p27Kip1, an Intrinsically Unstructured Protein with Scaffold Properties
Abstract
1. A Premise: Unfolding and Scaffolding Might Be Two Strictly Connected Features of Proteins
2. p27, a Multifaced Growth-Modulating IUP
2.1. p27 as a CDK Regulator
2.2. p27 Is an Intermediate Component of Key Growth Regulatory Loops
3. p27, a Platform Where Assembling Different Complexes and Cellular Structures
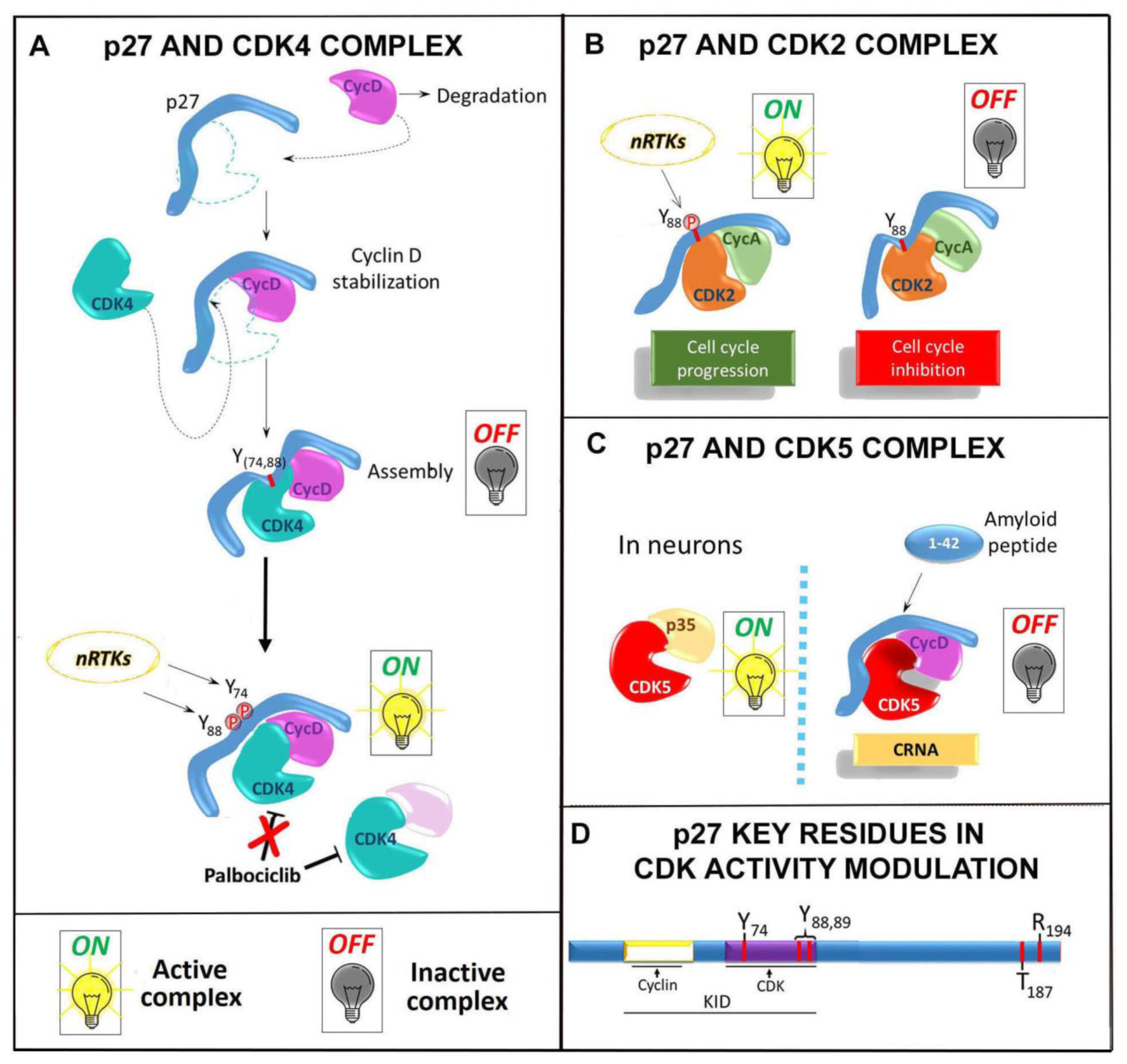
3.1. p27 and Cyclin/CDKs: Complex Formation and Control of Function
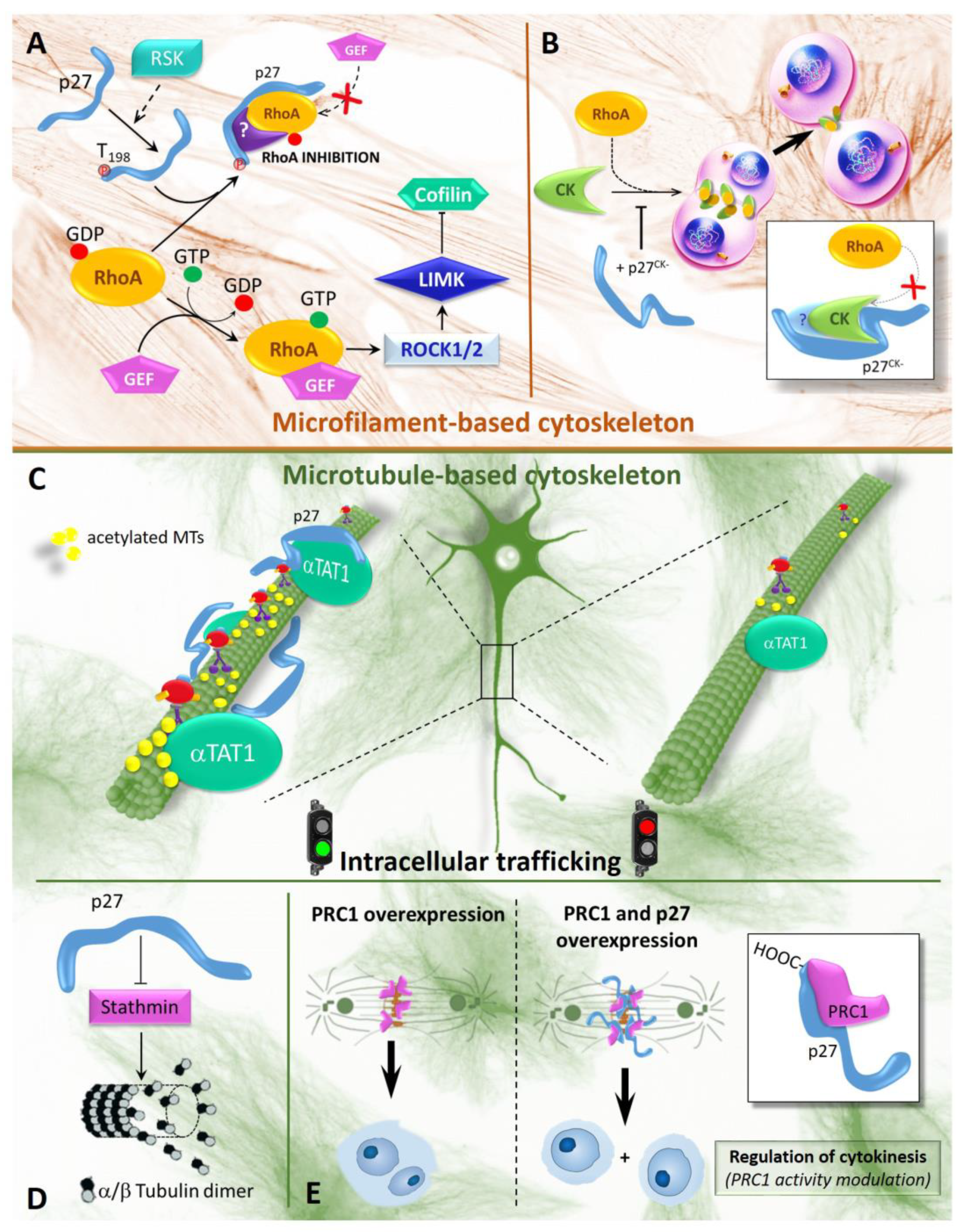
3.2. p27 as a Key Modulator of Cytoskeletal Dynamics and Cell Motility
3.3. p27, Lysosomal Function and Autophagy
3.4. p27 as a Platform for Transcriptional Complexes
4. Perspectives and Conclusions. p27, Human Diseases and Drug Design
Author Contributions
Funding
Conflicts of Interest
References
- Toker, D.; Sommer, F.T.; D’Esposito, M. A simple method for detecting chaos in nature. Commun. Biol. 2020, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Stark, J.; Hardy, K. Chaos: Useful at Last? Science 2003, 301, 1192–1193. [Google Scholar] [CrossRef]
- Newton, I.S. Philosophiae Naturalis Principia Mathematica, 1st ed.; Streater, J., Ed.; Royal Society of London: London, UK, 1687. [Google Scholar]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Minezaki, Y.; Homma, K.; Kinjo, A.R.; Nishikawa, K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006, 359, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Galea, C.A.; Wang, Y.; Sivakolundu, S.G.; Kriwacki, R.W. Regulation of Cell Division by Intrinsically Unstructured Proteins; Intrinsic Flexibility, Modularity and Signaling Conduits. Biochemistry 2008, 47, 7598–7609. [Google Scholar] [CrossRef]
- Sugase, K.; Dyson, H.J.; Wright, P.E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 2007, 447, 1021–1025. [Google Scholar] [CrossRef]
- Robustelli, P.; Piana, S.; Shaw, D.E. Mechanism of Coupled Folding-upon-Binding of an Intrinsically Disordered Protein. J. Am. Chem. Soc. 2020, 142, 11092–11101. [Google Scholar] [CrossRef]
- Piovesan, D.; Tabaro, F.; Mičetić, I.; Necci, M.; Quaglia, F.; Oldfield, C.J.; Aspromonte, M.C.; Davey, N.E.; Davidović, R.; Dosztányi, Z.; et al. DisProt 7.0: A major update of the database of disordered proteins. Nucleic Acids Res. 2017, 45, 219–227. [Google Scholar] [CrossRef]
- Good, M.C.; Zalatan, J.G.; Lim, W.A. Scaffold proteins: Hubs for controlling the flow of cellular information. Science 2011, 332, 680–686. [Google Scholar] [CrossRef]
- Langeberg, L.K.; Scott, J.D. Signalling scaffolds and local organization of cellular behavior. Nat. Rev. Mol. Cell Biol. 2015, 16, 235–244. [Google Scholar] [CrossRef]
- Shaw, A.S.; Filbert, E.L. Scaffold proteins and immune-cell signalling. Nat. Rev. Immunol. 2009, 9, 47–56. [Google Scholar] [CrossRef]
- Oh, K.; Yi, G.S. Prediction of scaffold proteins based on protein interaction and domain architectures. BMC Bioinform. 2016, 17 (Suppl. S6), 220. [Google Scholar] [CrossRef]
- Reed, B.J.; Locke, M.N.; Gardner, R.G. A conserved deubiquitinating enzyme uses intrinsically disordered regions to scaffold multiple protein interaction sites. J. Biol. Chem. 2015, 290, 20601–20612. [Google Scholar] [CrossRef]
- Polyak, K.; Kato, J.Y.; Solomon, M.J.; Sherr, C.J.; Massague, J.; Roberts, J.M.; Koff, A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994, 8, 9–22. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995, 9, 1149–1163. [Google Scholar] [CrossRef]
- Payne, S.R.; Zhang, S.; Tsuchiya, K.; Moser, R.; Gurley, K.E.; Longton, G.; DeBoer, J.; Kemp, C.J. p27kip1 deficiency impairs G2/M arrest in response to DNA damage, leading to an increase in genetic instability. Mol. Cell Biol. 2008, 28, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Baghdassarian, N.; Peiretti, A.; Devaux, E.; Bryon, P.A.; Ffrench, M. Involvement of p27Kip1 in the G1- and S/G2-phase lengthening mediated by glucocorticoids in normal human lymphocytes. Cell Growth Differ. 1999, 10, 405–412. [Google Scholar] [PubMed]
- Serres, M.P.; Kossatz, U.; Chi, Y.; Roberts, J.M.; Malek, N.P.; Besson, A. p27(Kip1) controls cytokinesis via the regulation of citron kinase activation. J. Clin. Investig. 2012, 122, 844–858. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.L.; Stampone, E.; Cervellera, C.; Borriello, A. Regulation of p27Kip1 and p57Kip2 functions by natural polyphenols. Biomolecules 2020, 10, 1316. [Google Scholar] [CrossRef] [PubMed]
- Chassot, A.A.; Lossaint, G.; Turchi, L.; Meneguzzi, G.; Fisher, D.; Ponzio, G.; Dulic, V. Confluence-induced cell cycle exit involves pre-mitotic CDK inhibition by p27(Kip1) and cyclin D1 downregulation. Cell Cycle 2008, 7, 2038–2046. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Olivier, P.; Diehl, J.A.; Fero, M.; Roussel, M.F.; Roberts, J.M.; Sherr, C.J. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999, 18, 1571–1583. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.A.; Jeffrey, P.D.; Patten, A.K.; Massagué, J.; Pavletich, N.P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 1996, 382, 325–331. [Google Scholar] [CrossRef]
- Grimmler, M.; Wang, Y.; Mundm, T.; Cilensek, Z.; Keidel, E.M.; Waddell, M.B.; Jäkel, H.; Kullmann, M.; Kriwacki, R.W.; Hengst, L. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell 2007, 128, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.L.; Senapati, S. Mechanism of p27 Unfolding for CDK2 Reactivation. Sci. Rep. 2016, 6, 26450. [Google Scholar] [CrossRef]
- Bencivenga, D.; Caldarelli, I.; Stampone, E.; Mancini, F.P.; Balestrieri, M.L.; Della Ragione, F.; Borriello, A. p27Kip1 and human cancers: A reappraisal of a still enigmatic protein. Cancer Lett. 2017, 403, 354–365. [Google Scholar] [CrossRef]
- Bencivenga, D.; Tramontano, A.; Borgia, A.; Negri, A.; Caldarelli, I.; Oliva, A.; Perrotta, S.; Della Ragione, F.; Borriello, A. p27Kip1 serine 10 phosphorylation determines its metabolism and interaction with cyclin-dependent kinases. Cell Cycle 2014, 13, 3768–3782. [Google Scholar] [CrossRef][Green Version]
- Chu, I.; Sun, J.; Arnaout, A.; Kahn, H.; Hanna, W.; Narod, S.; Sun, P.; Tan, C.K.; Hengst, L.; Slingerland, J. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell 2007, 128, 281–294. [Google Scholar] [CrossRef]
- Scott, J.D.; Pawson, T. Cell signaling in space and time: Where proteins come together and when they’re apart. Science 2009, 326, 1220–1224. [Google Scholar] [CrossRef]
- Besson, A.; Gurian-West, M.; Chen, X.; Kelly-Spratt, K.S.; Kemp, C.J.; Roberts, J.M. A pathway in quiescent cells that controls p27Kip1 stability, subcellular localization, and tumor suppression. Genes Dev. 2006, 20, 47–64. [Google Scholar] [CrossRef]
- Fabris, L.; Berton, S.; Pellizzari, I.; Segatto, I.; D’Andrea, S.; Armenia, J.; Bomben, R.; Schiappacassi, M.; Gattei, V.; Philips, M.R.; et al. p27kip1 controls H-Ras/MAPK activation and cell cycle entry via modulation of MT stability. Proc. Natl. Acad. Sci. USA 2015, 112, 13916–13921. [Google Scholar] [CrossRef] [PubMed]
- Abankwa, D.; Gorfe, A.A.; Hancock, J.F. Ras nanoclusters: Molecular structure and assembly. Semin. Cell Dev. Biol. 2007, 18, 599–607. [Google Scholar] [CrossRef][Green Version]
- Brazil, D.P.; Hemmings, B.A. Ten years of protein kinase B signalling: A hard Akt to follow. Trends Biochem. Sci. 2001, 26, 657–664. [Google Scholar] [CrossRef]
- Jacinto, E.; Facchinetti, V.; Liu, D.; Soto, N.; Wei, S.; Jung, S.Y.; Huang, Q.; Qin, J.; Su, B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006, 127, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Viglietto, G.; Motti, M.L.; Bruni, P.; Melillo, R.M.; D’Alessio, A.; Califano, D.; Vinci, F.; Chiappetta, G.; Tsichlis, P.; Bellacosa, A.; et al. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat. Med. 2002, 8, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Sato, S.; Katayama, K.; Tsuruo, T. Akt-dependent phosphorylation of p27Kip1 promotes binding to 14-3-3 and cytoplasmic localization. J. Biol. Chem. 2002, 277, 28706–28713. [Google Scholar] [CrossRef]
- Peng, M.; Wang, J.; Zhang, D.; Jin, H.; Li, J.; Wu, X.R.; Huang, C. PHLPP2 stabilization by p27 mediates its inhibition of bladder cancer invasion by promoting autophagic degradation of MMP2 protein. Oncogene 2018, 37, 5735–5748. [Google Scholar] [CrossRef]
- Brognard, J.; Sierecki, E.; Gao, T.; Newton, A.C. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell 2007, 25, 917–931. [Google Scholar] [CrossRef]
- Soos, T.J.; Kiyokawa, H.; Yan, J.S.; Rubin, M.S.; Giordano, A.; DeBlasio, A.; Bottega, S.; Wong, B.; Mendelsohn, J.; Koff, A. Formation of p27-CDK complexes during the human mitotic cell cycle. Cell Growth Differ. 1996, 7, 135–146. [Google Scholar] [PubMed]
- LaBaer, J.; Garrett, M.D.; Stevenson, L.; Slingerland, J.M.; Sandhu, C.; Chou, H.S.; Fattaey, A.; Harlow, E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997, 11, 847–862. [Google Scholar] [CrossRef]
- Bagui, T.K.; Mohapatra, S.; Haura, E.; Pledger, W.J. p27Kip1 and p21Cip1 are not required for the formation of active D cyclin-cdk4 complexes. Mol. Cell Biol. 2003, 23, 7285–7290. [Google Scholar] [CrossRef] [PubMed]
- Larrea, M.D.; Liang, J.; da Silva, T.; Hong, F.; Shao, S.H.; Han, K.; Dumont, D.; Slingerland, J.M. Phosphorylation of p27Kip1 regulates assembly and activation of cyclin D1-Cdk4. Mol. Cell Biol. 2008, 28, 6462–6472. [Google Scholar] [CrossRef]
- Ou, L.; Ferreira, A.M.; Otieno, S.; Xiao, L.; Bashford, D.; Kriwacki, R.W. Incomplete folding upon binding mediates Cdk4/cyclin D complex activation by tyrosine phosphorylation of inhibitor p27 protein. J. Biol. Chem. 2011, 286, 30142–30151. [Google Scholar] [CrossRef] [PubMed]
- Guiley, K.Z.; Stevenson, J.W.; Lou, K.; Barkovich, K.J.; Kumarasamy, V.; Wijeratne, T.U.; Bunch, K.L.; Tripathi, S.; Knudsen, E.S.; Witkiewicz, A.K.; et al. p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition. Science 2019, 366, eaaw2106. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Tsiperson, V.; Gottesman, S.R.S.; Somma, J.; Blain, S.W. Dual inhibition of CDK4 and CDK2 via targeting p27 tyrosine phosphorylation induces a potent and durable response in breast cancer cells. Mol. Cancer Res. 2018, 16, 361–377. [Google Scholar] [CrossRef]
- Li, J.; Vervoorts, J.; Carloni, P.; Rossetti, G.; Lüscher, B. Structural prediction of the interaction of the tumor suppressor p27Kip1 with cyclin A/CDK2 identifies a novel catalytically relevant determinant. BMC Bioinform. 2017, 18, 15. [Google Scholar] [CrossRef]
- Jeffrey, P.D.; Russo, A.A.; Polyak, K.; Gibbs, E.; Hurwitz, J.; Massagué, J.; Pavletich, N.P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 1995, 376, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Lagace, D.C.; Benavides, D.R.; Kansy, J.W.; Mapelli, M.; Greengard, P.; Bibb, J.A.; Eisch, A.J. Cdk5 is essential for adult hippocampal neurogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 18567–18571. [Google Scholar] [CrossRef]
- Ko, J.; Humbert, S.; Bronson, R.T.; Takahashi, S.; Kulkarni, A.B.; Li, E.; Tsai, L.H. p35 and p39 are essential for cyclin-dependent kinase 5 function during neurodevelopment. J. Neurosci. 2001, 21, 6758–6771. [Google Scholar] [CrossRef]
- Tanaka, T.; Veeranna, T.; Ohshima, T.; Rajan, P.; Amin, N.D.; Cho, A.; Sreenath, T.; Pant, H.C.; Brady, R.O.; Kulkarni, A.B. Neuronal cyclin-dependent kinase 5 activity is critical for survival. J. Neurosci. 2001, 21, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.T.; Wan, C.; Yang, W.; Cai, Z. Role of Cdk5 in Amyloid-beta Pathology of Alzheimer’s Disease. Curr. Alzheimer Res. 2019, 16, 1206–1215. [Google Scholar] [CrossRef]
- Modi, P.K.; Komaravelli, N.; Singh, N.; Sharma, P. Interplay between MEK-ERK signaling, cyclin D1, and cyclin-dependent kinase 5 regulates cell cycle reentry and apoptosis of neurons. Mol. Biol. Cell 2012, 23, 3722–3730. [Google Scholar] [CrossRef]
- Jaiswal, S.; Sharma, P. Role and regulation of p27 in neuronal apoptosis. J. Neurochem. 2017, 140, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Godden-Kent, D.; Talbot, S.J.; Boshoff, C.; Chang, Y.; Moore, P.; Weiss, R.A.; Mittnacht, S. The cyclin encoded by Kaposi’s sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J. Virol. 1997, 71, 4193–4198. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A.; Gloghini, A.; Bontempo, D.; Monini, P.; Tirelli, U.; Volpe, R.; Browning, P.J.; Gaidano, G. Proliferation in HHV-8-positive primary effusion lymphomas is associated with expression of HHV-8 cyclin but independent of p27(Kip1). Am. J. Pathol. 2000, 156, 1209–1215. [Google Scholar] [CrossRef]
- Järviluoma, A.; Koopal, S.; Räsänen, S.; Mäkelä, T.P.; Ojala, P.M. KSHV viral cyclin binds to p27Kip1 in primary effusion lymphomas. Blood 2004, 104, 3349–3354. [Google Scholar] [CrossRef] [PubMed]
- Sarek, G.; Järviluoma, A.; Ojala, P.M. KSHV viral cyclin inactivates p27Kip1 through Ser10 and Thr187 phosphorylation in proliferating primary effusion lymphomas. Blood 2006, 107, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, T.; Dehghani, F. The Cytoskeleton-A Complex Interacting Meshwork. Cells 2019, 8, 362. [Google Scholar] [CrossRef]
- Desai, A.; Mitchison, T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, O.C.; Schaefer, A.W.; Mandato, C.A.; Forscher, P.; Bement, W.M.; Waterman-Storer, C.M. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 2003, 5, 599–609. [Google Scholar] [CrossRef]
- Loreng, T.D.; Smith, E.F. The Central Apparatus of Cilia and Eukaryotic Flagella. Cold Spring Harb. Perspect. Biol. 2017, 9, a028118. [Google Scholar] [CrossRef]
- Nagahara, H.; Vocero-Akbani, A.M.; Snyder, E.L.; Ho, A.; Latham, D.G.; Lissy, N.A.; Becker-Hapak, M.; Ezhevsky, S.A.; Dowdy, S.F. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat. Med. 1998, 4, 1449–1452. [Google Scholar] [CrossRef] [PubMed]
- McAllister, S.S.; Becker-Hapak, M.; Pintucci, G.; Pagano, M.; Dowdy, S.F. Novel p27(Kip1) C-terminal scatter domain mediates Rac-dependent cell migration independent of cell cycle arrest functions. Mol. Cell Biol. 2003, 23, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Besson, A.; Gurian-West, M.; Schmidt, A.; Hall, A.; Roberts, J.M. p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 2004, 18, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 20, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, M.; Ishizaki, T.; Boku, S.; Watanabe, N.; Fujita, A.; Iwamatsu, A.; Obinata, T.; Ohashi, K.; Mizuno, K.; Narumiya, S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 1999, 285, 895–898. [Google Scholar] [CrossRef]
- Lappalainen, P.; Drubin, D.G. Cofilin promotes rapid actin filament turnover in vivo. Nature 1997, 388, 78–82. [Google Scholar] [CrossRef]
- Phillips, A.H.; Ou, L.; Gay, A.; Besson, A.; Kriwacki, R.W. Mapping Interactions between p27 and RhoA that Stimulate Cell Migration. J. Mol. Biol. 2018, 430, 751–758. [Google Scholar] [CrossRef]
- Larrea, M.D.; Hong, F.; Wander, S.A.; da Silva, T.G.; Helfman, D.; Lannigan, D.; Smith, J.A.; Slingerland, J.M. RSK1 drives p27Kip1 phosphorylation at T198 to promote RhoA inhibition and increase cell motility. Proc. Natl. Acad. Sci. USA 2009, 106, 9268–9273. [Google Scholar] [CrossRef]
- Chen, L.; Rolls, M.M. Microtubule deacetylation sets the stage for successful axon regeneration. EMBO J. 2012, 31, 3033–3035. [Google Scholar] [CrossRef]
- Godena, V.K.; Brookes-Hocking, N.; Moller, A.; Shaw, G.; Oswald, M.; Sancho, R.M.; Miller, C.C.; Whitworth, A.J.; De Vos, K.J. Increasing microtubule acetylation rescues axonal transport and locomotor deficits caused by LRRK2 Roc-COR domain mutations. Nat. Commun. 2014, 5, 5245. [Google Scholar] [CrossRef]
- Arrazola Sastre, A.; Luque Montoro, M.; Lacerda, H.M.; Llavero, F.; Zugaza, J.L. Small GTPases of the Rab and Arf Families: Key regulators of intracellular trafficking in neurodegeneration. Int. J. Mol. Sci. 2021, 22, 4425. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, X.J. Tubulin acetylation: Responsible enzymes, biological functions and human diseases. Cell Mol. Life Sci. 2015, 72, 4237–4255. [Google Scholar] [CrossRef] [PubMed]
- Morelli, G.; Even, A.; Gladwyn-Ng, I.; Le Bail, R.; Shilian, M.; Godin, J.D.; Peyre, E.; Hassan, B.A.; Besson, A.; Rigo, J.M.; et al. p27Kip1 Modulates Axonal Transport by Regulating α-Tubulin Acetyltransferase 1 Stability. Cell Rep. 2018, 23, 2429–2442. [Google Scholar] [CrossRef] [PubMed]
- Fero, M.L.; Rivkin, M.; Tasch, M.; Porter, P.; Carow, C.E.; Firpo, E.; Polyak, K.; Tsai, L.H.; Broudy, V.; Perlmutter, R.M.; et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 1996, 85, 733–744. [Google Scholar] [CrossRef]
- Soppa, U.; Schumacher, J.; Florencio Ortiz, V.; Pasqualon, T.; Tejedor, F.J.; Becker, W. The Down syndrome-related protein kinase DYRK1A phosphorylates p27(Kip1) and Cyclin D1 and induces cell cycle exit and neuronal differentiation. Cell Cycle 2014, 13, 2084–2100. [Google Scholar] [CrossRef]
- Godin, J.D.; Thomas, N.; Laguesse, S.; Malinouskaya, L.; Close, P.; Malaise, O.; Purnelle, A.; Raineteau, O.; Campbell, K.; Fero, M.; et al. p27(Kip1) is a microtubule-associated protein that promotes microtubule polymerization during neuron migration. Dev. Cell 2012, 23, 729–744. [Google Scholar] [CrossRef]
- Nguyen, L.; Besson, A.; Heng, J.I.; Schuurmans, C.; Teboul, L.; Parras, C.; Philpott, A.; Roberts, J.M.; Guillemot, F. p27Kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006, 20, 1511–1524. [Google Scholar] [CrossRef]
- Kawauchi, T.; Chihama, K.; Nabeshima, Y.; Hoshino, M. Cdk5 phosphorylates and stabilizes p27Kip1 contributing to actin organization and cortical neuronal migration. Nat. Cell Biol. 2006, 1, 17–26. [Google Scholar] [CrossRef]
- Akhshi, T.K.; Wernike, D.; Piekny, A. Microtubules and actin crosstalk in cell migration and division. Cytoskeleton (Hoboken) 2014, 71, 1–23. [Google Scholar] [CrossRef]
- Wojnacki, J.; Quassollo, G.; Marzolo, M.P.; Cáceres, A. Rho GTPases at the crossroad of signaling networks in mammals: Impact of Rho-GTPases on microtubule organization and dynamics. Small GTPases. 2014, 5, e28430. [Google Scholar] [CrossRef]
- Perchey, R.T.; Serres, M.P.; Nowosad, A.; Creff, J.; Callot, C.; Gay, A.; Manenti, S.; Margolis, R.L.; Hatzoglou, A.; Besson, A. p27Kip1 regulates the microtubule bundling activity of PRC1. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1630–1639. [Google Scholar] [CrossRef]
- Baldassarre, G.; Belletti, B.; Nicoloso, M.S.; Schiappacassi, M.; Vecchione, A.; Spessotto, P.; Morrione, A.; Canzonieri, V.; Colombatti, A. p27(Kip1)-stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell 2005, 7, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Jeannot, P.; Nowosad, A.; Perchey, R.; Callot, C.; Bennana, E.; Katsube, T.; Mayeux, P.; Guillonneau, F.; Manenti, S.; Besson, A. p27Kip1 promotes invadopodia turnover and invasion through the regulation of the PAK1/Cortactin pathway. Elife 2017, 6, e22207. [Google Scholar] [CrossRef]
- Liang, J.; Shao, S.H.; Xu, Z.X.; Hennessy, B.; Ding, Z.; Larrea, M.; Kondo, S.; Dumont, D.J.; Gutterman, J.U.; Walker, C.; et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 2007, 9, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Campos, T.; Ziehe, J.; Palma, M.; Escobar, D.; Tapia, J.C.; Pincheira, R.; Castro, A.F. Rheb promotes cancer cell survival through p27Kip1-dependent activation of autophagy. Mol. Carcinog. 2016, 55, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Campos, T.; Ziehe, J.; Fuentes-Villalobos, F.; Riquelme, O.; Peña, D.; Troncoso, R.; Lavandero, S.; Morin, V.; Pincheira, R.; Castro, A.F. Rapamycin requires AMPK activity and p27 expression for promoting autophagy-dependent Tsc2-null cell survival. Biochim. Biophys. Acta. 2016, 1863, 1200–1207. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Billin, A.N.; Campbell, M.E.; Russell, A.J.; Huffman, K.M.; Kraus, W.-E. The AMPK/p27Kip1 Axis Regulates Autophagy/Apoptosis Decisions in Aged Skeletal Muscle Stem Cells. Stem Cell Rep. 2018, 11, 425–439. [Google Scholar] [CrossRef]
- Nowosad, A.; Jeannot, P.; Callot, C.; Creff, J.; Perchey, R.T.; Joffre, C.; Codogno, P.; Manenti, S.; Besson, A. p27 controls Ragulator and mTOR activity in amino acid-deprived cells to regulate the autophagy-lysosomal pathway and coordinate cell cycle and cell growth. Nat. Cell Biol. 2020, 22, 1076–1090. [Google Scholar] [CrossRef] [PubMed]
- Nowosad, A.; Besson, A. CDKN1B/p27 regulates autophagy via the control of Ragulator and MTOR activity in amino acid-deprived cells. Autophagy 2020, 16, 2297–2298. [Google Scholar] [CrossRef]
- Bachs, O.; Gallastegui, E.; Orlando, S.; Bigas, A.; Morante-Redolat, J.M.; Serratosa, J.; Fariñas, I.; Aligué, R.; Pujol, M.J. Role of p27Kip1 as a transcriptional regulator. Oncotarget 2018, 9, 26259–26278. [Google Scholar] [CrossRef]
- Razavipour, S.F.; Harikumar, K.B.; Slingerland, J.M. p27 as a transcriptional regulator: New roles in development and cancer. Cancer Res. 2020, 80, 3451–3458. [Google Scholar] [CrossRef]
- Yoon, H.; Kim, M.; Jang, K.; Shin, M.; Besser, A.; Xiao, X.; Zhao, D.; Wander, S.A.; Briegel, K.; Morey, L.; et al. p27 transcriptionally coregulates cJun to drive programs of tumor progression. Proc. Natl. Acad. Sci. USA 2019, 116, 7005–7014. [Google Scholar] [CrossRef] [PubMed]
- Mateo, F.; Vidal-Laliena, M.; Canela, N.; Zecchin, A.; Martínez-Balbás, M.; Agell, N.; Giacca, M.; Pujol, M.J.; Bachs, O. The transcriptional co-activator PCAF regulates cdk2 activity. Nucleic Acids Res. 2009, 37, 7072–7084. [Google Scholar] [CrossRef]
- Perearnau, A.; Orlando, S.; Islam, A.B.M.M.K.; Gallastegui, E.; Martínez, J.; Jordan, A.; Bigas, A.; Aligué, R.; Pujol, M.J.; Bachs, O. p27Kip1, PCAF and PAX5 cooperate in the transcriptional regulation of specific target genes. Nucleic Acids Res. 2017, 45, 5086–5099. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pippa, R.; Paramio, J.; Bigas, A.; Bachs, O. p27Kip1 represses transcription by direct interaction with p130/E2F4 at the promoters of target genes. Oncogene 2012, 31, 4207–4220. [Google Scholar] [CrossRef] [PubMed]
- Fero, M.L.; Randel, E.; Gurley, K.E.; Roberts, J.M.; Kemp, C.J. The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature 1998, 396, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Pellegata, N.S.; Quintanilla-Martinez, L.; Siggelkow, H.; Samson, E.; Bink, K.; Höfler, H.; Fend, F.; Graw, J.; Atkinson, M.J. Germ-line mutations in p27Kip1 cause a multiple endocrine neoplasia syndrome in rats and humans. Proc. Natl. Acad. Sci. USA 2006, 103, 15558–15563. [Google Scholar] [CrossRef] [PubMed]
- Molatore, S.; Kügler, A.; Irmler, M.; Wiedemann, T.; Neff, F.; Feuchtinger, A.; Beckers, J.; Robledo, M.; Roncaroli, F.; Pellegata, N.S. Characterization of neuroendocrine tumors in heterozygous mutant MENX rats: A novel model of invasive medullary thyroid carcinoma. Endocr. Relat. Cancer 2018, 25, 145–162. [Google Scholar] [CrossRef]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef]
- Wander, S.A.; Zhao, D.; Slingerland, J.M. p27: A barometer of signaling deregulation and potential predictor of response to targeted therapies. Clin. Cancer Res. 2011, 1, 12–18. [Google Scholar] [CrossRef]
- Dietrich, S.; Hüllein, J.; Lee, S.C.; Hutter, B.; Gonzalez, D.; Jayne, S.; Dyer, M.J.; Oleś, M.; Else, M.; Liu, X.; et al. Recurrent CDKN1B (p27) mutations in hairy cell leukemia. Blood 2015, 126, 1005–1008. [Google Scholar] [CrossRef]
- Francis, J.M.; Kiezun, A.; Ramos, A.H.; Serra, S.; Pedamallu, C.S.; Qian, Z.R.; Banck, M.S.; Kanwar, R.; Kulkarni, A.A.; Karpathakis, A.; et al. Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nat. Genet. 2013, 45, 1483–1486. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, J.E.; Sherman, S.K.; Li, G.; Choi, A.B.; Bellizzi, A.M.; O’Dorisio, T.M.; Howe, J.R. Somatic alterations of CDKN1B are associated with small bowel neuroendocrine tumors. Cancer Genet. 2015, 208, 564–570. [Google Scholar] [CrossRef]
- Costa-Guda, J.; Marinoni, I.; Molatore, S.; Pellegata, N.S.; Arnold, A. Somatic mutation and germline sequence abnormalities in CDKN1B, encoding p27Kip1, in sporadic parathyroid adenomas. J. Clin. Endocrinol. Metab. 2011, 96, E701–E706. [Google Scholar] [CrossRef]
- Pellegata, N.S. MENX and MEN4. Clinics (Sao Paulo) 2012, 67 (Suppl. S1), 13–18. [Google Scholar] [CrossRef]
- Goncalves, G.A. p27kip1 as a key regulator of endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2018, 221, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Rowan, S.; Chang, M.L.; Reznikov, N.; Taylor, A. Disassembly of the lens fiber cell nucleus to create a clear lens: The p27 descent. Exp. Eye Res. 2017, 156, 72–78. [Google Scholar] [CrossRef]
- Zhou, N.; Huan, Q.; Cheng, W.; Ge, Y.; Li, D.; Wang, J. p27kip1 haploinsufficiency preserves myocardial function in the early stages of myocardial infarction via Atg5-mediated autophagy flux restoration. Mol. Med. Rep. 2019, 20, 3840–3848. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wang, X.; Huang, J.; Cui, S.; Zhang, L. CDKN1B mediates apoptosis of neuronal cells and inflammation induced by oxyhemoglobin via mir-502-5p after subarachnoid hemorrhage. J. Mol. Neurosci. 2020, 70, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Gashaw, I.; Ellinghaus, P.; Sommer, A.; Asadullah, K. What makes a good drug target? Drug Discov. Today 2012, 17, S24–S30. [Google Scholar] [CrossRef]
- Iconaru, L.I.; Das, S.; Nourse, A.; Shelat, A.A.; Zuo, J.; Kriwacki, R.W. Small molecule sequestration of the intrinsically disordered protein, p27Kip1, within soluble oligomers. J. Mol. Biol. 2021, 433, 167120. [Google Scholar] [CrossRef]
- Erkizan, H.V.; Kong, Y.; Merchant, M.; Schlottmann, S.; Barber-Rotenberg, J.S.; Yuan, L.; Abaan, O.D.; Chou, T.H.; Dakshanamurthy, S.; Brown, M.L.; et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing’s sarcoma. Nat. Med. 2009, 15, 750–756. [Google Scholar] [CrossRef]
- Selvanathan, S.P.; Graham, G.T.; Erkizan, H.V.; Dirksen, U.; Natarajan, T.G.; Dakic, A.; Yu, S.; Liu, X.; Paulsen, M.T.; Ljungman, M.E.; et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. USA 2015, 112, E1307–E1316. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Giap, C.; Lazo, J.S.; Prochownik, E.V. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 2003, 22, 6151–6159. [Google Scholar] [CrossRef] [PubMed]
- Follis, A.V.; Hammoudeh, D.I.; Wang, H.; Prochownik, E.V.; Metallo, S.J. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem. Biol. 2008, 15, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Hammoudeh, D.I.; Follis, A.V.; Prochownik, E.V.; Metallo, S.J. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J. Am. Chem. Soc. 2009, 131, 7390–7401. [Google Scholar] [CrossRef] [PubMed]
- Tóth, G.; Gardai, S.J.; Zago, W.; Bertoncini, C.W.; Cremades, N.; Roy, S.L.; Tambe, M.A.; Rochet, J.C.; Galvagnion, C.; Skibinski, G.; et al. Targeting the intrinsically disordered structural ensemble of α-synuclein by small molecules as a potential therapeutic strategy for Parkinson’s disease. PLoS ONE 2014, 9, e87133. [Google Scholar] [CrossRef] [PubMed]
- Convertino, M.; Vitalis, A.; Caflisch, A. Disordered binding of small molecules to Aβ(12-28). J. Biol. Chem. 2011, 286, 41578–41588. [Google Scholar] [CrossRef] [PubMed]
- Khoo, K.H.; Verma, C.S.; Lane, D.P. Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014, 13, 217–236. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Lee, C.; Kim, B.; Lee, S.H.; Han, K.H. Rescuing p53 from mdm2 by a pre-structured motif in intrinsically unfolded SUMO specific protease 4. BMB Rep. 2017, 50, 485–486. [Google Scholar] [CrossRef]
- Tsytlonok, M.; Sanabria, H.; Wang, Y.; Felekyan, S.; Hemmen, K.; Phillips, A.H.; Yun, M.K.; Waddell, M.B.; Park, C.G.; Vaithiyalingam, S.; et al. Dynamic anticipation by Cdk2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation. Nat. Commun. 2019, 10, 1676. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bencivenga, D.; Stampone, E.; Roberti, D.; Della Ragione, F.; Borriello, A. p27Kip1, an Intrinsically Unstructured Protein with Scaffold Properties. Cells 2021, 10, 2254. https://doi.org/10.3390/cells10092254
Bencivenga D, Stampone E, Roberti D, Della Ragione F, Borriello A. p27Kip1, an Intrinsically Unstructured Protein with Scaffold Properties. Cells. 2021; 10(9):2254. https://doi.org/10.3390/cells10092254
Chicago/Turabian StyleBencivenga, Debora, Emanuela Stampone, Domenico Roberti, Fulvio Della Ragione, and Adriana Borriello. 2021. "p27Kip1, an Intrinsically Unstructured Protein with Scaffold Properties" Cells 10, no. 9: 2254. https://doi.org/10.3390/cells10092254
APA StyleBencivenga, D., Stampone, E., Roberti, D., Della Ragione, F., & Borriello, A. (2021). p27Kip1, an Intrinsically Unstructured Protein with Scaffold Properties. Cells, 10(9), 2254. https://doi.org/10.3390/cells10092254








