson, I.B.H.; Paschinger, K. Glycosylation at an evolutionary nexus: The brittle star Ophiactis savignyi expresses both vertebrate and invertebrate N-glycomic features. J. Biol. Chem. 2020, 295, 3173–3188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanbeselaere, J.; Jin, C.; Eckmair, B.; Wilson, I.B.H.; Paschinger, K. Sulfated and sialylated N-glycans in the echinoderm Holothuria atra reflect its marine habitat and phylogeny. J. Biol. Chem. 2020, 295, 3159–3172. [Google Scholar] [CrossRef]
- Pasten, C.; Rosa, R.; Ortiz, S.; Gonzalez, S.; Garcia-Arraras, J.E. Characterization of proteolytic activities during intestinal regeneration of the sea cucumber, Holothuria glaberrima. Int. J. Dev. Biol. 2012, 56, 681–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, L.-J.; Zhan, C.-L.; Cai, Q.-F.; Weng, L.; Du, C.-H.; Liu, G.-M.; Su, W.-J.; Cao, M.-J. Purification, characterization, cDNA cloning and in vitro expression of a serine proteinase from the intestinal tract of sea cucumber (Stichopus japonicus) with collagen degradation activity. J. Agric. Food Chem. 2014, 62, 4769–4777. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.Y.; Chang, X.N.; Bao, S.S.; Song, L.; Zhu, B.W.; Dong, X.P.; Zong, Y.; Li, D.M.; Zhang, M.M.; Liu, Y.X.; et al. Purification and partial characterisation of a cathepsin L-like proteinase from sea cucumber (Stichopus japonicus) and its tissue distribution in body wall. Food Chem. 2014, 158, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Angerer, L.; Hussain, S.; Wei, Z.; Livingston, B.T. Sea urchin metalloproteases: A genomic survey of the BMP-1/tolloid-like, MMP and ADAM families. Dev. Biol. 2006, 300, 267–281. [Google Scholar] [CrossRef] [Green Version]
- Lamash, N.E.; Dolmatov, I.Y. Proteases from the regenerating gut of the holothurian Eupentacta fraudatrix. PLoS ONE 2013, 8, e58433. [Google Scholar] [CrossRef] [Green Version]
- Miao, T.; Wan, Z.; Sun, L.; Li, X.; Xing, L.; Bai, Y.; Wang, F.; Yang, H. Extracellular matrix remodeling and matrix metalloproteinases (ajMMP-2 like and ajMMP-16 like) characterization during intestine regeneration of sea cucumber Apostichopus japonicus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 212, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Shulga, A.P.; Lamash, N.E. Proteinases with gelatinase activity and their role in ambulacrum regeneration in holothurians Eupentacta fraudatrix (D’yakonov and Baranova, 1958) and Cucumaria japonica (Semper, 1868) (Echinodermata: Holothuroidea). Russ. J. Mar. Biol. 2020, 46, 461–471. [Google Scholar] [CrossRef]
- Yuan, Z.; Dahms, H.U.; Han, L.L.; Li, Q.Y.; Zhang, Q.Z.; Wu, R.J.; Tan, J.; Zou, X.Y.; Hou, L. Cloning and characterization of a trypsin-like serine protease gene, a novel regeneration-related gene from Apostichopus japonicus. Gene 2012, 502, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Mashanov, V.S.; Zueva, O.R.; García-Arrarás, J.E. Expression of Wnt9, TCTP, and Bmp1/Tll in sea cucumber visceral regeneration. Gene Expr. Patterns 2012, 12, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Green, K.A.; Lund, L.R. ECM degrading proteases and tissue remodelling in the mammary gland. BioEssays 2005, 27, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a005058. [Google Scholar] [CrossRef]
- Laronha, H.; Caldeira, J. Structure and function of human matrix metalloproteinases. Cells 2020, 9, 1076. [Google Scholar] [CrossRef]
- Brkic, M.; Sriram, B.; Libert, C.; Vandenbroucke, R. Friends or foes: Matrix metalloproteinases and their multifaceted roles in neurodegenerative diseases. Mediat. Inflamm 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Khalil, R.A. Matrix metalloproteinases, vascular remodeling, and vascular disease. Adv. Pharmacol. 2018, 81, 241–330. [Google Scholar]
- Parks, W.C. Matrix metalloproteinases in repair. Wound Repair Regen 1999, 7, 423–432. [Google Scholar] [CrossRef]
- Woessner, J.F., Jr. Matrix metalloproteinases and their inhibitors in connective tissue remodeling. FASEB J. 1991, 5, 2145–2154. [Google Scholar] [CrossRef] [Green Version]
- Becher, N.; Hein, M.; Uldbjerg, N.; Danielsen, C.C. Balance between matrix metalloproteinases (MMP) and tissue inhibitors of metalloproteinases (TIMP) in the cervical mucus plug estimated by determination of free non-complexed TIMP. Reprod. Biol. Endocrinol. 2008, 6, 45. [Google Scholar] [CrossRef] [Green Version]
- Brew, K.; Nagase, H. The tissue inhibitors of metalloproteinases (TIMPs): An ancient family with structural and functional diversity. BBA Mol. Cell Res. 2010, 1803, 55–71. [Google Scholar] [CrossRef] [Green Version]
- Murphy, G. Tissue inhibitors of metalloproteinases. Genome Biol. 2011, 12, 233. [Google Scholar] [CrossRef] [PubMed]
- Clouse, R.M.; Linchangco, G.V.; Kerr, A.M.; Reid, R.W.; Janies, D.A. Phylotranscriptomic analysis uncovers a wealth of tissue inhibitor of metalloproteinases variants in echinoderms. R. Soc. Open Sci. 2015, 2, 150377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolmatov, I.Y.; Kalacheva, N.V.; Tkacheva, E.S.; Shulga, A.P.; Zavalnaya, E.G.; Shamshurina, E.V.; Boyko, A.V.; Girich, A.S.; Eliseikina, M.G. Expression of Piwi, MMP, TIMP, and Sox during gut regeneration in holothurian Eupentacta fraudatrix (Holothuroidea, Dendrochirotida). Genes 2021, 12, 1292. [Google Scholar] [CrossRef]
- Dolmatov, I.Y.; Shulga, A.P.; Ginanova, T.T.; Eliseikina, M.G.; Lamash, N.E. Metalloproteinase inhibitor GM6001 delays regeneration in holothurians. Tissue Cell 2019, 59, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, J.L.; Rosa, R.; Ruiz, D.L.; García-Arrarás, J.E. Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima. Dev. Biol. 2002, 250, 181–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, A.R.; Barbaglio, A.; Oliveira, M.J.; Ribeiro, C.C.; Wilkie, I.C.; Candia Carnevali, M.D.; Barbosa, M.A. Matrix metalloproteinases in a sea urchin ligament with adaptable mechanical properties. PLoS ONE 2012, 7, e49016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wart, H.E.; Birkedal-Hansen, H. The cysteine switch: A principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. USA 1990, 87, 5578–5582. [Google Scholar] [CrossRef] [Green Version]
- Murphy, G.; Nagase, H. Progress in matrix metalloproteinase research. Mol. Asp. Med. 2008, 29, 290–308. [Google Scholar] [CrossRef] [Green Version]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef] [Green Version]
- Fanjul-Fernández, M.; Folgueras, A.R.; Cabrera, S.; López-Otín, C. Matrix metalloproteinases: Evolution, gene regulation and functional analysis in mouse models. BBA Mol. Cell Res. 2010, 1803, 3–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flood, J.; Mayne, J.; Robinson, J.J. Identification and characterization of gelatin-cleavage activities in the apically located extracellular matrix of the sea urchin embryo. Biochem. Cell Biol. 2000, 78, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Mayne, J.; Robinson, J.J. Localization and functional role of a 41 kDa collagenase/gelatinase activity expressed in the sea urchin embryo. Dev. Growth Differ. 2002, 44, 345–356. [Google Scholar] [CrossRef]
- Robinson, J.J.; Mayne, J. The effects of Ca2+ and Mg2+ on the major gelatinase activities present in the sea urchin embryo. Biochem. Biophys. Res. Commun. 1998, 243, 326–330. [Google Scholar] [CrossRef]
- Overall, C.M. Molecular determinants of metalloproteinase substrate specificity: Matrix metalloproteinase substrate binding domains, modules, and exosites. Mol. Biotechnol. 2002, 22, 51–86. [Google Scholar] [CrossRef]
- Fernandez-Catalan, C.; Bode, W.; Huber, R.; Turk, D.; Calvete, J.J.; Lichte, A.; Tschesche, H.; Maskos, K. Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor. EMBO J. 1998, 17, 5238–5248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woskowicz, A.M.; Weaver, S.A.; Shitomi, Y.; Ito, N.; Itoh, Y. MT-LOOP-dependent localization of membrane type I matrix metalloproteinase (MT1-MMP) to the cell adhesion complexes promotes cancer cell invasion. J. Biol. Chem. 2013, 288, 35126–35137. [Google Scholar] [CrossRef] [Green Version]
- Itoh, Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015, 44–46, 207–223. [Google Scholar] [CrossRef]
- Pei, D. Leukolysin/MMP25/MT6-MMP: A novel matrix metalloproteinase specifically expressed in the leukocyte lineage. Cell Res. 1999, 9, 291–303. [Google Scholar] [CrossRef] [Green Version]
- Itoh, Y.; Kajita, M.; Kinoh, H.; Mori, H.; Okada, A.; Seiki, M. Membrane type 4 matrix metalloproteinase (MT4-MMP, MMP-17) is a glycosylphosphatidylinositol-anchored proteinase. J. Biol. Chem. 1999, 274, 34260–34266. [Google Scholar] [CrossRef] [Green Version]
- Kojima, S.; Itoh, Y.; Matsumoto, S.; Masuho, Y.; Seiki, M. Membrane-type 6 matrix metalloproteinase (MT6-MMP, MMP-25) is the second glycosyl-phosphatidyl inositol (GPI)-anchored MMP. FEBS Lett. 2000, 480, 142–146. [Google Scholar] [CrossRef] [Green Version]
- Galea, C.A.; Nguyen, H.M.; George Chandy, K.; Smith, B.J.; Norton, R.S. Domain structure and function of matrix metalloprotease 23 (MMP23): Role in potassium channel trafficking. Cell. Mol. Life Sci. 2014, 71, 1191–1210. [Google Scholar] [CrossRef]
- Velasco, G.; Pendás, A.M.; Fueyo, A.; Knäuper, V.; Murphy, G.; López-Otín, C. Cloning and characterization of human MMP-23, a new matrix metalloproteinase predominantly expressed in reproductive tissues and lacking conserved domains in other family members. J. Biol. Chem. 1999, 274, 4570–4576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huxley-Jones, J.; Clarke, T.K.; Beck, C.; Toubaris, G.; Robertson, D.L.; Boot-Handford, R.P. The evolution of the vertebrate metzincins; insights from Ciona intestinalis and Danio rerio. BMC Evol. Biol. 2007, 7, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grinthal, A.; Guidotti, G. CD39, NTPDase 1, is attached to the plasma membrane by two transmembrane domains. Why? Purinergic Signal 2006, 2, 391–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanfear, R.; Frandsen, P.; Wright, A.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Nomura, K.; Suzuki, N. Stereo-specific inhibition of sea-urchin envelysin (hatchin enzyme) by a synthetic autoinhibitor peptide with a cysteine-switch consensus sequence. FEBS Lett. 1993, 321, 84–88. [Google Scholar] [CrossRef] [Green Version]
- Nomura, K.; Tanaka, H.; Kikkawa, Y.; Yamaguchi, M.; Suzuki, N. The specificity of sea-urchin hatching enzyme (envelysin) places it in the mammalian matrix metalloproteinase family. Biochemistry 1991, 30, 6115–6123. [Google Scholar] [CrossRef] [PubMed]
- Quigley, J.P.; Braithwaite, R.S.; Armstrong, P.B. Matrix metalloproteases of the developing sea urchin embryo. Differentiation 1993, 54, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Roe, J.L.; Lennarz, W.J. Biosynthesis and secretion of the hatching enzyme during sea urchin embryogenesis. J. Biol. Chem. 1990, 265, 8704–8711. [Google Scholar] [CrossRef]
- Lepage, T.; Gache, C. Purification and characterization of the sea urchin embryo hatching enzyme. J. Biol. Chem. 1989, 264, 4787–4793. [Google Scholar] [CrossRef]
- Lepage, T.; Gache, C. Early expression of a collagenase-like hatching enzyme gene in the sea urchin embryo. EMBO J. 1990, 9, 3003–3012. [Google Scholar] [CrossRef]
- Lepage, T.; Sardet, C.; Gache, C. Spatial expression of the hatching enzyme gene in the sea urchin embryo. Dev. Biol. 1992, 150, 23–32. [Google Scholar] [CrossRef]
- Ghiglione, C.; Lhomond, G.; Lepage, T.; Gache, C. Structure of the sea urchin hatching enzyme gene. Eur. J. Biochem. 1994, 219, 845–854. [Google Scholar] [CrossRef]
- Nomura, K.; Shimizu, T.; Kinoh, H.; Sendai, Y.; Inomata, M.; Suzuki, N. Sea urchin hatching enzyme (envelysin): cDNA cloning and deprivation of protein substrate specificity by autolytic degradation. Biochemistry 1997, 36, 7225–7238. [Google Scholar] [CrossRef]
- Li, Z.J.; Kim, S.M. A novel hatching enzyme from starfish Asterias amurensis: Purification, characterization, and cleavage specificity. Appl. Biochem. Biotechnol. 2013, 169, 1386–1396. [Google Scholar] [CrossRef]
- Li, Z.J.; Kim, S.M. Structural identification and proteolytic effects of the hatching enzyme from starfish Asterias amurensis. Protein Pept. Lett. 2014, 21, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Mayne, J.; Robinson, J.J. Purification and metal ion requirements of a candidate matrix metalloproteinase: A 41 kDa gelatinase activity in the sea urchin embryo. Biochem. Cell Biol. 1996, 74, 211–218. [Google Scholar] [CrossRef]
- Mayne, J.; Robinson, J.J. Calcium-protein interactions in the extracellular environment: Calcium binding, activation, and immunolocalization of a collagenase/gelatinase activity expressed in the sea urchin embryo. J. Cell. Biochem. 1998, 71, 546–558. [Google Scholar] [CrossRef]
- Mayne, J.; Robinson, J.J. Comparative analysis of the kinetic parameters and thermal stability of two matrix metalloproteinases expressed in the developing sea urchin embryo. Int. J. Biochem. Cell Biol. 1999, 31, 717–724. [Google Scholar] [CrossRef]
- Mayne, J.; Robinson, J.J. Comparative analysis of the structure and thermal sea stability of sea urchin peristome and rat tail tendon collagen. J. Cell. Biochem. 2002, 84, 567–574. [Google Scholar] [CrossRef]
- Ranganathan, L.; Rimsay, R.; Robinson, J.J. Zymogen activation and characterization of a major gelatin-cleavage activity localized to the sea urchin extraembryonic matrix. J. Cell. Biochem. 2004, 93, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.J. Characterization of a metalloproteinase: A late stage specific gelatinase activity in the sea urchin embryo. J. Cell. Biochem. 1997, 66, 337–345. [Google Scholar] [CrossRef]
- Robinson, J.J. Proteolytic processing of a sea urchin, ECM-localized protein into lower mol mass species possessing collagen-cleavage activity. J. Cell. Biochem. 2006, 99, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.J.; Mayne, J. Characterisation of a 41 kDa collagenase/gelatinase activity expressed in the sea urchin embryo. Zygote 2000, 8, S37–S38. [Google Scholar] [CrossRef]
- Robinson, J.J.; Sharpe, C.; Calloway, C. Identification and partial characterization of two inducible gelatin-cleavage activities localized to the sea urchin extraembryonic matrix, the hyaline layer. BBA Mol. Cell Res. 2003, 1621, 67–75. [Google Scholar] [CrossRef]
- Ingersoll, E.P.; Pendharkar, N.C. Characterization and expression of two matrix metalloproteinase genes during sea urchin development. Gene Expr. Patterns 2005, 5, 727–732. [Google Scholar] [CrossRef]
- Ingersoll, E.P.; Wilt, F.H. Matrix metalloproteinase inhibitors disrupt spicule formation by primary mesenchyme cells in the sea urchin embryo. Dev. Biol. 1998, 196, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Mann, K.; Wilt, F.H.; Poustka, A.J. Proteomic analysis of sea urchin (Strongylocentrotus purpuratus) spicule matrix. Proteome Sci. 2010, 8, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgulis, M.; Winter, M.R.; Shternhell, L.; Gildor, T.; Ben-Tabou de-Leon, S. VEGF signaling activates the matrix metalloproteinases, MmpL7 and MmpL5 at the sites of active skeletal growth and MmpL7 regulates skeletal elongation. Dev. Biol. 2021, 473, 80–89. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Liu, Y.X.; Zhou, D.Y.; Liu, X.Y.; Dong, X.P.; Li, D.M.; Shahidi, F. The role of matrix metalloprotease (MMP) to the autolysis of sea cucumber (Stichopus japonicus). J. Sci. Food Agric. 2019, 99, 5752–5759. [Google Scholar] [CrossRef] [PubMed]
- Quispe-Parra, D.J.; Medina-Feliciano, J.G.; Cruz-González, S.; Ortiz-Zuazaga, H.; García-Arrarás, J.E. Transcriptomic analysis of early stages of intestinal regeneration in Holothuria glaberrima. Sci. Rep. 2021, 11, 346. [Google Scholar] [CrossRef]
- Okamoto, T.; Akaike, T.; Sawa, T.; Miyamoto, Y.; van der Vliet, A.; Maeda, H. Activation of matrix metalloproteinases by peroxynitrite-induced protein S-glutathiolation via disulfide S-oxide formation. J. Biol. Chem. 2001, 276, 29596–29602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, Q.; Malinovskii, V.; Huang, W.; Hu, Y.; Chung, L.; Nagase, H.; Bode, W.; Maskos, K.; Brew, K. Residue 2 of TIMP-1 is a major determinant of affinity and specificity for matrix metalloproteinases but effects of substitutions do not correlate with those of the corresponding P1’ residue of substrate. J. Biol. Chem. 1999, 274, 10184–10189. [Google Scholar] [CrossRef] [Green Version]
- Tipper, J.P.; Lyons-Levy, G.; Atkinson, M.A.; Trotter, J.A. Purification, characterization and cloning of tensilin, the collagen-fibril binding and tissue-stiffening factor from Cucumaria frondosa dermis. Matrix Biol. 2003, 21, 625–635. [Google Scholar] [CrossRef]
- Wilkie, I.C. Mutable collagenous tissue: Overview and biotechnological perspective. In Progress in Molecular and Subcellular Biology. Subseries Marine Molecular Biotechnology; Matranga, V., Ed.; Springer: Heidelberg, Germany, 2005; pp. 221–250. [Google Scholar]
- Miller, A.K.; Kerr, A.M.; Paulay, G.; Reich, M.; Wilson, N.G.; Carvajal, J.I.; Rouse, G.W. Molecular phylogeny of extant Holothuroidea (Echinodermata). Mol. Phylogenet Evol. 2017, 111, 110–131. [Google Scholar] [CrossRef] [Green Version]
- Stetler-Stevenson, W.G. Tissue inhibitors of metalloproteinases in cell signaling: Metalloproteinase-independent biological activities. Sci. Signal. 2008, 1, re6. [Google Scholar] [CrossRef] [Green Version]
- Shabelnikov, S.V.; Bobkov, D.E.; Sharlaimova, N.S.; Petukhova, O.A. Injury affects coelomic fluid proteome of the common starfish, Asterias rubens. J. Exp. Biol. 2019, 222. [Google Scholar] [CrossRef] [Green Version]
- Dolmatov, I.Y. Regeneration of the aquapharyngeal complex in the holothurian Eupentacta fraudatrix (Holothuroidea, Dendrochirota). Monogr. Dev. Biol. 1992, 23, 40–50. [Google Scholar] [PubMed]
- Mashanov, V.S.; García-Arrarás, J.E. Gut regeneration in holothurians: A snapshot of recent developments. Biol. Bull. 2011, 221, 93–109. [Google Scholar] [CrossRef]
- Chirco, R.; Liu, X.W.; Jung, K.K.; Kim, H.R. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2006, 25, 99–113. [Google Scholar] [CrossRef]
- Crocker, S.J.; Pagenstecher, A.; Campbell, I.L. The TIMPs tango with MMPs and more in the central nervous system. J. Neurosci. Res. 2004, 75, 1–11. [Google Scholar] [CrossRef]
- Rivera, S.; Khrestchatisky, M.; Kaczmarek, L.; Rosenberg, G.A.; Jaworski, D.M. Metzincin proteases and their inhibitors, foes or friends in nervous system physiology? J. Neurosci. 2010, 30, 15337–15357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verstappen, J.; Von den Hoff, J.W. Tissue inhibitors of metalloproteinases (TIMPs): Their biological functions and involvement in oral disease. J. Dent. Res. 2006, 85, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Lambert, E.; Dasse, E.; Haye, B.; Petitfrere, E. TIMPs as multifacial proteins. Crit. Rev. Oncol. Hematol. 2004, 49, 187–198. [Google Scholar] [CrossRef]
- Hayakawa, T.; Yamashita, K.; Ohuchi, E.; Shinagawa, A. Cell growth-promoting activity of tissue inhibitor of metalloproteinases-2 (TIMP-2). J. Cell Sci. 1994, 107, 2373–2379. [Google Scholar] [CrossRef]
- Ricca, T.I.; Liang, G.; Suenaga, A.P.M.; Han, S.W.; Jones, P.A.; Jasiulionis, M.G. Tissue inhibitor of metalloproteinase 1 expression associated with gene demethylation confers anoikis resistance in early phases of melanocyte malignant transformation. Transl. Oncol. 2009, 2, 329–340. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Goldberg, I.D.; Shi, Y.E. Complex roles of tissue inhibitors of metalloproteinases in cancer. Oncogene 2002, 21, 2245–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, D.-W.; Li, H.; Guedez, L.; Wingfield, P.T.; Diaz, T.; Salloum, R.; Wei, B.-y.; Stetler-Stevenson, W.G. TIMP-2 mediated inhibition of angiogenesis. Cell 2003, 114, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Jung, K.-K.; Liu, X.-W.; Chirco, R.; Fridman, R.; Kim, H.-R.C. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J. 2006, 25, 3934–3942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Kang, Y.; Huang, L.; Wu, L.; Peng, Y. TIMP1 preserves the blood–brain barrier through interacting with CD63/integrin β1 complex and regulating downstream FAK/RhoA signaling. Acta Pharm. Sin. B 2020, 10, 987–1003. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dolmatov, I.Y.; Nizhnichenko, V.A.; Dolmatova, L.S. Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases in Echinoderms: Structure and Possible Functions. Cells 2021, 10, 2331. https://doi.org/10.3390/cells10092331
Dolmatov IY, Nizhnichenko VA, Dolmatova LS. Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases in Echinoderms: Structure and Possible Functions. Cells. 2021; 10(9):2331. https://doi.org/10.3390/cells10092331
Chicago/Turabian StyleDolmatov, Igor Yu., Vladimir A. Nizhnichenko, and Lyudmila S. Dolmatova. 2021. "Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases in Echinoderms: Structure and Possible Functions" Cells 10, no. 9: 2331. https://doi.org/10.3390/cells10092331
APA StyleDolmatov, I. Y., Nizhnichenko, V. A., & Dolmatova, L. S. (2021). Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases in Echinoderms: Structure and Possible Functions. Cells, 10(9), 2331. https://doi.org/10.3390/cells10092331





