FDA-Approved Drug Screening for Compounds That Facilitate Hematopoietic Stem and Progenitor Cells (HSPCs) Expansion in Zebrafish
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Maintenance
2.2. Drug Treatment
2.3. Whole-Mount In Situ Hybridization (WISH) and Quantification
2.4. Fluorescence Immunohistochemistry Staining
2.5. TUNEL Assay and EdU Incorporation
2.6. Fluorescence-Activated Cell Sorting (FACS) Analysis
2.7. Quantification and Statistics
3. Results
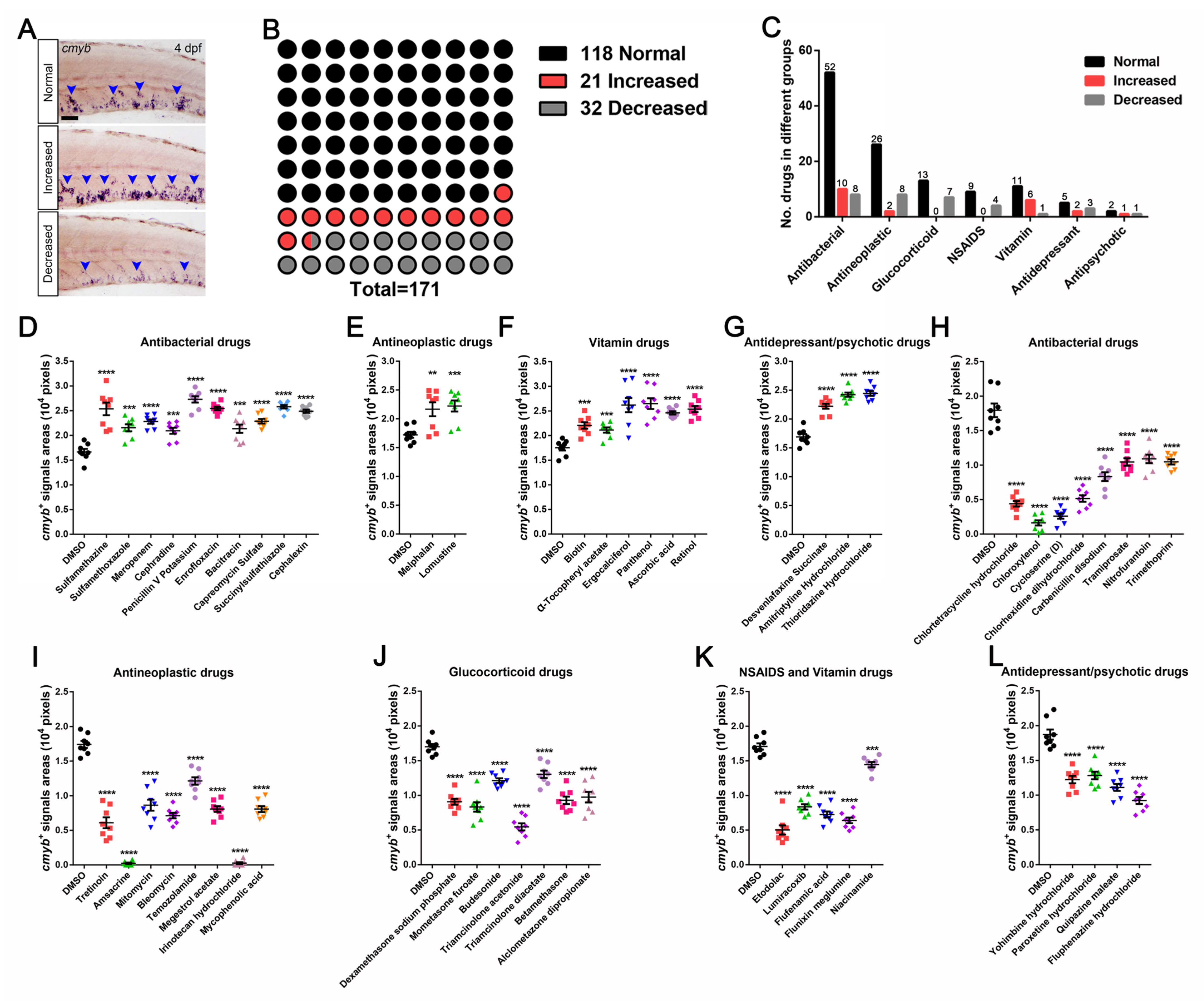
3.1. A Wide-Range Drug Screen for HSPCs Expansion in Zebrafish Using an FDA-Approved Library
3.2. Identification and Characterization of Drugs in Controlling HSPCs Homeostasis
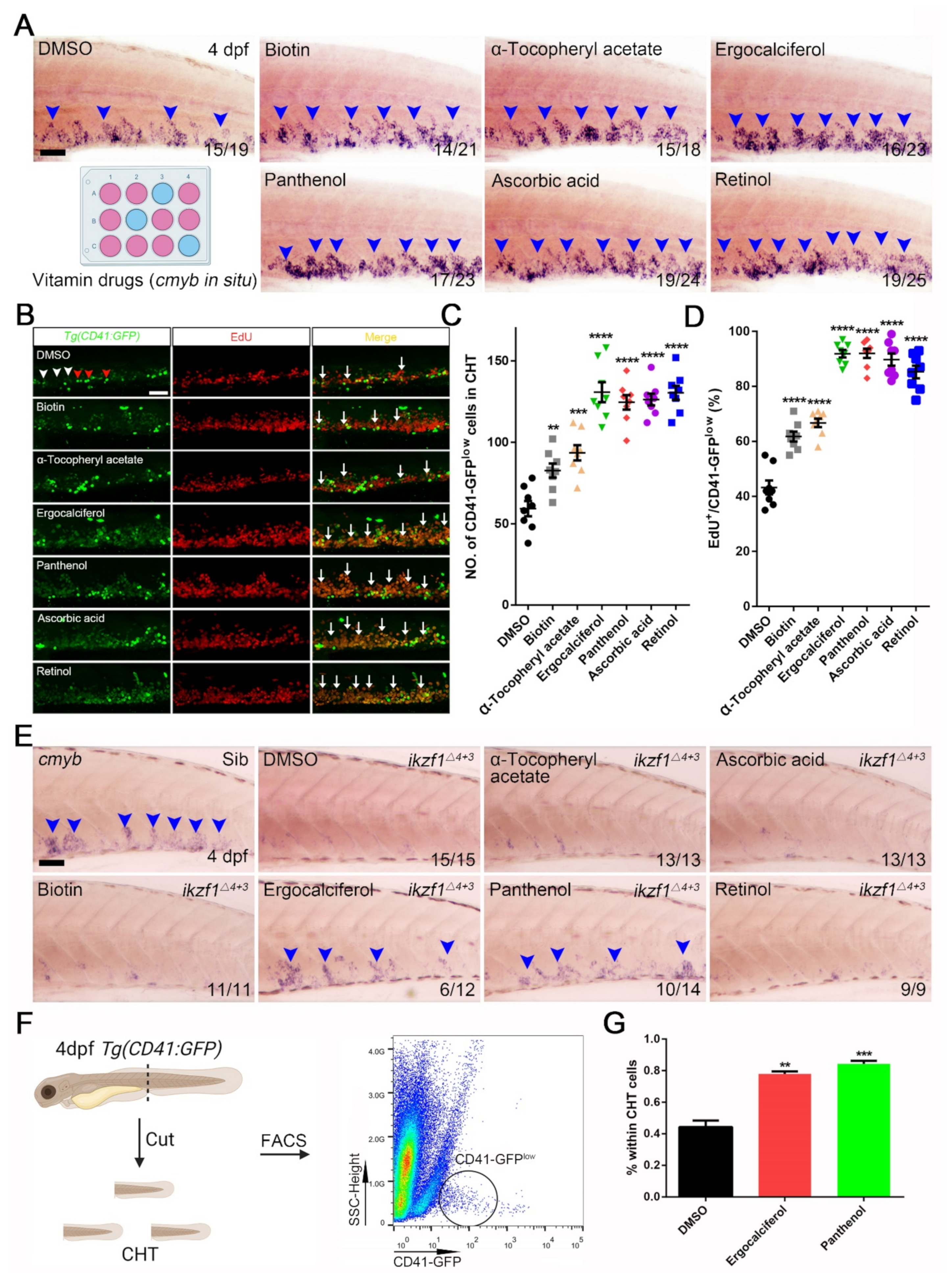
3.3. The Contribution of Vitamin Drugs to HSPCs Expansion and Mitigation of the Hematopoietic Phenotypes in Ikzf1−/− Mutants by Ergocalciferol and Panthenol
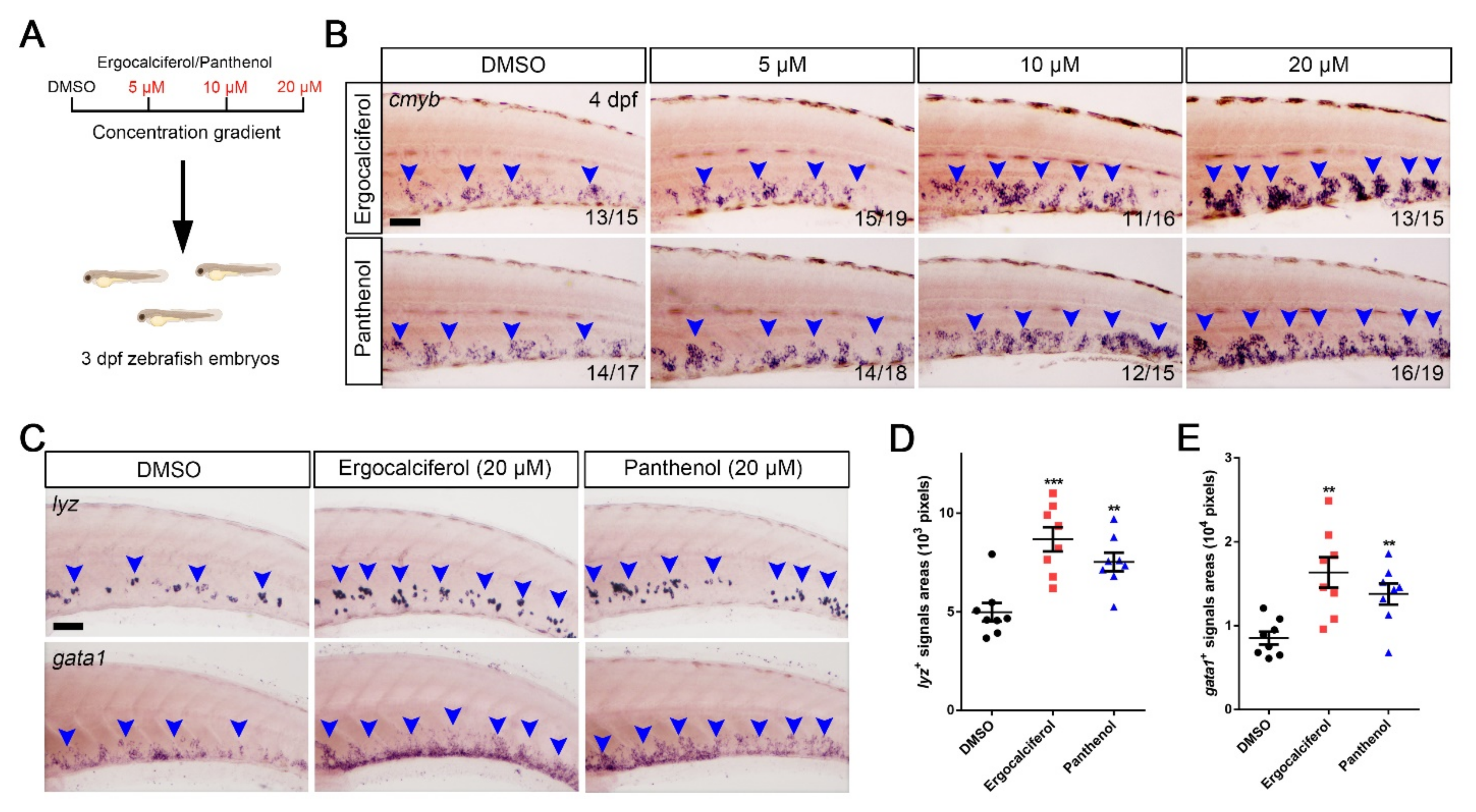
3.4. The Dose-Dependent Effects of Ergocalciferol and Panthenol on HSPCs Expansion
3.5. Comparison of the Analogs Effects of Ergocalciferol and Panthenol
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| NO | Chemical Name | CAS Number | cmyb+ Signals Areas (Pixels; Mean ± SEM, n = 8) | Significance (** p< 0.01, *** p < 0.001, **** p < 0.0001) |
|---|---|---|---|---|
| Control | DMSO | 67-68-5 | 17,530 ± 552 | |
| 1 | Sulfamethazine | 57-68-1 | 25,360 ± 1281 | YES **** |
| 2 | Sulfamethoxazole | 723-46-6 | 21,540 ± 699 | YES *** |
| 3 | Meropenem | 96036-03-2 | 22,790 ± 443 | YES **** |
| 4 | Cephradine | 38821-53-3 | 20,940 ± 616 | YES *** |
| 5 | Penicillin V Potassium | 132-98-9 | 27,290 ± 647 | YES **** |
| 6 | Enrofloxacin | 93106-60-6 | 25,450 ± 355 | YES **** |
| 7 | Bacitracin | 1405-87-4 | 21,350 ± 841 | YES *** |
| 8 | Capreomycin Sulfate | 11003-38-6 | 22,860 ± 454 | YES **** |
| 9 | Succinylsulfathiazole | 116-43-8 | 25,800 ± 341 | YES **** |
| 10 | Cephalexin | 23325-78-2 | 24,890 ± 348 | YES **** |
| 11 | Melphalan | 148-82-3 | 21,650 ± 1205 | YES ** |
| 12 | Lomustine | 13010-47-4 | 22,230 ± 963 | YES *** |
| 13 | Biotin | 22879-79-4 | 22,050 ± 651 | YES *** |
| 14 | α-Tocopheryl acetate | 7695-91-2 | 21,130 ± 528 | YES *** |
| 15 | Ergocalciferol | 50-14-6 | 26,190 ± 1457 | YES **** |
| 16 | Panthenol | 81-13-0 | 26,490 ± 1084 | YES **** |
| 17 | Ascorbic acid | 50-81-7 | 24,610 ± 345 | YES **** |
| 18 | Retinol | 68-26-8 | 25,380 ± 622 | YES **** |
| 19 | Desvenlafaxine Succinate | 386750-22-7 | 22,210 ± 437 | YES **** |
| 20 | Amitriptyline Hydrochloride | 549-18-8 | 24,230 ± 362 | YES **** |
| 21 | Thioridazine Hydrochloride | 1257-76-7 | 24,450 ± 421 | YES **** |
| 22 | Chlortetracycline hydrochloride | 64-72-2 | 4400 ± 3402 | YES **** |
| 23 | Chloroxylenol | 133-53-9 | 1625 ± 380 | YES **** |
| 24 | Cycloserine | 68-41-7 | 2600 ± 369 | YES **** |
| 25 | Chlorhexidine dihydrochloride | 3697-42-5 | 5163 ± 479 | YES **** |
| 26 | Carbenicillin disodium | 4697-36-3 | 8325 ± 638 | YES **** |
| 27 | Tramiprosate | 3687-18-1 | 10,450 ± 543 | YES **** |
| 28 | Nitrofurantoin | 67-20-9 | 10,900 ± 614 | YES **** |
| 29 | Trimethoprim | 738-70-5 | 10,480 ± 376 | YES **** |
| 30 | Tretinoin | 302-79-4 | 6088 ± 783 | YES **** |
| 31 | Amsacrine | 51264-14-3 | 238 ± 107 | YES **** |
| 32 | Mitomycin | 50-07-7 | 8625 ± 802 | YES **** |
| 33 | Bleomycin | 11056-06-7 | 7125 ± 394 | YES **** |
| 34 | Temozolamide | 85622-93-1 | 12,130 ± 531 | YES **** |
| 35 | Megestrol acetate | 595-33-5 | 8075 ± 420 | YES **** |
| 36 | Irinotecan hydrochloride | 100286-90-6 | 263 ± 145 | YES **** |
| 37 | Mycophenolic acid | 24280-93-1 | 8063 ± 431 | YES **** |
| 38 | Dexamethasone sodium phosphate | 2392-39-4 | 9075 ± 433 | YES **** |
| 39 | Mometasone furoate | 83919-23-7 | 8325 ± 676 | YES **** |
| 40 | Budesonide | 51333-22-3 | 12,140 ± 332 | YES **** |
| 41 | Triamcinolone acetonide | 76-25-5 | 5463 ± 512 | YES **** |
| 42 | Triamcinolone diacetate | 67-78-7 | 13,040 ± 532 | YES **** |
| 43 | Betamethasone | 378-44-9 | 9288 ± 539 | YES **** |
| 44 | Alclometasone Dipropionate | 66734-13-2 | 9738 ± 767 | YES **** |
| 45 | Etodolac | 41340-25-4 | 5025 ± 663 | YES **** |
| 46 | Lumiracoxib | 220991-20-8 | 8350 ± 385 | YES **** |
| 47 | Flufenamic acid | 530-78-9 | 7250 ± 436 | YES **** |
| 48 | Flunixin meglumine | 42461-84-7 | 6400 ± 400 | YES **** |
| 49 | Niacinamide | 98-92-0 | 14,450 ± 383 | YES ** |
| 50 | Yohimbine hydrochloride | 65-19-0 | 12,240 ± 535 | YES **** |
| 51 | Paroxetine hydrochloride | 78246-49-8 | 12,830 ± 546 | YES **** |
| 52 | Quipazine maleate | 5786-68-5 | 11,100 ± 502 | YES **** |
| 53 | Fluphenazine hydrochloride | 146-56-5 | 9238 ± 509 | YES **** |
| 54 | Lasalocid sodium | 25999-20-6 | 17,450 ± 402 | NO |
| 55 | Sulfadimethoxine | 122-11-2 | 17,810 ± 393 | NO |
| 56 | Sulfamonomethoxine | 1220-83-3 | 17,730 ± 510 | NO |
| 57 | Sulfamethoxypyridazine | 80-35-3 | 17,240 ± 565 | NO |
| 58 | Oxolinic acid | 59587-08-5 | 18,000 ± 503 | NO |
| 59 | Ofloxacin | 82419-36-1 | 17,380 ± 440 | NO |
| 60 | Difloxacin hydrochloride | 91296-86-5 | 17,490 ± 540 | NO |
| 61 | Erythromycin | 114-07-8 | 16,990 ± 685 | NO |
| 62 | Azlocillin sodium | 37091-65-9 | 18,160 ± 421 | NO |
| 63 | Clarithromycin | 81103-11-9 | 18,200 ± 381 | NO |
| 64 | Oleandomycin phosphate | 7060-74-4 | 17,700 ± 683 | NO |
| 65 | Rifaximin | 17-36-7 | 16,530 ± 430 | NO |
| 66 | Rolitetracycline | 751-97-3 | 17,100 ± 274 | NO |
| 67 | Teicoplanin | 89139-42-4 | 17,300 ± 287 | NO |
| 68 | Sulfathiazole | 72-14-0 | 17,150 ± 340 | NO |
| 69 | Spectinomycin hydrochloride | 21736-83-4 | 18,180 ± 717 | NO |
| 70 | Piperacillin sodium | 89785-84-2 | 18,860 ± 720 | NO |
| 71 | Demeclocycline hydrochloride | 64-73-3 | 18,240 ± 432 | NO |
| 72 | Dihydrostreptomycin sulfate | 5490-27-7 | 17,450 ± 388 | NO |
| 73 | Doxycycline hydrochloride | 10592-13-9 | 17,590 ± 429 | NO |
| 74 | Erythromycin Ethylsuccinate | 1264-62-6 | 17,540 ± 420 | NO |
| 75 | Streptomycin sulfate | 3810-74-0 | 19,680 ± 1142 | NO |
| 76 | Tetracycline hydrochloride | 64-75-5 | 15,760 ± 449 | NO |
| 77 | Cloxacillin sodium | 7081-44-9 | 17,780 ± 367 | NO |
| 78 | Cinoxacin | 28657-80-9 | 17,090 ± 383 | NO |
| 79 | Cloxiquine | 130-16-5 | 17,500 ± 329 | NO |
| 80 | Colistimethate sodium | 8068-37-9 | 17,000 ± 411 | NO |
| 81 | Rifampicin | 13292-46-1 | 17,980 ± 291 | NO |
| 82 | Penicillin G potassium | 113-98-4 | 18,080 ± 635 | NO |
| 83 | Polymyxin B sulfate | 1405-20-5 | 18,560 ± 680 | NO |
| 84 | Gemifloxacin mesylate | 210353-53-0 | 17,560 ± 320 | NO |
| 85 | Norfloxacin | 70458-96-7 | 17,780 ± 388 | NO |
| 86 | Cefmenoxime hydrochloride | 75738-58-8 | 17,960 ± 343 | NO |
| 87 | Cefamandole nafate | 42540-40-9 | 17,850 ± 377 | NO |
| 88 | Ampicillin sodium | 69-52-3 | 17,760 ± 438 | NO |
| 89 | Novobiocin sodium | 1476-53-5 | 17,830 ± 727 | NO |
| 90 | Tobramycin | 32986-56-4 | 17,990 ± 276 | NO |
| 91 | Amoxicillin | 26787-78-0 | 18,430 ± 882 | NO |
| 92 | Daptomycin | 103060-53-3 | 16,590 ± 447 | NO |
| 93 | Sulfacetamide | 144-80-9 | 17,350 ± 247 | NO |
| 94 | Chloramphenicol | 56-75-7 | 17,830 ± 308 | NO |
| 95 | Chloramphenicol succinate sodium | 982-57-0 | 19,280 ± 342 | NO |
| 96 | Cefotaxime sodium | 64485-93-4 | 18,940 ± 264 | NO |
| 97 | Sulfapyridine | 144-83-2 | 19,410 ± 670 | NO |
| 98 | Hydrastine | 118-08-1 | 17,660 ± 332 | NO |
| 99 | Tyrothricin | 1404-88-2 | 17,750 ± 302 | NO |
| 100 | Vancomycin hydrochloride | 1404-93-9 | 19,230 ± 332 | NO |
| 101 | Merbromin | 129-16-8 | 18,610 ± 854 | NO |
| 102 | Sulfisoxazole | 127-69-5 | 17,750 ± 288 | NO |
| 103 | Bekanamycin sulfate | 29701-07-3 | 18,080 ± 310 | NO |
| 104 | Cefaclor | 53994-73-3 | 19,550 ± 1041 | NO |
| 105 | Sulfabenzamide | 127-71-9 | 16,130 ± 608 | NO |
| 106 | Sodium phenylbutyrate | 1716-12-7 | 19,100 ± 437 | NO |
| 107 | Mechlorethamine | 51-75-2 | 17,190 ± 299 | NO |
| 108 | Oxaliplatin | 61825-94-3 | 19,130 ± 416 | NO |
| 109 | Sirolimus | 53123-88-9 | 19,900 ± 1037 | NO |
| 110 | Pipobroman | 54-91-1 | 19,940 ± 754 | NO |
| 111 | Thioguanine | 154-42-7 | 19,250 ± 452 | NO |
| 112 | Ifosfamide | 3778-73-2 | 19,540 ± 552 | NO |
| 113 | Puromycin dihydrochloride | 58-58-2 | 19,510 ± 994 | NO |
| 114 | Letrozole | 112809-51-5 | 17,800 ± 294 | NO |
| 115 | Doxorubicin | 23214-92-8 | 16,440 ± 737 | NO |
| 116 | Semustine | 13909-09-6 | 16,830 ± 376 | NO |
| 117 | Mitoxantrone hydrochloride | 70476-82-3 | 18,100 ± 192 | NO |
| 118 | Azacitidine | 320-67-2 | 18,430 ± 607 | NO |
| 119 | Toremifene citrate | 89778-27-8 | 19,480 ± 661 | NO |
| 120 | Anastrozole | 120511-73-1 | 17,500 ± 403 | NO |
| 121 | Paclitaxel | 33069-62-4 | 17,860 ± 356 | NO |
| 122 | Streptomycin | 3810-74-0 | 18,580 ± 532 | NO |
| 123 | Flutamide | 13311-84-7 | 18,980 ± 1173 | NO |
| 124 | Urethane | 51-79-6 | 19,660 ± 377 | NO |
| 125 | Tamoxifen citrate | 54965-24-1 | 19,340 ± 320 | NO |
| 126 | Docetaxel | 114977-28-5 | 19,440 ± 848 | NO |
| 127 | Casanthranol | 8024-48-4 | 20,730 ± 1352 | NO |
| 128 | Azathioprine | 446-86-6 | 17,080 ± 247 | NO |
| 129 | Chlorambucil | 305-03-3 | 17,500 ± 400 | NO |
| 130 | Testosterone | 58-22-0 | 18,000 ± 275 | NO |
| 131 | Uracil | 66-22-8 | 19,130 ± 495 | NO |
| 132 | Betamethasone valerate | 2152-44-5 | 19,600 ± 730 | NO |
| 133 | Hydrocortisone | 50-23-7 | 18,940 ± 557 | NO |
| 134 | Cortisone acetate | 50-04-4 | 19,700 ± 738 | NO |
| 135 | Dapsone | 80-08-0 | 17,290 ± 565 | NO |
| 136 | Prednisolone | 50-24-8 | 17,410 ± 467 | NO |
| 137 | Prednisone | 53-03-2 | 17,440 ± 235 | NO |
| 138 | Prednisolone succinate | 2920-86-7 | 17,800 ± 303 | NO |
| 139 | Prednisolone sodium phosphate | 125-02-0 | 16,840 ± 249 | NO |
| 140 | Clobetasol propionate | 25122-46-7 | 19,530 ± 741 | NO |
| 141 | Fluticasone propionate | 80474-14-2 | 19,250 ± 465 | NO |
| 142 | Flumethasone | 2135-17-3 | 17,480 ± 205 | NO |
| 143 | Methylprednisolone sodium succinate | 2375-03-3 | 17,640 ± 365 | NO |
| 144 | Betamethasone sodium phosphate | 151-73-5 | 19,790 ± 457 | NO |
| 145 | Firocoxib | 189954-96-9 | 17,750 ± 230 | NO |
| 146 | Tenoxicam | 59804-37-4 | 18,760 ± 907 | NO |
| 147 | Mesalamine | 89-57-6 | 20,650 ± 1220 | NO |
| 148 | Tolmetin sodium | 64490-92-2 | 17,440 ± 357 | NO |
| 149 | Ketorolac tromethamine | 74103-07-4 | 17,780 ± 350 | NO |
| 150 | Dexibuprofen | 51146-56-6 | 19,350 ± 700 | NO |
| 151 | Oxyphenbutazone | 129-20-4 | 18,910 ± 566 | NO |
| 152 | Meloxicam sodium | 71125-39-8 | 18,530 ± 506 | NO |
| 153 | Fenoprofen | 31879-05-7 | 18,160 ± 522 | NO |
| 154 | β-Carotene | 7235-40-7 | 17,410 ± 341 | NO |
| 155 | Retinyl palmitate | 79-81-2 | 17,460 ± 326 | NO |
| 156 | Niacin | 59-67-6 | 19,850 ± 850 | NO |
| 157 | Adenine | 73-24-5 | 18,950 ± 529 | NO |
| 158 | Cyanocobalamin | 68-19-9 | 17,160 ± 314 | NO |
| 159 | Phytonadione | 84-80-0 | 18,410 ± 427 | NO |
| 160 | Ethopabate | 59-06-3 | 17,940 ± 757 | NO |
| 161 | Riboflavin | 83-88-5 | 19,800 ± 800 | NO |
| 162 | Niacinamide | 98-92-0 | 17,760 ± 361 | NO |
| 163 | Thiamine | 70-16-6 | 17,450 ± 446 | NO |
| 164 | Folic acid | 59-30-3 | 18,810 ± 476 | NO |
| 165 | Amoxapine | 14028-44-5 | 19,330 ± 678 | NO |
| 166 | Cotinine | 486-56-6 | 18,540 ± 488 | NO |
| 167 | Trazodone hydrochloride | 25332-39-2 | 17,930 ± 503 | NO |
| 168 | Tranylcypromine sulfate | 13492-01-8 | 18,240 ± 831 | NO |
| 169 | Tryptophan | 73-22-3 | 19,680 ± 955 | NO |
| 170 | Trifluoperazine hydrochloride | 117-89-5 | 18,960 ± 811 | NO |
| 171 | Promazine hydrochloride | 53-60-1 | 17,550 ± 404 | NO |
References
- Bai, T.; Li, J.; Sinclair, A.; Imren, S.; Merriam, F.; Sun, F.; O’Kelly, M.B.; Nourigat, C.; Jain, P.; Delrow, J.J.; et al. Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel. Nat. Med. 2019, 25, 1566–1575. [Google Scholar] [CrossRef]
- Bari, S.; Seah, K.K.H.; Poon, Z.; Cheung, A.M.S.; Fan, X.; Ong, S.-Y.; Li, S.; Koh, L.P.; Hwang, W.Y.K. Expansion and homing of umbilical cord blood hematopoietic stem and progenitor cells for clinical transplantation. Biol. Blood Marrow Transplant. 2015, 21, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Sica, R.A.; Terzioglu, M.K.; Mahmud, D.; Mahmud, N. Mechanistic Basis of ex Vivo Umbilical Cord Blood Stem Progenitor Cell Expansion. Stem Cell Rev. Rep. 2020, 16, 628–638. [Google Scholar] [CrossRef]
- Gao, S.; Liu, F. Fetal liver: An ideal niche for hematopoietic stem cell expansion. Sci. China Life Sci. 2018, 61, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Coskun, S.; Hirschi, K.K. Establishment and regulation of the HSC niche: Roles of osteoblastic and vascular compartments. Birth Defects Res. C Embryo Today 2010, 90, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, D. Hematopoietic cytokines. Blood 2008, 111, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Krosl, J.; Mamo, A.; Chagraoui, J.; Wilhelm, B.T.; Girard, S.; Louis, I.; Lessard, J.; Perreault, C.; Sauvageau, G. A mutant allele of the Swi/Snf member BAF250a determines the pool size of fetal liver hemopoietic stem cell populations. Blood 2010, 116, 1678–1684. [Google Scholar] [CrossRef] [PubMed]
- Pineault, N.; Abu-Khader, A. Advances in umbilical cord blood stem cell expansion and clinical translation. Exp. Hematol. 2015, 43, 498–513. [Google Scholar] [CrossRef]
- Mendelson, A.; Frenette, P.S. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 2014, 20, 833–846. [Google Scholar] [CrossRef]
- Chaurasia, P.; Gajzer, D.C.; Schaniel, C.; D’Souza, S.; Hoffman, R. Epigenetic reprogramming induces the expansion of cord blood stem cells. J. Clin. Investig. 2014, 124, 2378–2395. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, N.; Petro, B.; Baluchamy, S.; Li, X.; Taioli, S.; Lavelle, D.; Quigley, J.G.; Suphangul, M.; Araki, H. Differential effects of epigenetic modifiers on the expansion and maintenance of human cord blood stem/progenitor cells. Biol. Blood Marrow Transplant. 2014, 20, 480–489. [Google Scholar] [CrossRef]
- Park, B.; Yoo, K.H.; Kim, C. Hematopoietic stem cell expansion and generation: The ways to make a breakthrough. Blood Res. 2015, 50, 194–203. [Google Scholar] [CrossRef]
- Nosengo, N. Can you teach old drugs new tricks? Nature 2016, 534, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Boatman, S.; Barrett, F.; Satishchandran, S.; Jing, L.; Shestopalov, I.; Zon, L.I. Assaying hematopoiesis using zebrafish. Blood Cells Mol. Dis. 2013, 51, 271–276. [Google Scholar] [CrossRef]
- Orkin, S.H.; Zon, L.I. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 2008, 132, 631–644. [Google Scholar] [CrossRef]
- Jagannathan-Bogdan, M.; Zon, L.I. Hematopoiesis. Development 2013, 140, 2463–2467. [Google Scholar] [CrossRef]
- Rowe, R.G.; Mandelbaum, J.; Zon, L.I.; Daley, G.Q. Engineering Hematopoietic Stem Cells: Lessons from Development. Cell Stem Cell 2016, 18, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Paik, E.J.; Zon, L.I. Hematopoietic development in the zebrafish. Int. J. Dev. Biol. 2010, 54, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Avagyan, S.; Zon, L.I. Fish to Learn: Insights into Blood Development and Blood Disorders from Zebrafish Hematopoiesis. Hum. Gene Ther. 2016, 27, 287–294. [Google Scholar] [CrossRef]
- Murphey, R.D.; Zon, L.I. Small molecule screening in the zebrafish. Methods 2006, 39, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Zon, L.I.; Peterson, R.T. In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 2005, 4, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Patton, E.E.; Zon, L.I.; Langenau, D.M. Zebrafish disease models in drug discovery: From preclinical modelling to clinical trials. Nat. Rev. Drug Discov. 2021, 20, 611–628. [Google Scholar] [CrossRef]
- North, T.E.; Goessling, W.; Walkley, C.; Lengerke, C.; Kopani, K.R.; Lord, A.M.; Weber, G.J.; Bowman, T.V.; Jang, I.-H.; Grosser, T.; et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 2007, 447, 1007–1011. [Google Scholar] [CrossRef]
- Hagedorn, E.J.; Durand, E.M.; Fast, E.M.; Zon, L.I. Getting more for your marrow: Boosting hematopoietic stem cell numbers with PGE2. Exp. Cell Res. 2014, 329, 220–226. [Google Scholar] [CrossRef]
- Taylor, A.M.; Macari, E.R.; Chan, I.T.; Blair, M.C.; Doulatov, S.; Vo, L.T.; Raiser, D.M.; Siva, K.; Basak, A.; Pirouz, M.; et al. Calmodulin inhibitors improve erythropoiesis in Diamond-Blackfan anemia. Sci. Transl. Med. 2020, 12, eabb5831. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-F.; Traver, D.; Zhu, H.; Dooley, K.; Paw, B.; Zon, L.I.; Handin, R.I. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood 2005, 106, 3803–3810. [Google Scholar] [CrossRef] [PubMed]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef]
- Chang, C.; Hu, M.; Zhu, Z.; Lo, L.J.; Chen, J.; Peng, J. liver-enriched gene 1a and 1b encode novel secretory proteins essential for normal liver development in zebrafish. PLoS ONE 2011, 6, e22910. [Google Scholar] [CrossRef]
- Ma, Z.; Zhu, P.; Shi, H.; Guo, L.; Zhang, Q.; Chen, Y.; Chen, S.; Zhang, Z.; Peng, J.; Chen, J. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 2019, 568, 259–263. [Google Scholar] [CrossRef]
- Zhao, F.; Shi, Y.; Huang, Y.; Zhan, Y.; Zhou, L.; Li, Y.; Wan, Y.; Li, H.; Huang, H.; Ruan, H.; et al. Irf8 regulates the progression of myeloproliferative neoplasm-like syndrome via Mertk signaling in zebrafish. Leukemia 2018, 32, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Li, C.; Zhang, Y.; Lin, C.; Zhang, Y.; Shu, L.; Luo, L.; Zhuo, J.; Li, L. Macrophage-Derived IL-1β Regulates Emergency Myelopoiesis via the NF-κB and C/ebpβ in Zebrafish. J. Immunol. 2020, 205, 2694–2706. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Chi, M.; Laplace-Builhé, B.; Travnickova, J.; Crawford, P.L.; Tejedor, G.; Phan, Q.T.; Duroux-Richard, I.; Levraud, J.-P.; Kissa, K.; Lutfalla, G.; et al. Identification of polarized macrophage subsets in zebrafish. eLife 2015, 4, e07288. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Gao, S.; Lv, J.; Li, W.; Liu, F. BLOS2 maintains hematopoietic stem cells in the fetal liver via repressing Notch signaling. Exp. Hematol. 2017, 51, 1–6.e2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xue, Y.; Liu, D.; Cui, G.; Ding, Y.; Ai, D.; Gao, S.; Zhang, Y.; Suo, S.; Wang, X.; Lv, P.; et al. A 3D Atlas of Hematopoietic Stem and Progenitor Cell Expansion by Multi-dimensional RNA-Seq Analysis. Cell Rep. 2019, 27, 1567–1578.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jin, H.; Li, L.; Wen, Z.; Qin, F.X.-F. cMyb regulates hematopoietic stem/progenitor cell mobilization during zebrafish hematopoiesis. Blood 2011, 118, 4093–4101. [Google Scholar] [CrossRef]
- Cortes, M.; Chen, M.J.; Stachura, D.L.; Liu, S.Y.; Kwan, W.; Wright, F.; Vo, L.T.; Theodore, L.; Esain, V.; Frost, I.M.; et al. Developmental Vitamin D Availability Impacts Hematopoietic Stem Cell Production. Cell Rep. 2016, 17, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Studzinski, G.P.; Harrison, J.S.; Wang, X.; Sarkar, S.; Kalia, V.; Danilenko, M. Vitamin D Control of Hematopoietic Cell Differentiation and Leukemia. J. Cell Biochem. 2015, 116, 1500–1512. [Google Scholar] [CrossRef]
- Hall, A.C.; Juckett, M.B. The role of vitamin D in hematologic disease and stem cell transplantation. Nutrients 2013, 5, 2206–2221. [Google Scholar] [CrossRef]
- Cabezas-Wallscheid, N.; Buettner, F.; Sommerkamp, P.; Klimmeck, D.; Ladel, L.; Thalheimer, F.B.; Pastor-Flores, D.; Roma, L.P.; Renders, S.; Zeisberger, P.; et al. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell 2017, 169, 807–823.e19. [Google Scholar] [CrossRef]
- Huang, Y.; Lu, Y.; He, Y.; Feng, Z.; Zhan, Y.; Huang, X.; Liu, Q.; Zhang, J.; Li, H.; Huang, H.; et al. Ikzf1 regulates embryonic T lymphopoiesis via Ccr9 and Irf4 in zebrafish. J. Biol. Chem. 2019, 294, 16152–16163. [Google Scholar] [CrossRef]
- Cross, M.; Mangelsdorf, I.; Wedel, A.; Renkawitz, R. Mouse lysozyme M gene: Isolation, characterization, and expression studies. Proc. Natl. Acad. Sci. USA 1988, 85, 6232–6236. [Google Scholar] [CrossRef]
- Monteiro, R.; Pouget, C.; Patient, R. The gata1/pu.1 lineage fate paradigm varies between blood populations and is modulated by tif1γ. Embo J. 2011, 30, 1093–1103. [Google Scholar]
- Seda, M.; Geerlings, M.; Lim, P.; Jeyabalan-Srikaran, J.; Cichon, A.-C.; Scambler, P.J.; Beales, P.L.; Hernandez, V.; Stoker, A.W.; Jenkins, D. An FDA-Approved Drug Screen for Compounds Influencing Craniofacial Skeletal Development and Craniosynostosis. Mol. Syndromol. 2019, 10, 98–114. [Google Scholar] [CrossRef]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef]
- Ni, X.; Hu, G.; Cai, X. The success and the challenge of all-trans retinoic acid in the treatment of cancer. Crit. Rev. Food Sci. Nutr. 2019, 59, S71–S80. [Google Scholar] [CrossRef]
- Tomita, A.; Kiyoi, H.; Naoe, T. Mechanisms of action and resistance to all-trans retinoic acid (ATRA) and arsenic trioxide (As2O3) in acute promyelocytic leukemia. Int. J. Hematol. 2013, 97, 717–725. [Google Scholar] [CrossRef]
- Borel, P.; Caillaud, D.; Cano, N.J. Vitamin D bioavailability: State of the art. Crit. Rev. Food Sci. Nutr. 2015, 55, 1193–1205. [Google Scholar] [CrossRef] [PubMed]
- Bissett, D.L. Common cosmeceuticals. Clin. Dermatol. 2009, 27, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Gorski, J.; Proksch, E.; Baron, J.M.; Schmid, D.; Zhang, L. Dexpanthenol in Wound Healing after Medical and Cosmetic Interventions (Postprocedure Wound Healing). Pharmaceuticals 2020, 13, 138. [Google Scholar] [CrossRef] [PubMed]
- Galandrin, S.; Oligny-Longpré, G.; Bouvier, M. The evasive nature of drug efficacy: Implications for drug discovery. Trends Pharmacol. Sci. 2007, 28, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Salahudeen, M.S.; Nishtala, P.S. An overview of pharmacodysnamic modelling, ligand-binding approach and its application in clinical practice. Saudi Pharm. J. 2017, 25, 165–175. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Z.; Lin, C.; Tu, L.; Su, M.; Song, C.; Liu, S.; Suryanto, M.E.; Hsiao, C.-D.; Li, L. FDA-Approved Drug Screening for Compounds That Facilitate Hematopoietic Stem and Progenitor Cells (HSPCs) Expansion in Zebrafish. Cells 2021, 10, 2149. https://doi.org/10.3390/cells10082149
Feng Z, Lin C, Tu L, Su M, Song C, Liu S, Suryanto ME, Hsiao C-D, Li L. FDA-Approved Drug Screening for Compounds That Facilitate Hematopoietic Stem and Progenitor Cells (HSPCs) Expansion in Zebrafish. Cells. 2021; 10(8):2149. https://doi.org/10.3390/cells10082149
Chicago/Turabian StyleFeng, Zhi, Chenyu Lin, Limei Tu, Ming Su, Chunyu Song, Shengnan Liu, Michael Edbert Suryanto, Chung-Der Hsiao, and Li Li. 2021. "FDA-Approved Drug Screening for Compounds That Facilitate Hematopoietic Stem and Progenitor Cells (HSPCs) Expansion in Zebrafish" Cells 10, no. 8: 2149. https://doi.org/10.3390/cells10082149
APA StyleFeng, Z., Lin, C., Tu, L., Su, M., Song, C., Liu, S., Suryanto, M. E., Hsiao, C.-D., & Li, L. (2021). FDA-Approved Drug Screening for Compounds That Facilitate Hematopoietic Stem and Progenitor Cells (HSPCs) Expansion in Zebrafish. Cells, 10(8), 2149. https://doi.org/10.3390/cells10082149







