The Role of Galanin during Bacterial Infection in Larval Zebrafish
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Maintenance
2.2. Bacteria Preparation
2.3. Infection
2.4. Treatment with NAX 5055
2.5. Imaging and Fluorescence Quantification
2.6. RNA Isolation and RT-qPCR
2.7. RNA-Seq Analysis
2.8. COPAS Analysis
2.9. Statistical Analysis
3. Results
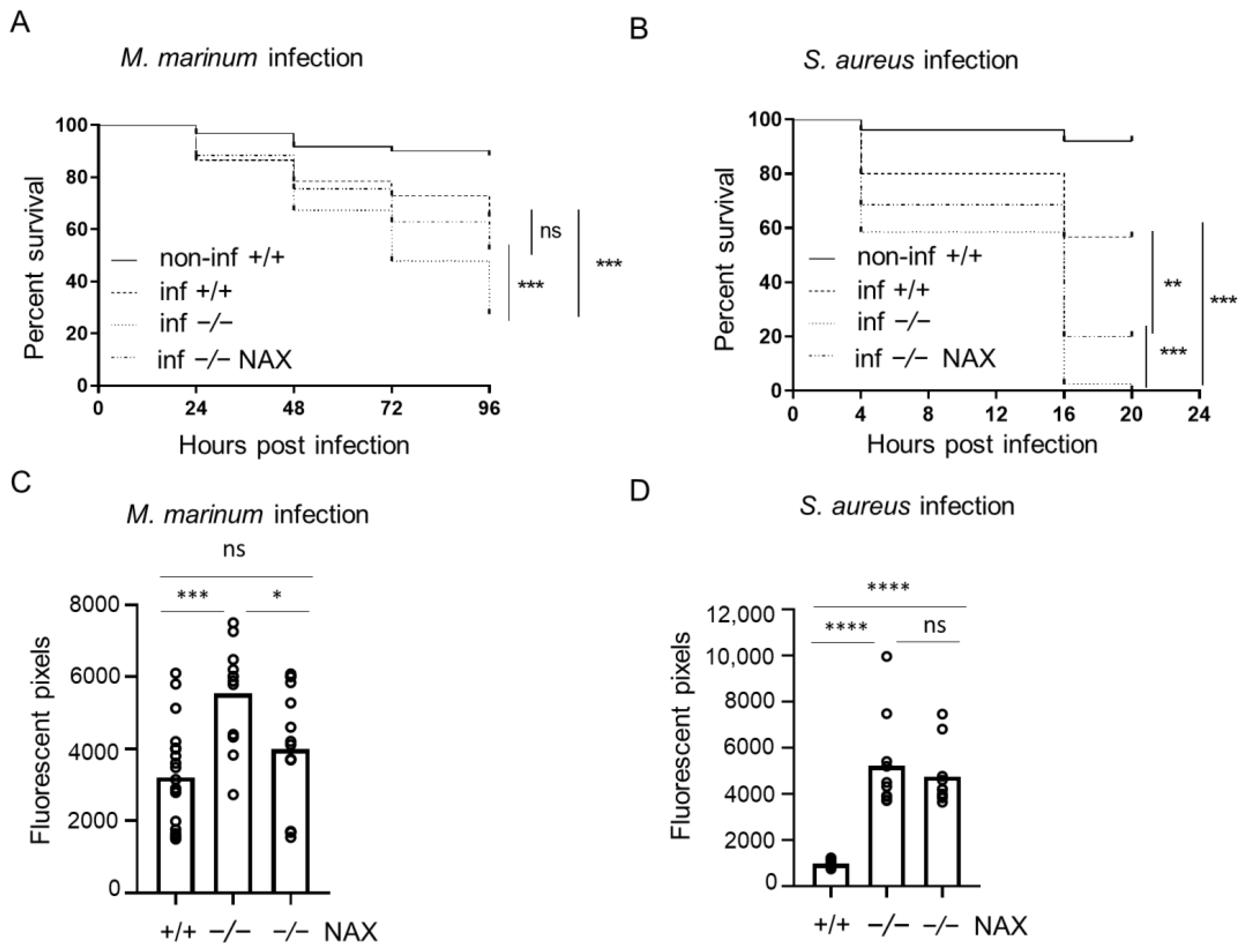
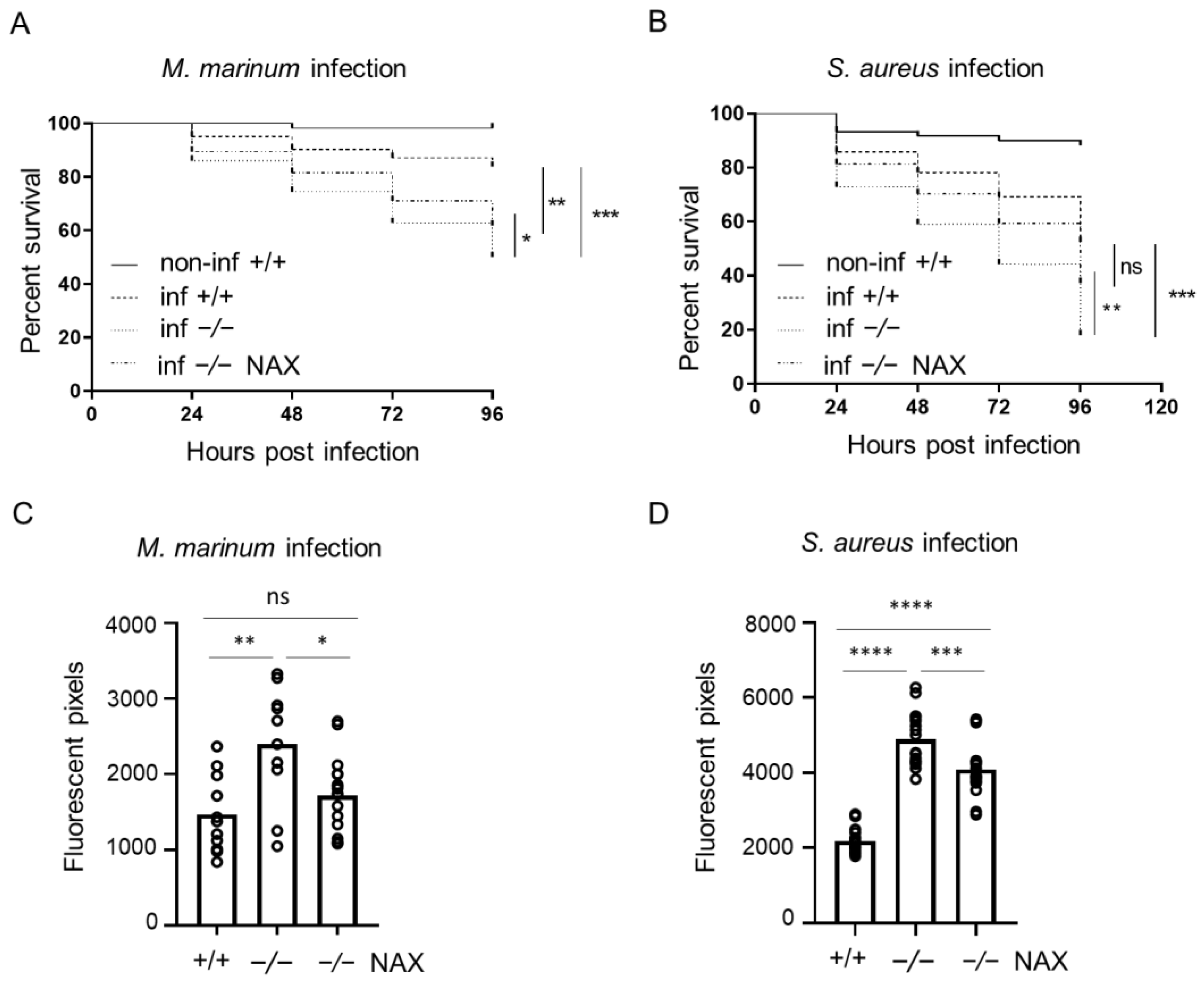
3.1. Bacterial Infection in Galanin-Deficient Zebrafish Larvae
3.2. NAX 5055 Treatment in the Infected Galanin Mutants
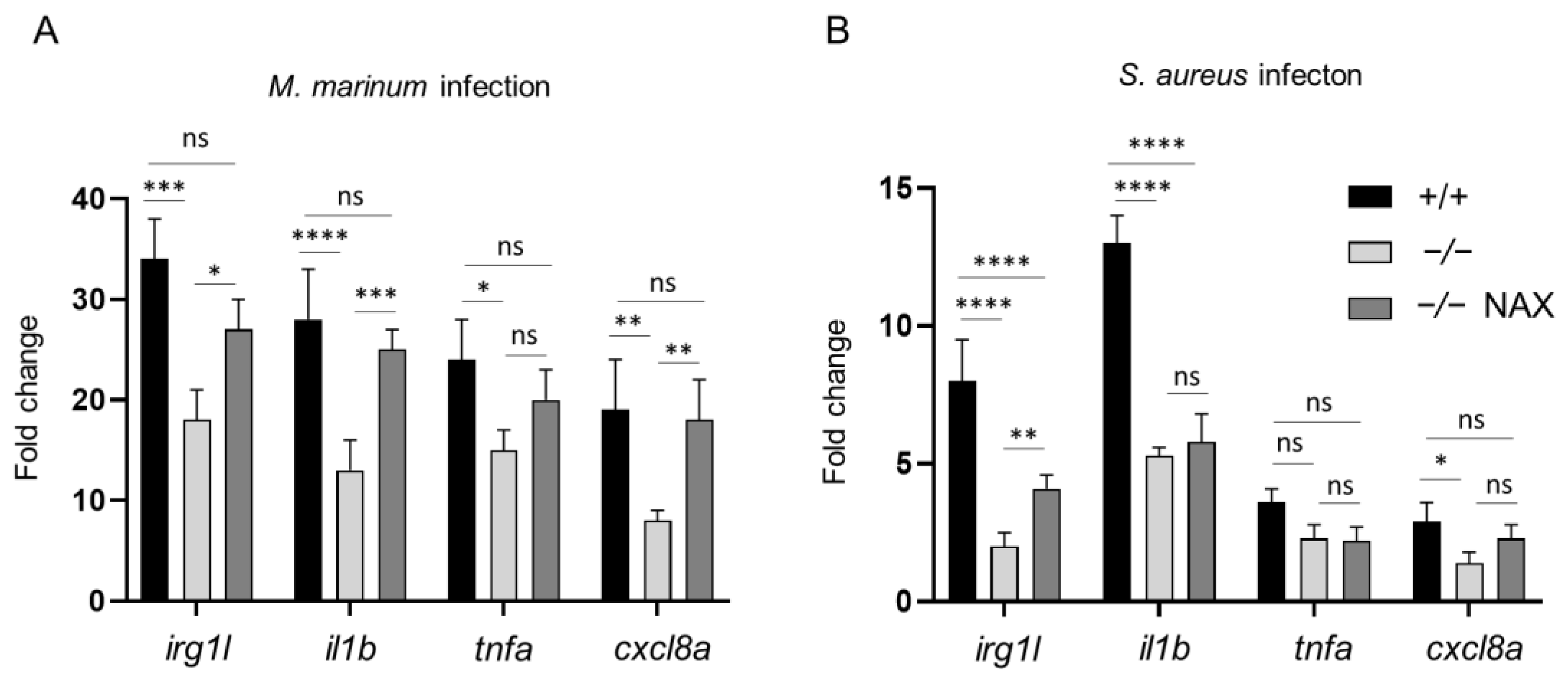
3.3. Galanin Deficiency and Immune-Related Gene Expression
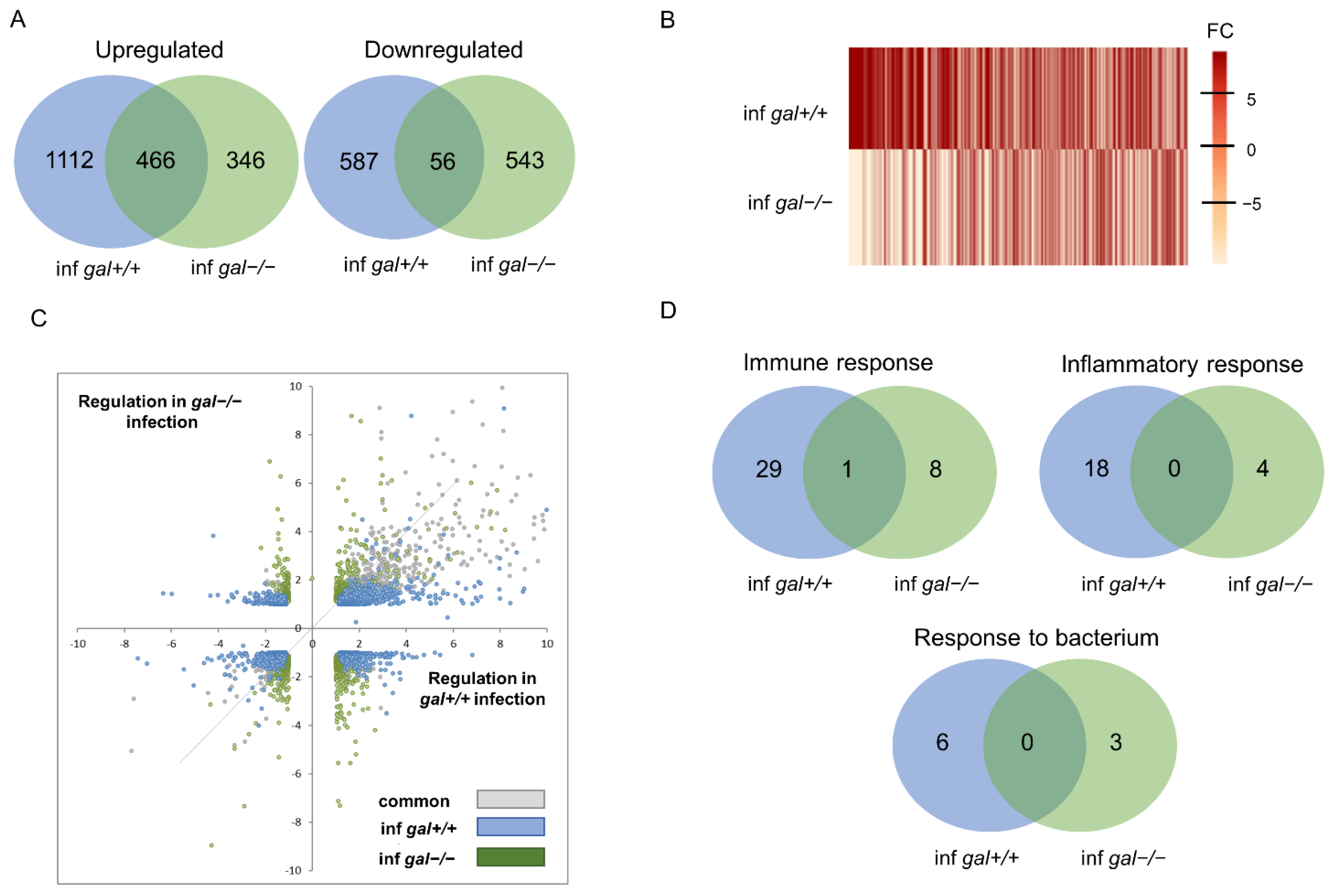
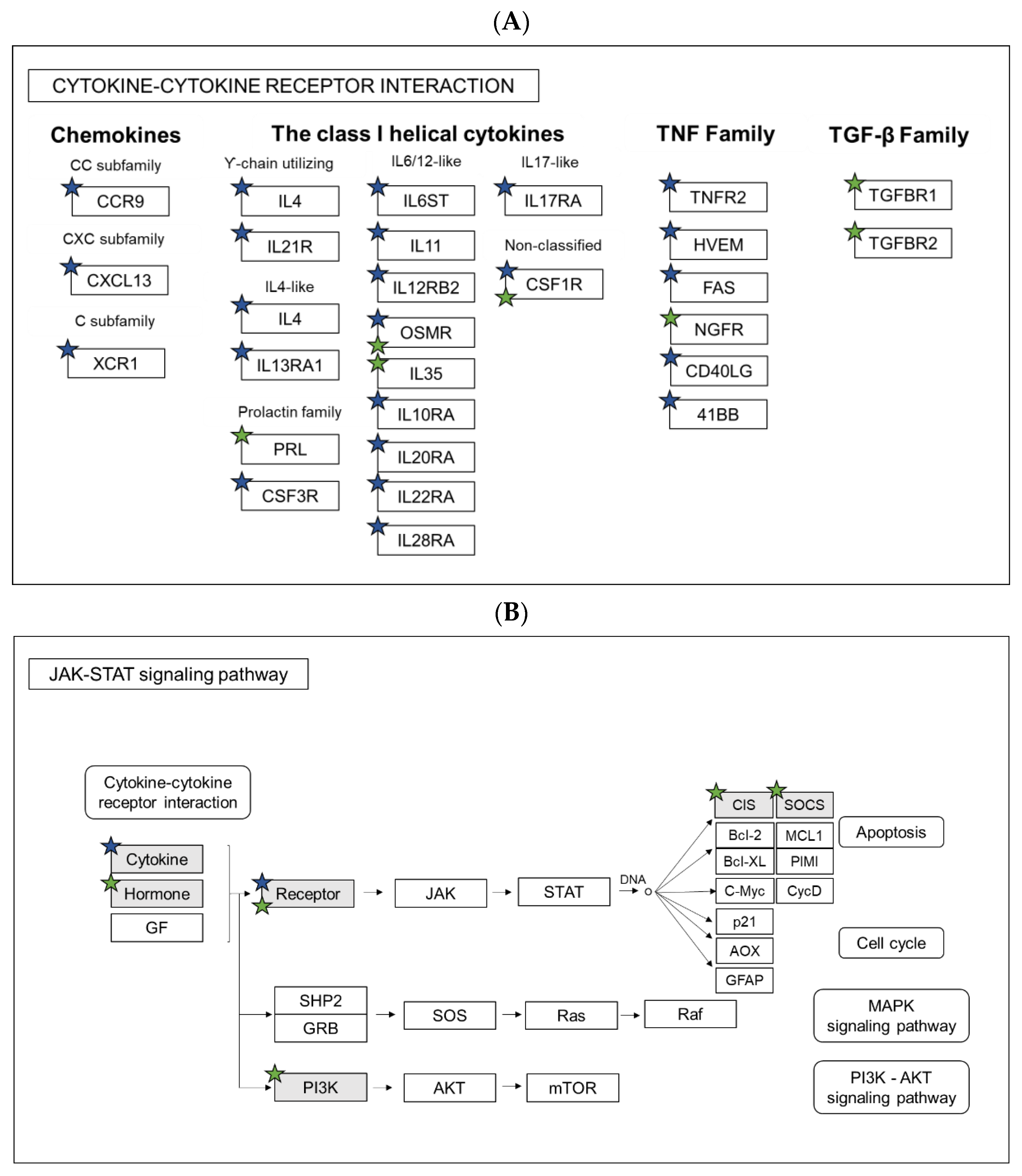
3.4. Transcriptomic Profiling of Galanin-Deficient Zebrafish Larvae Infected with M. marinum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gonzalez-Rey, E.; Chorny, A.; Delgado, M. Regulation of immune tolerance by anti-inflammatory neuropeptides. Nat. Rev. Immunol. 2007, 7, 52–63. [Google Scholar] [CrossRef]
- Rivest, S. Interactions between the immune and neuroendocrine systems. Prog. Brain Res. 2010, 181, 43–53. [Google Scholar]
- Sterner-Kock, A.; Braun, R.K.; van der Vliet, A.; Schrenzel, M.D.; McDonald, R.J.; Kabbur, M.B.; Vulliet, P.R.; Hyde, D.M. Substance P primes the formation of hydrogen peroxide and nitric oxide in human neutrophils. J. Leukoc. Biol. 1999, 65, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Kinhult, J.; Egesten, A.; Uddman, R.; Cardell, L.O. PACAP enhances the expression of CD11b, CD66b andCD63 in human neutrophils. Peptides 2002, 23, 1735–1739. [Google Scholar] [CrossRef]
- Monneret, G.; Arpin, M.; Venet, F.; Maghni, K.; Debard, A.L.; Pachot, A.; Lepape, A.; Bienvenu, J. Calcitonin gene related peptide and N-procalcitonin modulate CD11b upregulation in lipopolysaccharide activated monocytes and neutrophils. Intensive Care Med. 2003, 29, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Dimitrijevic, M.; Stanojevic, S.; Micic, S.; Vujic, V.; Kovacevic-Jovanovic, V.; Mitic, K.; von Horsten, S.; Kosec, D. Neuropeptide Y (NPY) modulates oxidative burst and nitric oxide production in carrageenan-elicited granulocytes from rat air pouch. Peptides 2006, 27, 3208–3215. [Google Scholar] [CrossRef]
- Tatemoto, K.; Rokaeus, A.; Jornvall, H.; McDonald, T.J.; Mutt, V. Galanin—A novel biologically active peptide from porcine intestine. FEBS Lett. 1983, 164, 124–128. [Google Scholar] [CrossRef]
- Kroeger, D.; Absi, G.; Gagliardi, C.; Bandaru, S.S.; Madara, J.C.; Ferrari, L.L.; Arrigoni, E.; Münzberg, H.; Scammell, T.E.; Saper, C.B.; et al. Galanin neurons in the ventrolateral pre-optic area promote sleep and heat loss in mice. Nat. Commun. 2018, 9, 4129. [Google Scholar] [CrossRef]
- Lang, R.; Gundlach, A.L.; Holmes, F.E.; Hobson, S.A.; Wynick, D.; Hökfelt, T.; Kofler, B. Physiology, signaling, and pharmacology of galanin peptides and receptors: Three decades of emerging diversity. Pharm. Rev. 2015, 67, 118–175. [Google Scholar] [CrossRef]
- Podlasz, P.; Wasowicz, K. Neurochemical characteristics of paracervical ganglion in the pig. Veterinární Med. 2008, 53, 135. [Google Scholar] [CrossRef]
- Podlasz, P.; Sallinen, V.; Chen, Y.C.; Kudo, H.; Fedorowska, N.; Panula, P. Galanin gene expression and effects of its knock-down on the development of the nervous system in larval zebrafish. J. Comp. Neurol. 2012, 520, 3846–3862. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Yu, M.; Guo, L.; Bo, P.; Zhang, Z.; Shi, M. Galanin and its receptors: A novel strategy for appetite control and obesity therapy. Peptides. 2012, 36, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Matkowskyj, K.A.; Nathaniel, R.; Prasad, R.; Weihrauch, D.; Rao, M.; Benya, R.V. Galanin contributes to the excess colonic fluid secretion observed in dextran sulfate sodium murine colitis. Inflamm. Bowel Dis. 2004, 10, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Talero, E.; Sánchez-Fidalgo, S.; Calvo, J.R.; Motilva, V. Chronic administration of galanin attenuates the TNBS-induced colitis in rats. Regul. Pept. 2007, 141, 96–104. [Google Scholar] [CrossRef]
- Schmidhuber, S.M.; Rauch, I.; Kofler, B.; Brain, S.D. Evidence that the modulatory effect of galanin on inflammatory edema formation is mediated by the galanin receptor 3 in the murine microvasculature. J. Mol. Neurosci. 2009, 37, 177–181. [Google Scholar] [CrossRef]
- Pilmane, M.; Shine, J.; Iismaa, T.P. Distribution of galanin immunoreactivity in the bronchi of humans with tuberculosis. Ann. N. Y. Acad. Sci. 1998, 863, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Jurkowski, W.; Yazdi, S.; Elofsson, A. Ligand binding properties of human galanin receptors. Mol. Membr. Biol. 2013, 30, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xu, Y.; Zhang, S. Evolution of galanin receptor genes: Insights from the deuterostome genomes. J. Biomol. Stuct. Dyn. 2010, 28, 97–106. [Google Scholar] [CrossRef]
- Eskova, A.; Frohnhöfer, H.G.; Nüsslein-Volhard, C.; Irion, U. Galanin Signaling in the Brain Regulates Color Pattern Formation in Zebrafish. Curr. Biol. 2020, 30, 298–303. [Google Scholar] [CrossRef]
- Li, L.; Wei, S.; Huang, Q.; Feng, D.; Zhang, S.; Liu, Z. A novel galanin receptor 1a gene in zebrafish: Tissue distribution, developmental expression and roles in nutrition regulation. Comp. Biochem. Physiology. Part B Biochem. Mol. Biol. 2013, 164, 159–167. [Google Scholar] [CrossRef]
- Kim, E.; Jeong, I.; Kim, S.; Kim, H.K.; Lee, D.W.; Kim, B.; Seong, J.Y.; Bae, Y.K.; Ryu, J.H.; Park, H.C. Distribution of galanin receptor 2b neurons and interaction with galanin in the zebrafish central nervous system. Neurosci. Lett. 2016, 628, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Varshney, G.K.; Pei, W.; LaFave, M.C.; Idol, J.; Xu, L.; Gallardo, V.; Carrington, B.; Bishop, K.; Jones, M.; Li, M.; et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015, 25, 1030–1042. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 7446, 498–503. [Google Scholar] [CrossRef]
- Panula, P.; Sallinen, V.; Sundvik, M.; Kolehmainen, J.; Torkko, V.; Tiittula, A.; Moshnyakov, M.; Podlasz, P. Modulatory neurotransmitter systems and behavior: Towards zebrafish models of neurodegenerative diseases. Zebrafish 2006, 3, 235–247. [Google Scholar] [CrossRef]
- Tulotta, C.; He, S.; van der Ent, W.; Chen, L.; Groenewoud, A.; Spaink, H.P.; Snaar-Jagalska, B.E. Imaging Cancer Angiogenesis and Metastasis in a Zebrafish Embryo Model. Adv. Exp. Med. Biol. 2016, 916, 239–263. [Google Scholar] [PubMed]
- Veneman, W.J.; Marín-Juez, R.; de Soneville, J.; Ordas, A.; Jong-Raadsen, S.; Meijer, A.H.; Spaink, H.P. Establishment and optimization of a high throughput setup to study Staphylococcus epidermidis and Mycobacterium marinum infection as a model for drug discovery. J. Vis. Exp. 2014, 88, e51649. [Google Scholar] [CrossRef] [PubMed]
- Habu, A.; Ohishi, T.; Mihara, S.; Ohkubo, R.; Hong, Y.M.; Mochizuki, T.; Yanaihara, N. Isolation and Sequence Determination of Galanin from the Pituitary of Yellowfin Tuna. Biomed. Res. 1994, 15, 357–362. [Google Scholar] [CrossRef][Green Version]
- World Health Organization. WHO Global Tuberculosis Report 2020; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Ramakrishnan, L. Revisiting the role of the granuloma in tuberculosis. Nat. Rev. Immunol. 2012, 12, 352–366. [Google Scholar] [CrossRef]
- Josse, J.; Velard, F.; Gangloff, S.C. Staphylococcus aureus vs. Osteoblast: Relationship and Consequences in Osteomyelitis. Front. Cell Infect. Microbiol. 2015, 26, 5:85. [Google Scholar] [CrossRef]
- Thomer, L.; Schneewind, O.; Missiakas, D. Pathogenesis of Staphylococcus aureus Bloodstream Infections. Annu. Rev. Pathol. 2016, 11, 343–364. [Google Scholar] [CrossRef]
- Keynan, Y.; Rubinstein, E. Staphylococcus aureus bacteremia, risk factors, complications, and management. Crit. Care Clin. 2013, 29, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Torraca, V.; Mostowy, S. Zebrafish Infection: From Pathogenesis to Cell Biology. Trends Cell Biol. 2018, 28, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.M.; Clay, H.; Lewis, J.L.; Ghori, N.; Herbomel, P.; Ramakrishnan, L. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 2002, 17, 693–702. [Google Scholar] [CrossRef]
- Prouty, M.G.; Correa, N.E.; Barker, L.P.; Jagadeeswaran, P.; Klose, K.E. Zebrafish-Mycobacterium marinum model for mycobacterial pathogenesis. FEMS Microbiol. Lett. 2003, 25, 177–182. [Google Scholar] [CrossRef]
- Novoa, B.; Figueras, A. Zebrafish: Model for the study of inflammation and the innate immune response to infectious diseases. Adv. Exp. Med. Biol. 2012, 946, 253–275. [Google Scholar] [PubMed]
- Meijer, A.H. Protection and pathology in TB: Learning from the zebrafish model. Semin. Immunopathol. 2016, 38, 261–273. [Google Scholar] [CrossRef]
- Prajsnar, T.K.; Cunliffe, V.T.; Foster, S.J.; Renshaw, S.A. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell Microbiol. 2008, 11, 2312–2325. [Google Scholar] [CrossRef]
- White, H.S.; Scholl, E.A.; Klein, B.D.; Flynn, S.P.; Pruess, T.H.; Green, B.R.; Zhang, L.; Bulaj, B. Developing novel antiepileptic drugs: Characterization of NAX 5055, a systemically-active galanin analog, in epilepsy models. Neurotherapeutics 2009, 6, 372–380. [Google Scholar] [CrossRef]
- Bulaj, G.; Green, B.R.; Lee, H.K.; Robertson, C.R.; White, K.; Zhang, L.; Sochanska, M.; Flynn, S.P.; Scholl, E.A.; Pruess, T.H.; et al. Design, synthesis and characterization of high-affinity, systemically-active galanin analogs with potent anticonvulsant activities. J. Med. Chem. 2008, 51, 8038–8047. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Kaini, P.; Sander, J.D.; Joung, J.K.; Peterson, R.T.; Yeh, J.R. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS ONE. 2013, 8, e68708. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Benard, E.L.; van der Sar, A.M.; Ellett, F.; Lieschke, G.J.; Spaink, H.P.; Meijer, A.H. Infection of Zebrafish Embryos with Intracellular Bacterial Pathogens. J. Vis. Exp. 2012, 61, 3781. [Google Scholar] [CrossRef] [PubMed]
- Boldock, E.; Surewaard, B.G.J.; Shamarina, D.; Na, M.; Fei, Y.; Ali, A.; Williams, A.; Pollitt, E.J.G.; Szkuta, P.; Morris, P.; et al. Human skin commensals augment Staphylococcus aureus pathogenesis. Nat. Microbiol. 2018, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Stoop, E.J.; Schipper, T.; Rosendahl Huber, S.K.; Nezhinsky, A.E.; Verbeek, F.J.; Gurcha, S.S.; Besra, G.S.; Vandenbroucke-Grauls, C.M.; Bitter, W.; van der Sar, A.M. Zebrafish embryo screen for mycobacterial genes involved in the initiation of granuloma formation reveals a newly identified ESX-1 component. Dis. Model. Mech. 2011, 4, 526–536. [Google Scholar] [CrossRef]
- Rotman, J.; van Gils, W.; Butler, D.; Spaink, H.P.; Meijer, A.H. Rapid screening of innate immune gene expression in zebrafish using reverse transcription—Multiplex ligation-dependent probe amplification. BMC Res. Notes. 2011, 15, 196. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Veneman, W.J.; de Sonneville, J.; van der Kolk, K.J.; Ordas, A.; Al-Ars, Z.; Meijer, A.H.; Spaink, H.P. Analysis of RNAseq datasets from a comparative infectious disease zebrafish model using GeneTiles bioinformatics. Immunogenetics 2015, 67, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Grant, A. Leukocytes and neurogenic inflammation. Inflammopharmacology 2011, 9, 403–420. [Google Scholar] [CrossRef]
- Rauch, I.; Lundström, L.; Hell, M.; Sperl, W.; Kofler, B. Galanin message-associated peptide suppresses growth and the budded-to-hyphal-form transition of Candida albicans. Antimicrob. Agents Chemother. 2007, 51, 4167–4170. [Google Scholar] [CrossRef]
- Locker, F.; Vidali, S.; Holub, B.S.; Stockinger, J.; Brunner, S.M.; Ebner, S.; Koller, A.; Trost, A.; Reitsamer, H.A.; Schwarzenbacher, D.; et al. Lack of Galanin Receptor 3 Alleviates Psoriasis by Altering Vascularization, Immune Cell Infiltration, and Cytokine Expression. J. Investig. Dermatol. 2018, 138, 199–207. [Google Scholar] [CrossRef]
- Fehr, A.; Eshwar, A.K.; Neuhauss, S.C.; Ruetten, M.; Lehner, A.; Vaughan, L. Evaluation of zebrafish as a model to study the pathogenesis of the opportunistic pathogen Cronobacter turicensis. Emerg. Microbes Infect. 2015, 5, e29. [Google Scholar]
- van der Sar, A.M.; Musters, R.J.; van Eeden, F.J.; Appelmelk, B.J.; Vandenbroucke-Grauls, C.M.; Bitter, W. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell Microbiol. 2003, 9, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.R. TNF-mediated inflammatory disease. J. Pathol. 2008, 214, 149–160. [Google Scholar] [CrossRef]
- Clay, H.; Volkman, H.E.; Ramakrishnan, L. Tumor necrosis factor signaling mediates resistance to mycobacteria by inhibiting bacterial growth and macrophage death. Immunity 2008, 29, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Tobin, D.M.; Vary, J.C., Jr.; Ray, J.P.; Walsh, G.S.; Dunstan, S.J.; Bang, N.D.; Hagge, D.A.; Khadge, S.; King, M.C.; Hawn, T.R.; et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 2010, 140, 717–730. [Google Scholar] [CrossRef]
- Roca, F.J.; Mulero, I.; López-Muñoz, A.; Sepulcre, M.P.; Renshaw, S.A.; Meseguer, J.; Mulero, V. Evolution of the inflammatory response in vertebrates: Fish TNF-alpha is a powerful activator of endothelial cells but hardly activates phagocytes. J. Immunol. 2008, 181, 5071–5081. [Google Scholar] [CrossRef]
- Di Paolo, N.C.; Shafiani, S.; Day, T.; Papayannopoulou, T.; Russell, D.W.; Iwakura, Y.; Sherman, D.; Urdahl, K.; Shayakhmetov, D.M. Interdependence between interleukin-1 and tumor necrosis factor regulates TNF-dependent control of Mycobacterium tuberculosis infection. Immunity 2015, 43, 1125–1136. [Google Scholar] [CrossRef]
- Oehlers, S.H.; Flores, M.V.; Hall, C.J.; O’Toole, R.; Swift, S.; Crosier, K.E.; Crosier, P.S. Expression of zebrafish cxcl8 (interleukin-8) and its receptors during development and in response to immune stimulation. Dev. Comp. Immunol. 2010, 34, 352–359. [Google Scholar] [CrossRef]
- van Soest, J.J.; Stockhammer, O.W.; Ordas, A.; Bloemberg, G.V.; Spaink, H.P.; Meijer, A.H. Comparison of static immersion and intravenous injection systems for exposure of zebrafish embryos to the natural pathogen Edwardsiella tarda. BMC Immunol. 2011, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Stockhammer, O.W.; Zakrzewska, A.; Hegedûs, Z.; Spaink, H.P.; Meijer, A.H. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J. Immunol. 2009, 182, 5641–5653. [Google Scholar] [CrossRef]
- de Oliveira, S.; Reyes-Aldasoro, C.C.; Candel, S.; Renshaw, S.A.; Mulero, V.; Calado, A. Cxcl8 (IL-8) mediates neutrophil recruitment and behavior in the zebrafish inflammatory response. J. Immunol. 2013, 190, 4349–4359. [Google Scholar] [CrossRef] [PubMed]
- Benard, E.L.; Rougeot, J.; Racz, P.I.; Spaink, H.P.; Meijer, A.H. Transcriptomic Approaches in the Zebrafish Model for Tuberculosis-Insights Into Host- and Pathogen-specific Determinants of the Innate Immune Response. Adv. Genet. 2016, 95, 217–251. [Google Scholar] [PubMed]
- Koller, A.; Brunner, S.M.; Bianchini, R.; Ramspacher, A.; Emberger, M.; Locker, F.; Schlager, S.; Kofler, B. Galanin is a potent modulator of cytokine and chemokine expression in human macrophages. Sci Rep. 2019, 9, 7237. [Google Scholar] [CrossRef]
- Locker, F.; Lang, A.A.; Koller, A.; Lang, R.; Bianchini, R.; Kofler, B. Galanin modulates human and murine neutrophil activation in vitro. Acta Physiol. 2015, 213, 595–602. [Google Scholar] [CrossRef]
- Schindler, C.W.; Darnell, J.E. Transcriptional responses to polypeptide ligands: The JAK-STAT pathway. Annu. Rev. Biochem. 1995, 64, 621–651. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Miyagi, C.; Carmany-Rampey, A.; Shimizu, T.; Fujii, R.; Schier, A.F.; Hirano, T. STAT3 controls cell movements during zebrafish gastrulation. Dev. Cell 2002, 2, 363–375. [Google Scholar] [CrossRef]
- Naylor, M.J.; Oakes, S.R.; Gardiner-Garden, M.; Harris, J.; Blazek, K.; Ho, T.W.; Li, F.C.; Wynick, D.; Walker, A.M.; Ormandy, C.J. Transcriptional Changes Underlying the Secretory Activation Phase of Mammary Gland Development. Mol. Endocrinol. 2005, 19, 1868–1883. [Google Scholar] [CrossRef]
- Tiwari, B.M.; Kannan, N.; Vemu, L.; Raghunand, T.R. The Mycobacterium tuberculosis PE proteins Rv0285 and Rv1386 modulate innate immunity and mediate bacillary survival in macrophages. PLoS ONE 2012, 12, e51686. [Google Scholar] [CrossRef] [PubMed]
| KEGG-Pathways | |||
|---|---|---|---|
| UPREGULATED | |||
| gal+/+ | gal−/− | ||
| TERM | COUNT | TERM | COUNT |
| Cytokine-cytokine receptor interaction | 18 | Protein processing in endoplasmic reticulum | 11 |
| Jak-STAT signaling pathway | 13 | ||
| Herpes simplex infection | 12 | ||
| Cell adhesion molecules (CAMs) | 11 | ||
| FoxO signaling pathway | 11 | ||
| p53 signaling pathway | 10 | ||
| Toll-like receptor signaling pathway | 10 | ||
| Tight junction | 8 | ||
| Insulin resistance | 8 | ||
| Apoptosis | 7 | ||
| Adipocytokine signaling pathway | 7 | ||
| RIG-I-like receptor signaling pathway | 6 | ||
| Cytosolic DNA-sensing pathway | 5 | ||
| Arachidonic acid metabolism | 5 | ||
| Steroid biosynthesis | 4 | ||
| DOWNREGULATED | |||
| Cell cycle | 12 | MAPK signaling pathway | 10 |
| Purine metabolism | 11 | Vascular smooth muscle contraction | 7 |
| Focal adhesion | 11 | Melanogenesis | 6 |
| ECM-receptor interaction | 10 | Cytokine-cytokine receptor interaction | 6 |
| Melanogenesis | 8 | Jak-STAT signaling pathway | 5 |
| DNA replication | 7 | Glycolysis/Gluconeogenesis | 4 |
| Pyrimidine metabolism | 6 | Steroid biosynthesis | 3 |
| Tyrosine metabolism | 5 | Galactose metabolism | 3 |
| Phototransduction | 4 | ||
| Retinol metabolism | 4 | ||
| Glutathione metabolism | 4 | ||
| Caffeine metabolism | 2 | ||
| GO: Biological processes | |||
| UPREGULATED | |||
| gal+/+ | gal−/− | ||
| TERM | COUNT | TERM | COUNT |
| Proteolysis | 46 | Immune response | 9 |
| Immune response | 30 | Cell redox homeostasis | 5 |
| Oxidation-reduction process | 27 | Protein folding | 4 |
| Inflammatory response | 18 | Inflammatory response | 4 |
| Regulation of cell proliferation | 15 | Response to endoplasmic reticulum stress | 3 |
| Cell adhesion | 15 | Gene silencing by RNA | 3 |
| Regulation of apoptotic process | 14 | Polyadenylation-dependent snorna 3′-end processing | 3 |
| Protein ubiquitination | 12 | CUT catabolic process | 3 |
| Innate immune response | 11 | Response to bacterium | 3 |
| Chemotaxis | 8 | Nuclear polyadenylation-dependent rrna catabolic process | 2 |
| Response to lipopolysaccharide | 8 | DNA methylation involved in gamete generation | 2 |
| Blood coagulation | 7 | Pirna metabolic process | 2 |
| Cytokine-mediated signaling pathway | 7 | Exonucleolytic nuclear-transcribed mrna catabolic process involved in deadenylation-dependent decay | 2 |
| Transmembrane receptor protein tyrosine kinase signaling pathway | 7 | U4 snrna 3′-end processing | 2 |
| Cell-matrix adhesion | 6 | Nuclear-transcribed mrna catabolic process, exonucleolytic, 3′-5′ | 2 |
| Response to bacterium | 6 | Exonucleolytic trimming to generate mature 3′-end of 5.8S rrna from tricistronic rrna transcript (SSU-rrna, 5.8S rrna, LSU-rrna) | 2 |
| Peptidyl-tyrosine autophosphorylation | 6 | Nuclear retention of pre-mrna with aberrant 3′-ends at the site of transcription | 2 |
| Cell chemotaxis | 6 | Glycogen metabolic process | 2 |
| Positive regulation of apoptotic process | 6 | Adrenal gland development | 2 |
| Positive regulation of MAPK cascade | 5 | Nuclear polyadenylation-dependent trna catabolic process | 2 |
| Regulation of cell growth | 5 | ||
| Epiboly involved in gastrulation with mouth forming second | 5 | ||
| Immune system process | 4 | ||
| Response to wounding | 4 | ||
| Circadian rhythm | 4 | ||
| Neutrophil chemotaxis | 4 | ||
| Regulation of cell migration | 4 | ||
| Definitive hemopoiesis | 4 | ||
| Positive regulation of hematopoietic progenitor cell differentiation | 3 | ||
| Activation of MAPK activity | 3 | ||
| Regulation of hematopoietic progenitor cell differentiation | 3 | ||
| Response to glucose | 3 | ||
| Response to cytokine | 3 | ||
| Intracellular sequestering of iron ion | 3 | ||
| Response to heat | 3 | ||
| Iron ion transport | 3 | ||
| Peptide cross-linking | 3 | ||
| Regulation of inflammatory response | 3 | ||
| Response to cadmium ion | 3 | ||
| Macrophage differentiation | 3 | ||
| Response to mechanical stimulus | 3 | ||
| Activation of innate immune response | 2 | ||
| Regulation of cysteine-type endopeptidase activity involved in apoptotic process | 2 | ||
| Plasminogen activation | 2 | ||
| DOWNREGULATED | |||
| Transport | 35 | Regulation of transcription, DNA-templated | 32 |
| Transmembrane transport | 15 | Transport | 30 |
| Visual perception | 14 | Oxidation-reduction process | 18 |
| Response to stimulus | 13 | Transmembrane transport | 17 |
| DNA replication | 10 | Regulation of cell growth | 6 |
| Cellular response to light stimulus | 8 | Steroid hormone mediated signaling pathway | 6 |
| Phototransduction | 8 | Potassium ion transmembrane transport | 5 |
| Protein-chromophore linkage | 7 | Single organismal cell-cell adhesion | 4 |
| DNA replication initiation | 6 | Neurotransmitter transport | 4 |
| Retina development in camera-type eye | 6 | One-carbon metabolic process | 4 |
| Melanosome organization | 5 | Heart contraction | 4 |
| Microtubule-based process | 5 | Muscle contraction | 3 |
| Developmental pigmentation | 5 | Carbohydrate transport | 3 |
| Erythrocyte differentiation | 4 | Glucose 6-phosphate metabolic process | 2 |
| Embryonic hemopoiesis | 4 | Sodium-dependent phosphate transport | 2 |
| Oxygen transport | 4 | Mitotic G1 DNA damage checkpoint | 2 |
| Mitotic cell cycle | 4 | ||
| Nucleobase-containing compound metabolic process | 4 | ||
| Positive regulation of cell proliferation | 4 | ||
| Spindle assembly | 3 | ||
| Skeletal muscle contraction | 3 | ||
| Response to light stimulus | 3 | ||
| Error-free translation synthesis | 2 | ||
| DNA strand elongation | 2 | ||
| Regulation of hematopoietic stem cell differentiation | 2 | ||
| Melanin biosynthetic process | 2 | ||
| Detection of chemical stimulus involved in sensory perception of bitter taste | 2 | ||
| Negative regulation of cysteine-type endopeptidase activity | 2 | ||
| Regulation of G2/M transition of mitotic cell cycle | 2 | ||
| Immune Response | |
|---|---|
| gal+/+ | gal−/− |
| CD74 molecule, major histocompatibility complex, class II invariant chain a(cd74a) | Fas ligand (TNF superfamily, member 6)(faslg) |
| CX chemokine ligand 34b, duplicate 11(cxl34b.11) | chemokine (C-C motif) ligand 19a, tandem duplicate 2(ccl19a.2) |
| Fas cell surface death receptor(fas) | chemokine CCL-C17a(LOC100002392) |
| chemokine (C-C motif) ligand 19a, tandem duplicate 1(ccl19a.1) | complement component 7a(c7a) |
| chemokine (C-C motif) ligand 36, duplicate 1(ccl36.1) | interleukin 10(il10) |
| chemokine (C-X-C motif) ligand 11, duplicate 1(cxcl11.1) | si:rp71-1i20.2(si:rp71-1i20.2) |
| chemokine (C-X-C motif) ligand 11, duplicate 7(cxcl11.7) | tumor necrosis factor (ligand) superfamily, member 10(tnfsf10) |
| chemokine (C-X-C motif) ligand 19(cxcl19) | uncharacterized LOC101882211(LOC101882211) |
| chemokine (C-X-C motif) ligand 8b, duplicate 1(cxcl8b.1) | zmp:0000000652(zmp:0000000652) |
| complement factor properdin(cfp) | |
| interleukin 4(il4) | |
| lymphocyte cytosolic protein 2a(lcp2a) | |
| negative regulator of reactive oxygen species(nrros) | |
| nuclear factor, interleukin 3 regulated(nfil3) | |
| proteoglycan 4b(prg4b) | |
| serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1(serpine1) | |
| si:ch211-137i24.12(si:ch211-137i24.12) | |
| si:ch73-27e22.6(si:ch73-27e22.6) | |
| si:ch73-27e22.7(si:ch73-27e22.7) | |
| si:ch73-44m9.1(si:ch73-44m9.1) | |
| si:dkey-19a16.4(si:dkey-19a16.4) | |
| si:dkey-253d23.4(si:dkey-253d23.4) | |
| si:rp71-36a1.1(si:rp71-36a1.1) | |
| titin-like(LOC101883412) | |
| tumor necrosis factor (ligand) superfamily, member 11(tnfsf11) | |
| tumor necrosis factor receptor superfamily, member 1B(tnfrsf1b) | |
| tumor necrosis factor, alpha-induced protein 3(tnfaip3) | |
| uncharacterized LOC101883645(LOC101883645) | |
| uncharacterized LOC101885444(LOC101885444) | |
| zgc:153759(zgc:153759) | |
| Inflammatory Response | |
| C5a anaphylatoxin chemotactic receptor(c5ar1) | chemokine (C-C motif) ligand 19a, tandem duplicate 2(ccl19a.2) |
| CD40 molecule, TNF receptor superfamily member 5(cd40) | interleukin 10(il10) |
| E74-like factor 3 (ets domain transcription factor, epithelial-specific) (elf3) | toll-like receptor 22(tlr22) |
| Fas cell surface death receptor(fas) | toll-like receptor 5b(tlr5b) |
| chemokine (C-C motif) ligand 19a, tandem duplicate 1(ccl19a.1) | |
| chemokine (C-X-C motif) ligand 11, duplicate 1(cxcl11.1) | |
| chemokine (C-X-C motif) ligand 11, duplicate 7(cxcl11.7) | |
| chemokine (C-X-C motif) ligand 19(cxcl19) | |
| colony stimulating factor 1 receptor, a(csf1ra) | |
| myeloid differentiation primary response 88(myd88) | |
| negative regulator of reactive oxygen species(nrros) | |
| nitric oxide synthase 2a, inducible(nos2a) | |
| prostaglandin E receptor 2b (subtype EP2)(ptger2b) | |
| serum/glucocorticoid regulated kinase 1(sgk1) | |
| toll-like receptor 5a(tlr5a) | |
| tumor necrosis factor receptor superfamily, member 1B(tnfrsf1b) | |
| v-rel avian reticuloendotheliosis viral oncogene homolog(rel) | |
| zgc:153759(zgc:153759) | |
| Response to Bacterium | |
| CCAAT/enhancer binding protein (C/EBP), beta(cebpb) | matrix metallopeptidase 9(mmp9) |
| coagulation factor V(f5) | toll-like receptor 22(tlr22) |
| hepcidin antimicrobial peptide(hamp) | transferrin-a(tfa) |
| immunoresponsive gene 1, like(irg1l) | |
| interleukin-1 receptor-associated kinase 4(irak4) | |
| leukocyte cell-derived chemotaxin 2 like(lect2l) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowik, N.; Prajsnar, T.K.; Przyborowska, A.; Rakus, K.; Sienkiewicz, W.; Spaink, H.P.; Podlasz, P. The Role of Galanin during Bacterial Infection in Larval Zebrafish. Cells 2021, 10, 2011. https://doi.org/10.3390/cells10082011
Nowik N, Prajsnar TK, Przyborowska A, Rakus K, Sienkiewicz W, Spaink HP, Podlasz P. The Role of Galanin during Bacterial Infection in Larval Zebrafish. Cells. 2021; 10(8):2011. https://doi.org/10.3390/cells10082011
Chicago/Turabian StyleNowik, Natalia, Tomasz K. Prajsnar, Anna Przyborowska, Krzysztof Rakus, Waldemar Sienkiewicz, Herman P. Spaink, and Piotr Podlasz. 2021. "The Role of Galanin during Bacterial Infection in Larval Zebrafish" Cells 10, no. 8: 2011. https://doi.org/10.3390/cells10082011
APA StyleNowik, N., Prajsnar, T. K., Przyborowska, A., Rakus, K., Sienkiewicz, W., Spaink, H. P., & Podlasz, P. (2021). The Role of Galanin during Bacterial Infection in Larval Zebrafish. Cells, 10(8), 2011. https://doi.org/10.3390/cells10082011







