Renalase: A Multi-Functional Signaling Molecule with Roles in Gastrointestinal Disease
Abstract
1. Introduction
2. Renalase Has Potential Roles in Disease: Findings from Preclinical Studies
2.1. Effects on Acute Injury
2.2. Effects on Cancer
2.3. The Cell Growth and Anti-Inflammatory Activities of RNLS Localize to a Specific Site
3. RNLS Forms, Conservation, Structure, and Expression
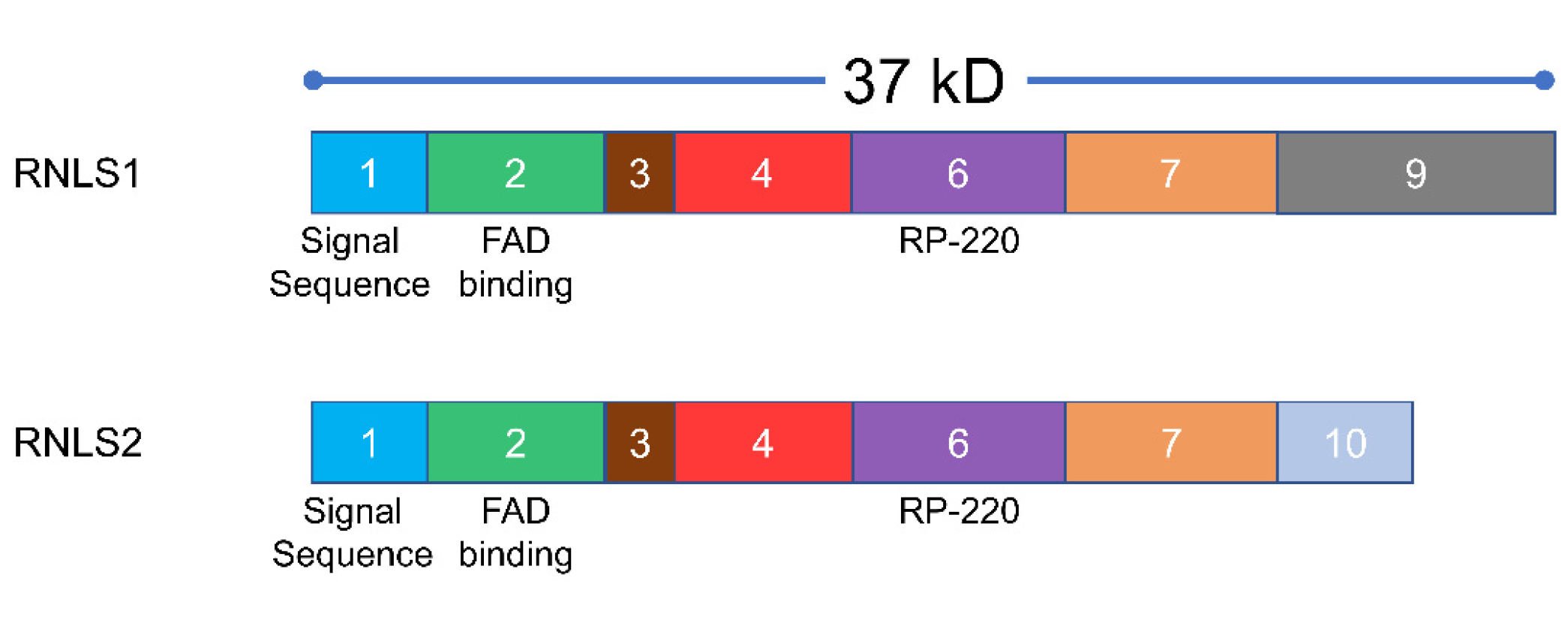
3.1. RNLS Isoforms
3.2. RNLS Sequence Conservation
3.3. RNLS Structure
4. RNLS Cell Biology
4.1. Cellular Distribution/Secretion
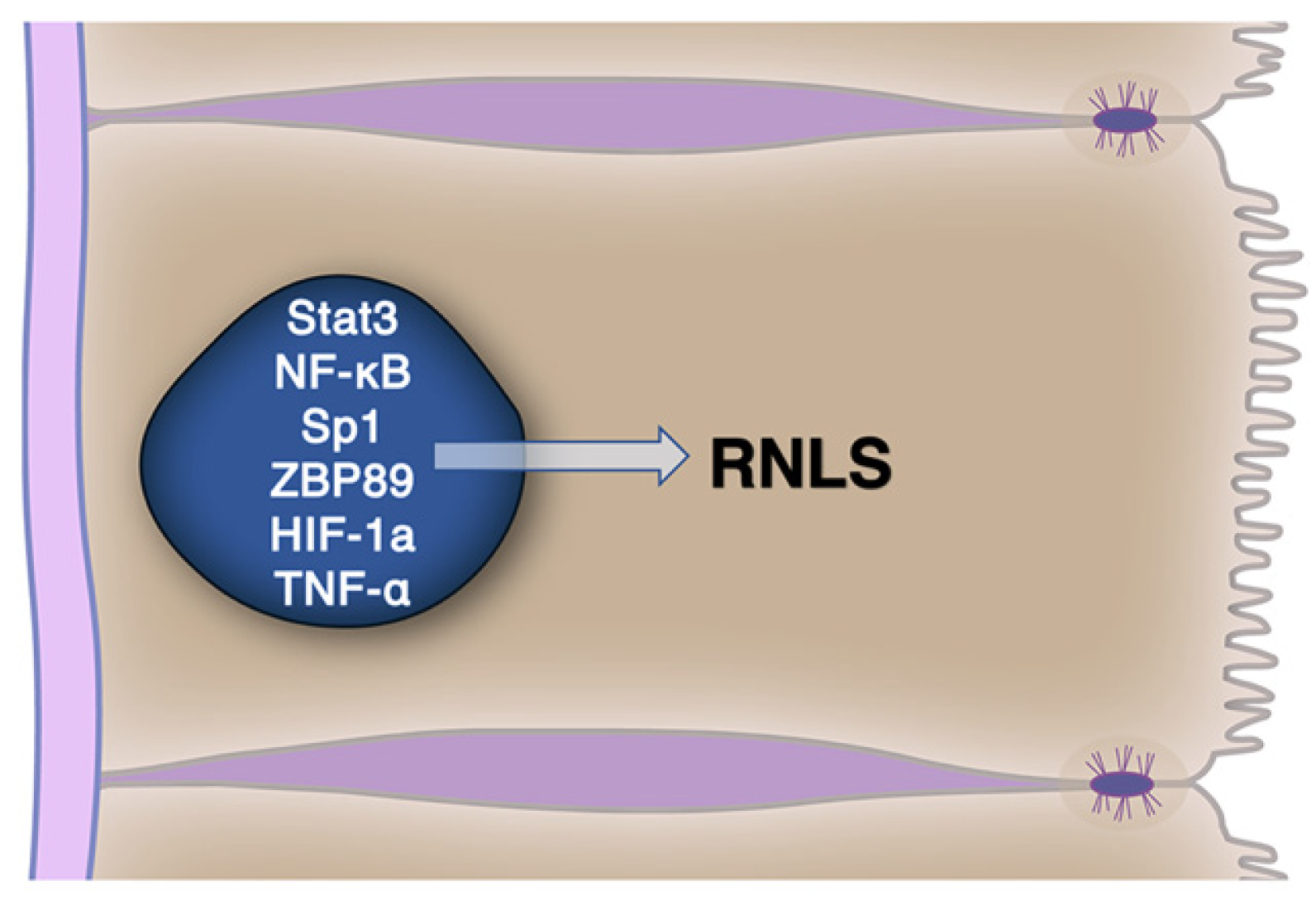
4.2. RNLS Cellular Expression Is Regulated
5. Renalase Signaling
5.1. Enzymatic Activities of RNLS
5.1.1. Enzymatic Activities and NAD
5.1.2. Other Potential Enzymatic Activities
6. Signaling by Extracellular RNLS
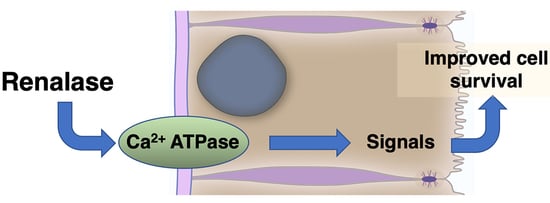
6.1. The Growth and Modulatory Effects of RNLS Are Found in a Specific RNLS Site
6.2. Extracellular RNLS Stimulates Intracellular Signaling
6.2.1. Identification of a Plasma Membrane Receptor for RNLS
6.2.2. RNLS Stimulates Distinct Intracellular Signals
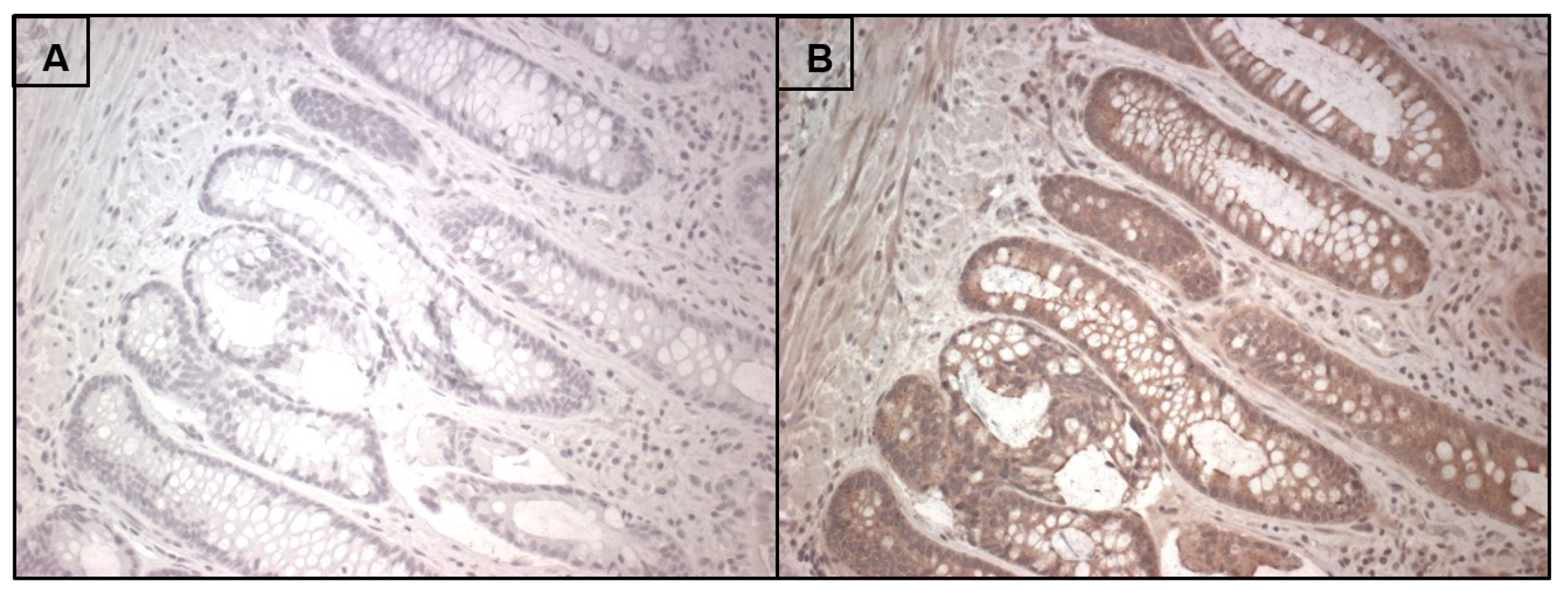
7. RNLS Presence in Human Intestine
8. Cell and Tissue-Specific RNLS Responses
8.1. Macrophages
8.2. Others
8.3. Cancer
9. Conclusions/Future Studies
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Tissue Specimens
Appendix A.2. Immunohistochemistry
References
- Xu, J.; Li, G.; Wang, P.; Velazquez, H.; Yao, X.; Li, Y.; Wu, Y.; Peixoto, A.; Crowley, S.; Desir, G.V. Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure. J. Clin. Investig. 2005, 115, 1275–1280. [Google Scholar] [CrossRef]
- Desir, G.V. Renalase is a novel renal hormone that regulates cardiovascular function. J. Am. Soc. Hypertens. 2007, 1, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Xu, J.; Wang, P.; Velazquez, H.; Li, Y.; Wu, Y.; Desir, G.V. Catecholamines regulate the activity, secretion, and synthesis of renalase. Circulation 2008, 117, 1277–1282. [Google Scholar] [CrossRef] [PubMed]
- Desir, G.V. Regulation of blood pressure and cardiovascular function by renalase. Kidney Int. 2009, 76, 366–370. [Google Scholar] [CrossRef]
- Lee, H.T.; Kim, J.Y.; Kim, M.; Wang, P.; Tang, L.; Baroni, S.; D’Agati, V.D.; Desir, G.V. Renalase protects against ischemic AKI. J. Am. Soc. Nephrol. JASN 2013, 24, 445–455. [Google Scholar] [CrossRef]
- Gorelick, F.S.; Lerch, M.M. Do Animal Models of Acute Pancreatitis Reproduce Human Disease? Cell Mol. Gastroenterol. Hepatol. 2017, 4, 251–262. [Google Scholar] [CrossRef]
- Kolodecik, T.R.; Reed, A.M.; Date, K.; Shugrue, C.A.; Patel, V.; Chung, S.L.; Desir, G.V.; Gorelick, F.S. The serum protein renalase reduces injury in experimental pancreatitis. J. Biol. Chem. 2017, 292, 21047–21059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Gu, J.; Guo, J.; Chen, K.; Li, H.; Wang, J. Renalase Attenuates Mouse Fatty Liver Ischemia/Reperfusion Injury through Mitigating Oxidative Stress and Mitochondrial Damage via Activating SIRT1. Oxid. Med. Cell. Longev. 2019, 2019, 7534285. [Google Scholar] [CrossRef]
- Aoki, K.; Yanazawa, K.; Tokinoya, K.; Sugasawa, T.; Suzuki, T.; Yoshida, Y.; Nakano, T.; Omi, N.; Kawakami, Y.; Takekoshi, K. Renalase is localized to the small intestine crypt and expressed upon the activation of NF-kappaB p65 in mice model of fasting-induced oxidative stress. Life Sci. 2021, 267, 118904. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Hollander, L.; MacPherson, D.; Wang, L.; Velazquez, H.; Chang, J.; Safirstein, R.; Cha, C.; Gorelick, F.; Desir, G.V. Inhibition of renalase expression and signaling has antitumor activity in pancreatic cancer. Sci. Rep. 2016, 6, 22996. [Google Scholar] [CrossRef]
- Hollander, L.; Guo, X.; Velazquez, H.; Chang, J.; Safirstein, R.; Kluger, H.; Cha, C.; Desir, G.V. Renalase Expression by Melanoma and Tumor-Associated Macrophages Promotes Tumor Growth through a STAT3-Mediated Mechanism. Cancer Res. 2016, 76, 3884–3894. [Google Scholar] [CrossRef]
- Wang, L.; Velazquez, H.; Moeckel, G.; Chang, J.; Ham, A.; Lee, H.T.; Safirstein, R.; Desir, G.V. Renalase prevents AKI independent of amine oxidase activity. J. Am. Soc. Nephrol. 2014, 25, 1226–1235. [Google Scholar] [CrossRef]
- Baroni, S.; Milani, M.; Pandini, V.; Pavesi, G.; Horner, D.; Aliverti, A. Is renalase a novel player in catecholaminergic signaling? The mystery of the catalytic activity of an intriguing new flavoenzyme. Curr. Pharm. Des. 2013, 19, 2540–2551. [Google Scholar] [CrossRef] [PubMed]
- Fedchenko, V.I.; Kaloshin, A.A.; Mezhevikina, L.M.; Buneeva, O.A.; Medvedev, A.E. Construction of the coding sequence of the transcription variant 2 of the human Renalase gene and its expression in the prokaryotic system. Int. J. Mol. Sci. 2013, 14, 12764–12779. [Google Scholar] [CrossRef] [PubMed]
- Desir, G.V.; Wang, L.; Peixoto, A.J. Human renalase: A review of its biology, function, and implications for hypertension. J. Am. Soc. Hypertens. 2012, 6, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Desir, G.V.; Peixoto, A.J. Renalase in hypertension and kidney disease. Nephrol. Dial. Transplant. 2014, 29, 22–28. [Google Scholar] [CrossRef]
- Yuan, S.; Guo, J.H.; Du, J.B.; Lin, H.H. Phylogenetic analyses of plastid-originated proteins imply universal endosymbiosis in ancestors of animals and fungi. Z. Nat. C J. Biosci. 2008, 63, 903–908. [Google Scholar] [CrossRef]
- Embley, T.M.; Martin, W. Eukaryotic evolution, changes and challenges. Nature 2006, 440, 623–630. [Google Scholar] [CrossRef]
- Yuan, S.; Sun, X.; Mu, L.C.; Lei, T.; Liu, W.J.; Wang, J.H.; Du, J.B.; Lin, H.H. Nuclear-localized plastid DNA fragments in protozoa, metazoa and fungi. Z. Nat. C J. Biosci. 2007, 62, 123–132. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef]
- Fedchenko, V.I.; Buneeva, O.A.; Kopylov, A.T.; Veselovsky, A.V.; Zgoda, V.G.; Medvedev, A.E. Human urinary renalase lacks the N-terminal signal peptide crucial for accommodation of its FAD cofactor. Int. J. Biol. Macromol. 2015, 78, 347–353. [Google Scholar] [CrossRef]
- Fedchenko, V.; Kopylov, A.; Kozlova, N.; Buneeva, O.; Kaloshin, A.; Zgoda, V.; Medvedev, A. Renalase Secreted by Human Kidney HEK293T Cells Lacks its N-Terminal Peptide: Implications for Putative Mechanisms of Renalase Action. Kidney Blood Press. Res. 2016, 41, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Dailey, T.A.; Dailey, H.A. Identification of an FAD superfamily containing protoporphyrinogen oxidases, monoamine oxidases, and phytoene desaturase. Expression and characterization of phytoene desaturase of Myxococcus xanthus. J. Biol. Chem. 1998, 273, 13658–13662. [Google Scholar] [CrossRef] [PubMed]
- Milani, M.; Ciriello, F.; Baroni, S.; Pandini, V.; Canevari, G.; Bolognesi, M.; Aliverti, A. FAD-binding site and NADP reactivity in human renalase: A new enzyme involved in blood pressure regulation. J. Mol. Biol. 2011, 411, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Pandini, V.; Ciriello, F.; Tedeschi, G.; Rossoni, G.; Zanetti, G.; Aliverti, A. Synthesis of human renalase1 in Escherichia coli and its purification as a FAD-containing holoprotein. Protein Expr. Purif. 2010, 72, 244–253. [Google Scholar] [CrossRef]
- Binda, C.; Hubalek, F.; Li, M.; Castagnoli, N.; Edmondson, D.E.; Mattevi, A. Structure of the human mitochondrial monoamine oxidase B: New chemical implications for neuroprotectant drug design. Neurology 2006, 67, S5–S7. [Google Scholar] [CrossRef] [PubMed]
- Bruzzone, S.; Moreschi, I.; Guida, L.; Usai, C.; Zocchi, E.; De Flora, A. Extracellular NAD+ regulates intracellular calcium levels and induces activation of human granulocytes. Biochem. J. 2006, 393, 697–704. [Google Scholar] [CrossRef]
- Tararina, M.A.; Allen, K.N. Bioinformatic Analysis of the Flavin-Dependent Amine Oxidase Superfamily: Adaptations for Substrate Specificity and Catalytic Diversity. J. Mol. Biol. 2020, 432, 3269–3288. [Google Scholar] [CrossRef]
- Morrison, C.S.; Paskaleva, E.E.; Rios, M.A.; Beusse, T.R.; Blair, E.M.; Lin, L.Q.; Hu, J.R.; Gorby, A.H.; Dodds, D.R.; Armiger, W.B.; et al. Improved soluble expression and use of recombinant human renalase. PLoS ONE 2020, 15, e0242109. [Google Scholar] [CrossRef]
- Moran, G.R.; Hoag, M.R. The enzyme: Renalase. Arch. Biochem. Biophys. 2017, 632, 66–76. [Google Scholar] [CrossRef]
- Nagata, R.; Fujihashi, M.; Sato, T.; Atomi, H.; Miki, K. Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants. Nat. Commun. 2018, 9, 1765. [Google Scholar] [CrossRef] [PubMed]
- Quelhas-Santos, J.; Soares-Silva, I.; Fernandes-Cerqueira, C.; Simões-Silva, L.; Ferreira, I.; Carvalho, C.; Coentrão, L.; Vaz, R.; Sampaio-Maia, B.; Pestana, M. Plasma and urine renalase levels and activity during the recovery of renal function in kidney transplant recipients. Exp. Biol. Med. 2014, 239, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Malyszko, J.; Zbroch, E.; Malyszko, J.S.; Koc-Zorawska, E.; Mysliwiec, M. Renalase, A Novel Regulator of Blood Pressure, Is Predicted by Kidney Function in Renal Transplant Recipients. Transplant. Proc. 2011, 43, 3004–3007. [Google Scholar] [CrossRef] [PubMed]
- Zbroch, E.; Koc-Zorawska, E.; Malyszko, J.; Malyszko, J.; Mysliwiec, M. Circulating levels of renalase, norepinephrine, and dopamine in dialysis patients. Ren. Fail. 2013, 35, 673–679. [Google Scholar] [CrossRef]
- Wang, F.; Li, J.; Xing, T.; Xie, Y.; Wang, N. Serum renalase is related to catecholamine levels and renal function. Clin. Exp. Nephrol. 2015, 19, 92–98. [Google Scholar] [CrossRef]
- Rybi–Szumińska, A.; Michaluk-Skutnik, J.; Osipiuk-Remża, B.; Kossakowska, A.; Wasilewska, A. Normal values for urine renalase excretion in children. Pediatric Nephrol. 2014, 29, 2191–2195. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, P.J.; Gupta, V.; Sasi, B.K.; Kalyani, A.; Natarajan, B.; Khan, A.A.; Sahu, B.S.; Mahapatra, N.R. Transcriptional Regulation of the Novel Monoamine Oxidase Renalase: Crucial Roles of Transcription Factors Sp1, STAT3, and ZBP89. Biochemistry 2014, 53, 6878–6892. [Google Scholar] [CrossRef]
- Wang, F.; Yin, J.; Lu, Z.; Zhang, G.; Li, J.; Xing, T.; Zhuang, S.; Wang, N. Limb ischemic preconditioning protects against contrast-induced nephropathy via renalase. EBioMedicine 2016, 9, 356–365. [Google Scholar] [CrossRef]
- Du, M.; Huang, K.; Huang, D.; Yang, L.; Gao, L.; Wang, X.; Huang, D.; Li, X.; Wang, C.; Zhang, F.; et al. Renalase is a novel target gene of hypoxia-inducible factor-1 in protection against cardiac ischaemia–reperfusion injury. Cardiovasc. Res. 2014, 105, 182–191. [Google Scholar] [CrossRef]
- Beaupre, B.A.; Hoag, M.R.; Moran, G.R. Renalase does not catalyze the oxidation of catecholamines. Arch. Biochem. Biophys. 2015, 579, 62–66. [Google Scholar] [CrossRef]
- Beaupre, B.A.; Hoag, M.R.; Roman, J.; Försterling, F.H.; Moran, G.R. Metabolic Function for Human Renalase: Oxidation of Isomeric Forms of β-NAD(P)H that Are Inhibitory to Primary Metabolism. Biochemistry 2015, 54, 795–806. [Google Scholar] [CrossRef]
- Desir, G.V.; Tang, L.; Wang, P.; Li, G.; Sampaio-Maia, B.; Quelhas-Santos, J.; Pestana, M.; Velazquez, H. Renalase lowers ambulatory blood pressure by metabolizing circulating adrenaline. J. Am. Heart Assoc. 2012, 1, e002634. [Google Scholar] [CrossRef] [PubMed]
- Desir, G.V. The Author Replies. Kidney Int. 2011, 79, 1380–1381. [Google Scholar] [CrossRef][Green Version]
- Farzaneh-Far, R.; Desir, G.V.; Na, B.; Schiller, N.B.; Whooley, M.A. A functional polymorphism in renalase (Glu37Asp) is associated with cardiac hypertrophy, dysfunction, and ischemia: Data from the heart and soul study. PLoS ONE 2010, 5, e13496. [Google Scholar] [CrossRef] [PubMed]
- Beaupre, B.A.; Hoag, M.R.; Carmichael, B.R.; Moran, G.R. Kinetics and equilibria of the reductive and oxidative half-reactions of human renalase with alpha-NADPH. Biochemistry 2013, 52, 8929–8937. [Google Scholar] [CrossRef] [PubMed]
- Beaupre, B.A.; Carmichael, B.R.; Hoag, M.R.; Shah, D.D.; Moran, G.R. Renalase is an alpha-NAD(P)H oxidase/anomerase. J. Am. Chem. Soc. 2013, 135, 13980–13987. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, J.; Velazquez, H.; Wang, P.; Li, G.; Liu, D.; Sampaio-Maia, B.; Quelhas-Santos, J.; Russell, K.; Russell, R.; et al. Renalase deficiency aggravates ischemic myocardial damage. Kidney Int. 2011, 79, 853–860. [Google Scholar] [CrossRef]
- Cantó, C.; Menzies, K.J.; Auwerx, J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef]
- Boomsma, F.; Tipton, K.F. Renalase, a catecholamine-metabolising enzyme. J. Neural Transm. 2007, 114, 775–776. [Google Scholar] [CrossRef][Green Version]
- Wang, L.; Velazquez, H.; Chang, J.; Safirstein, R.; Desir, G.V. Identification of a receptor for extracellular renalase. PLoS ONE 2015, 10, e0122932. [Google Scholar] [CrossRef]
- Wang, Y.; Safirstein, R.; Velazquez, H.; Guo, X.-J.; Hollander, L.; Chang, J.; Chen, T.-M.; Mu, J.-J.; Desir, G.V. Extracellular renalase protects cells and organs by outside-in signalling. J. Cell. Mol. Med. 2017, 21, 1260–1265. [Google Scholar] [CrossRef]
- Potts, L.; Phillips, C.; Hwang, M.; Fulcher, S.; Choi, H. Rescue of human corneal epithelial cells after alkaline insult using renalase derived peptide, RP-220. Int. J. Ophthalmol. 2019, 12, 1667–1673. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Zhou, J.; Li, W.; Zhou, T.; Chen, Q.; Yang, F.; Wei, T. Plasma membrane calcium ATPase 4b inhibits nitric oxide generation through calcium-induced dynamic interaction with neuronal nitric oxide synthase. Protein Cell 2013, 4, 286–298. [Google Scholar] [CrossRef]
- Armesilla, A.L.; Williams, J.C.; Buch, M.H.; Pickard, A.; Emerson, M.; Cartwright, E.J.; Oceandy, D.; Vos, M.D.; Gillies, S.; Clark, G.J.; et al. Novel Functional Interaction between the Plasma Membrane Ca2+ Pump 4b and the Proapoptotic Tumor Suppressor Ras-associated Factor 1 (RASSF1)*. J. Biol. Chem. 2004, 279, 31318–31328. [Google Scholar] [CrossRef] [PubMed]
- Naffa, R.; Vogel, L.; Hegedus, L.; Paszty, K.; Toth, S.; Kelemen, K.; Singh, N.; Remenyi, A.; Kallay, E.; Cserepes, M.; et al. P38 MAPK Promotes Migration and Metastatic Activity of BRAF Mutant Melanoma Cells by Inducing Degradation of PMCA4b. Cells 2020, 9, 1209. [Google Scholar] [CrossRef] [PubMed]
- Gerasimenko, J.V.; Gerasimenko, O.V.; Petersen, O.H. The role of Ca2+ in the pathophysiology of pancreatitis. J. Physiol. 2014, 592, 269–280. [Google Scholar] [CrossRef]
- Tran, Q.-K.; VerMeer, M.; Burgard, M.A.; Hassan, A.B.; Giles, J. Hetero-oligomeric Complex between the G Protein-coupled Estrogen Receptor 1 and the Plasma Membrane Ca2+-ATPase 4b. J. Biol. Chem. 2015, 290, 13293–13307. [Google Scholar] [CrossRef]
- Tokinoya, K.; Sekine, N.; Aoki, K.; Ono, S.; Kuji, T.; Sugasawa, T.; Yoshida, Y.; Takekoshi, K. Effects of renalase deficiency on liver fibrosis markers in a nonalcoholic steatohepatitis mouse model. Mol. Med. Rep. 2021, 23, 210. [Google Scholar] [CrossRef]
- Kim, E.; DeMarco, S.J.; Marfatia, S.M.; Chishti, A.H.; Sheng, M.; Strehler, E.E. Plasma membrane Ca2+ ATPase isoform 4b binds to membrane-associated guanylate kinase (MAGUK) proteins via their PDZ (PSD-95/Dlg/ZO-1) domains. J. Biol. Chem. 1998, 273, 1591–1595. [Google Scholar] [CrossRef]
- Schuh, K.; Uldrijan, S.; Telkamp, M.; Rothlein, N.; Neyses, L. The plasmamembrane calmodulin-dependent calcium pump: A major regulator of nitric oxide synthase I. J. Cell Biol. 2001, 155, 201–205. [Google Scholar] [CrossRef]
- Schuh, K.; Quaschning, T.; Knauer, S.; Hu, K.; Kocak, S.; Roethlein, N.; Neyses, L. Regulation of vascular tone in animals overexpressing the sarcolemmal calcium pump. J. Biol. Chem. 2003, 278, 41246–41252. [Google Scholar] [CrossRef] [PubMed]
- DeMarco, S.J.; Strehler, E.E. Plasma membrane Ca2+-atpase isoforms 2b and 4b interact promiscuously and selectively with members of the membrane-associated guanylate kinase family of PDZ (PSD95/Dlg/ZO-1) domain-containing proteins. J. Biol. Chem. 2001, 276, 21594–21600. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fuentes, E.J. Emerging Themes in PDZ Domain Signaling: Structure, Function, and Inhibition. Int. Rev. Cell Mol. Biol. 2019, 343, 129–218. [Google Scholar] [PubMed]
- Williams, J.C.; Armesilla, A.L.; Mohamed, T.M.A.; Hagarty, C.L.; McIntyre, F.H.; Schomburg, S.; Zaki, A.O.; Oceandy, D.; Cartwright, E.J.; Buch, M.H.; et al. The Sarcolemmal Calcium Pump, α-1 Syntrophin, and Neuronal Nitric-oxide Synthase Are Parts of a Macromolecular Protein Complex*. J. Biol. Chem. 2006, 281, 23341–23348. [Google Scholar] [CrossRef]
- Kim, E.; Sheng, M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004, 5, 771–781. [Google Scholar] [CrossRef]
- Gonzalez de Valdivia, E.; Broselid, S.; Kahn, R.; Olde, B.; Leeb-Lundberg, L.M.F. G protein-coupled estrogen receptor 1 (GPER1)/GPR30 increases ERK1/2 activity through PDZ motif-dependent and -independent mechanisms. J. Biol. Chem. 2017, 292, 9932–9943. [Google Scholar] [CrossRef]
- Terry, L.E.; VerMeer, M.; Giles, J.; Tran, Q.K. Suppression of store-operated Ca(2+) entry by activation of GPER: Contribution to a clamping effect on endothelial Ca(2+) signaling. Biochem. J. 2017, 474, 3627–3642. [Google Scholar] [CrossRef]
- Hegedüs, L.; Padányi, R.; Molnár, J.; Pászty, K.; Varga, K.; Kenessey, I.; Sárközy, E.; Wolf, M.; Grusch, M.; Hegyi, Z.; et al. Histone Deacetylase Inhibitor Treatment Increases the Expression of the Plasma Membrane Ca(2+) Pump PMCA4b and Inhibits the Migration of Melanoma Cells Independent of ERK. Front. Oncol. 2017, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Ribiczey, P.; Tordai, A.; Andrikovics, H.; Filoteo, A.G.; Penniston, J.T.; Enouf, J.; Enyedi, A.; Papp, B.; Kovács, T. Isoform-specific up-regulation of plasma membrane Ca2+ATPase expression during colon and gastric cancer cell differentiation. Cell Calcium 2007, 42, 590–605. [Google Scholar] [CrossRef]
- El-Yazbi, A.F.; Cho, W.J.; Schulz, R.; Daniel, E.E. Calcium extrusion by plasma membrane calcium pump is impaired in caveolin-1 knockout mouse small intestine. Eur. J. Pharm. 2008, 591, 80–87. [Google Scholar] [CrossRef]
- Guo, X.; Wang, L.; Velazquez, H.; Safirstein, R.; Desir, G.V. Renalase: Its role as a cytokine, and an update on its association with type 1 diabetes and ischemic stroke. Curr. Opin. Nephrol. Hypertens. 2014, 23, 513–518. [Google Scholar] [CrossRef]
- Cai, E.P.; Ishikawa, Y.; Zhang, W.; Leite, N.C.; Li, J.; Hou, S.; Kiaf, B.; Hollister-Lock, J.; Yilmaz, N.K.; Schiffer, C.A.; et al. Genome-scale in vivo CRISPR screen identifies RNLS as a target for beta cell protection in type 1 diabetes. Nat. Metab. 2020, 2, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.; Chakraborty, S.; Paul Chowdhury, B.; Banerjee, S.; Halder, K.; Majumder, S.; Majumdar, S.; Sen, P.C. Regulation of PKC mediated signaling by calcium during visceral leishmaniasis. PLoS ONE 2014, 9, e110843. [Google Scholar]
- Akkoc, R.F.; Aydin, S.; Goksu, M.; Ozcan Yildirim, S.; Eroksuz, Y.; Ogeturk, M.; Ugur, K.; Dagli, A.F.; Yakar, B.; Sahin, I.; et al. Can renalase be a novel candidate biomarker for distinguishing renal tumors? Biotech. Histochem. 2020, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Han, P.; Wang, J.; Sun, H.; Shao, M. Renalase overexpression in ER-positive breast cancer. Int. J. Clin. Exp. Pathol. 2018, 11, 1297–1307. [Google Scholar] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pointer, T.C.; Gorelick, F.S.; Desir, G.V. Renalase: A Multi-Functional Signaling Molecule with Roles in Gastrointestinal Disease. Cells 2021, 10, 2006. https://doi.org/10.3390/cells10082006
Pointer TC, Gorelick FS, Desir GV. Renalase: A Multi-Functional Signaling Molecule with Roles in Gastrointestinal Disease. Cells. 2021; 10(8):2006. https://doi.org/10.3390/cells10082006
Chicago/Turabian StylePointer, Thomas C., Fred S. Gorelick, and Gary V. Desir. 2021. "Renalase: A Multi-Functional Signaling Molecule with Roles in Gastrointestinal Disease" Cells 10, no. 8: 2006. https://doi.org/10.3390/cells10082006
APA StylePointer, T. C., Gorelick, F. S., & Desir, G. V. (2021). Renalase: A Multi-Functional Signaling Molecule with Roles in Gastrointestinal Disease. Cells, 10(8), 2006. https://doi.org/10.3390/cells10082006