PopulusPtERF85 Balances Xylem Cell Expansion and Secondary Cell Wall Formation in Hybrid Aspen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Cloning and Generation of Transgenic Trees
2.3. Histochemical Staining
2.4. Wood Density Measurements
2.5. Transmission Electron Microscopy
2.6. Quantification of Xylem Cells, Fiber Diameter and Cell Wall Thickness
2.7. RNA Extraction
2.8. RNA-Seq Sample Preparation and Statistical Analysis
2.9. Validation of RNA-Seq Results by qPCR
2.10. Bioinformatic and Statistical Analysis
3. Results
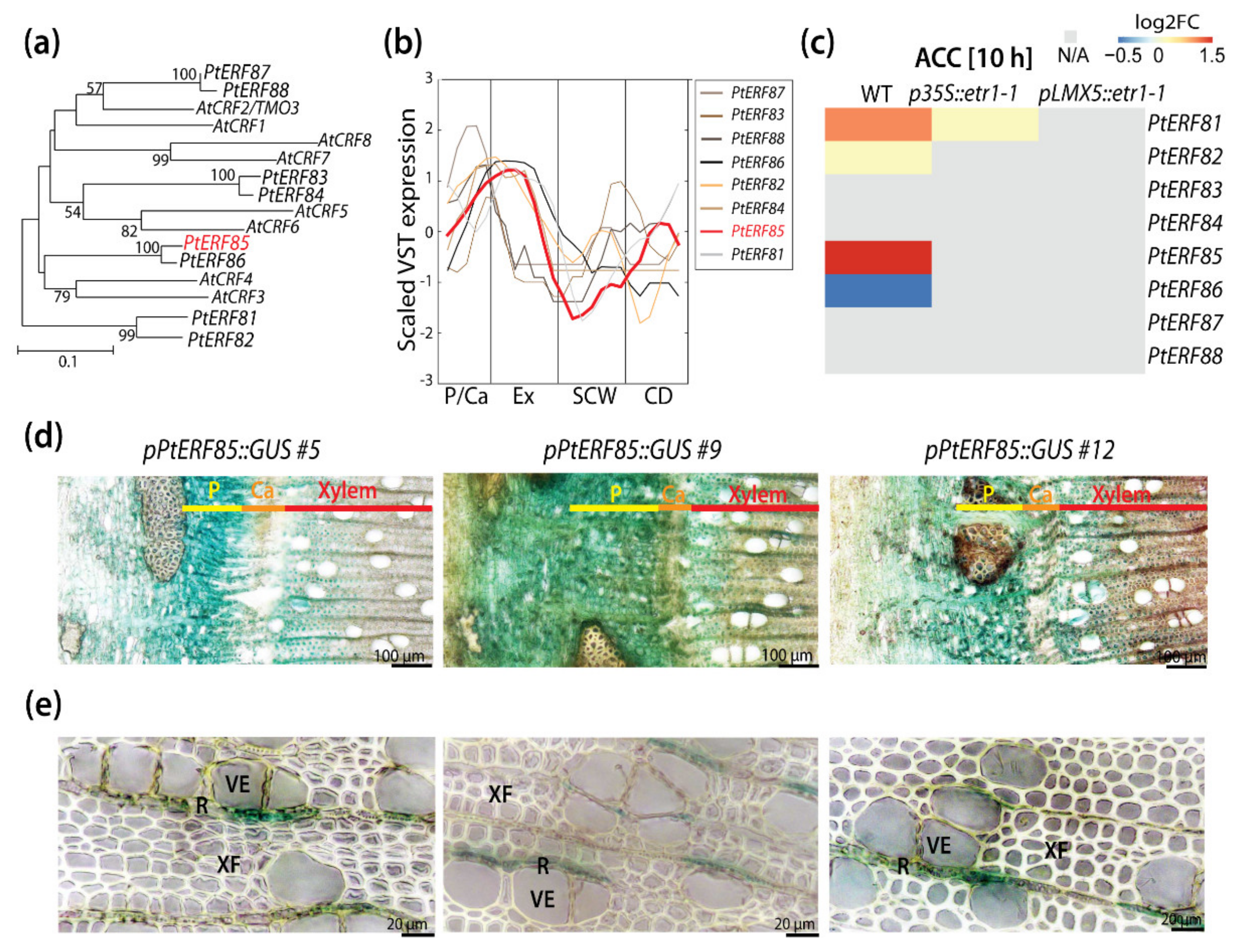
3.1. PtERF85 Is Expressed in the Cambium and Expanding Xylem
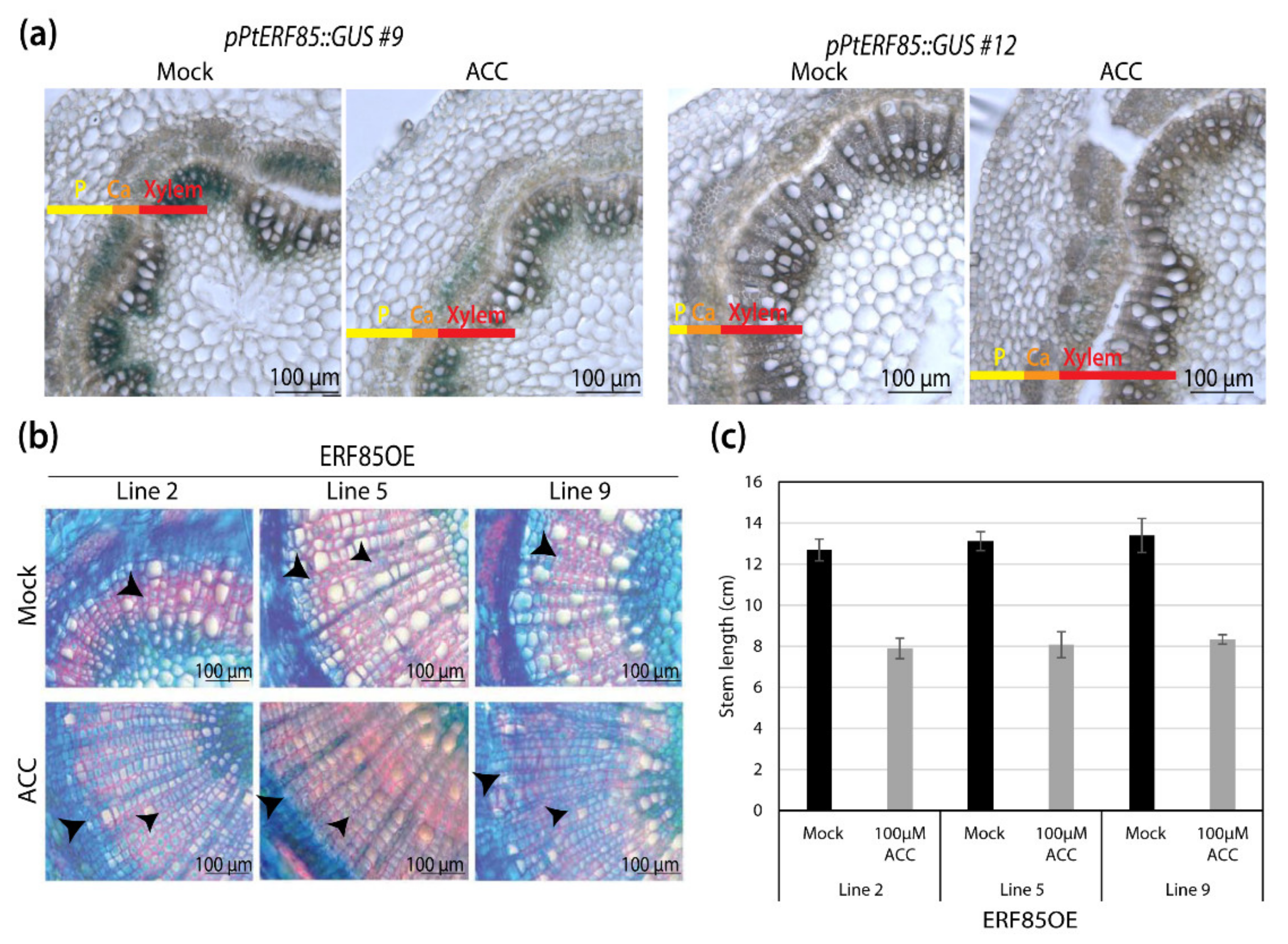
3.2. Ectopic PtERF85 Induction Did Not Change Ethylene Response in Hybrid Aspen Wood
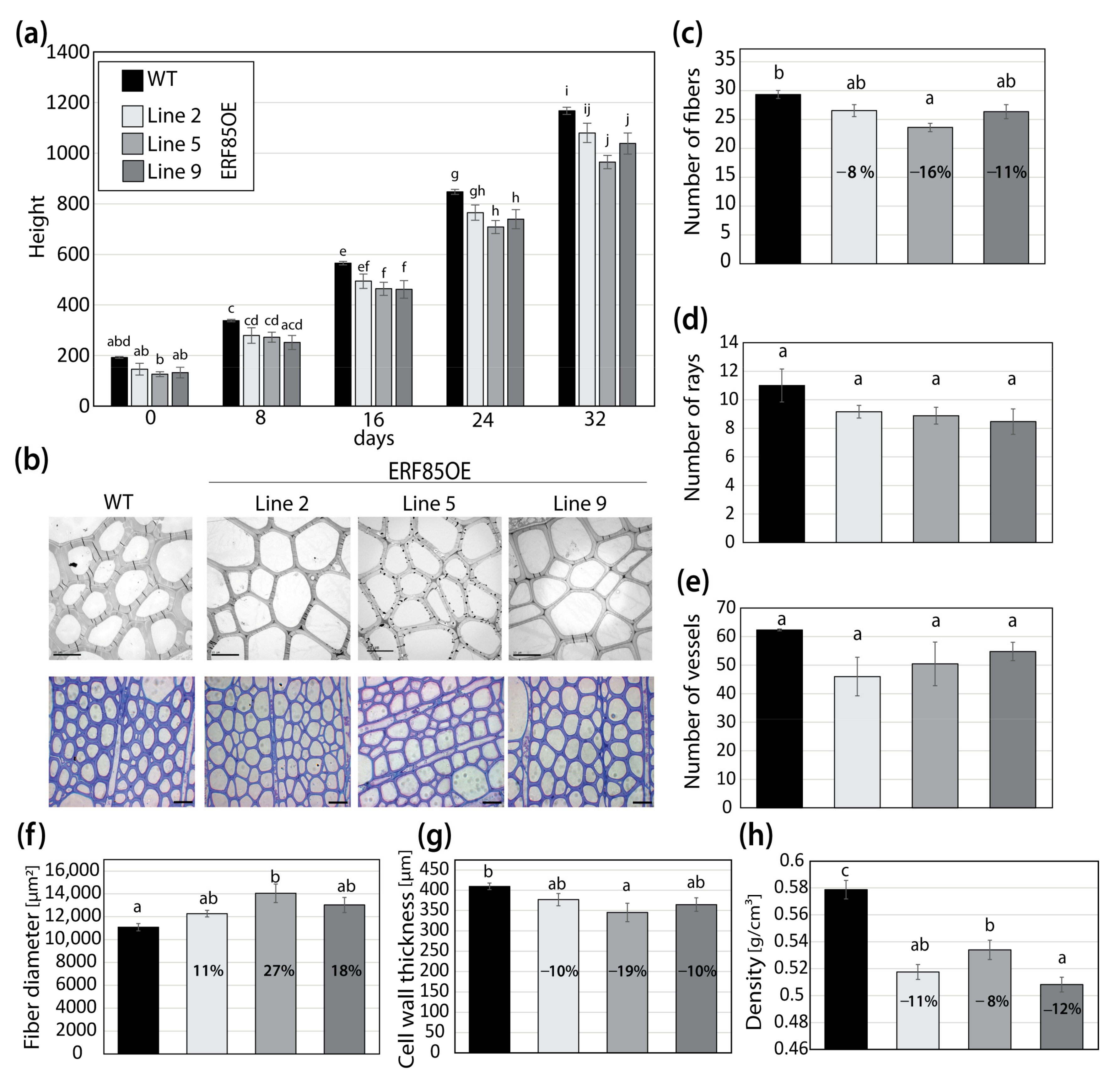
3.3. ERF85OE Lines Showed Increased Xylem Fiber Diameter
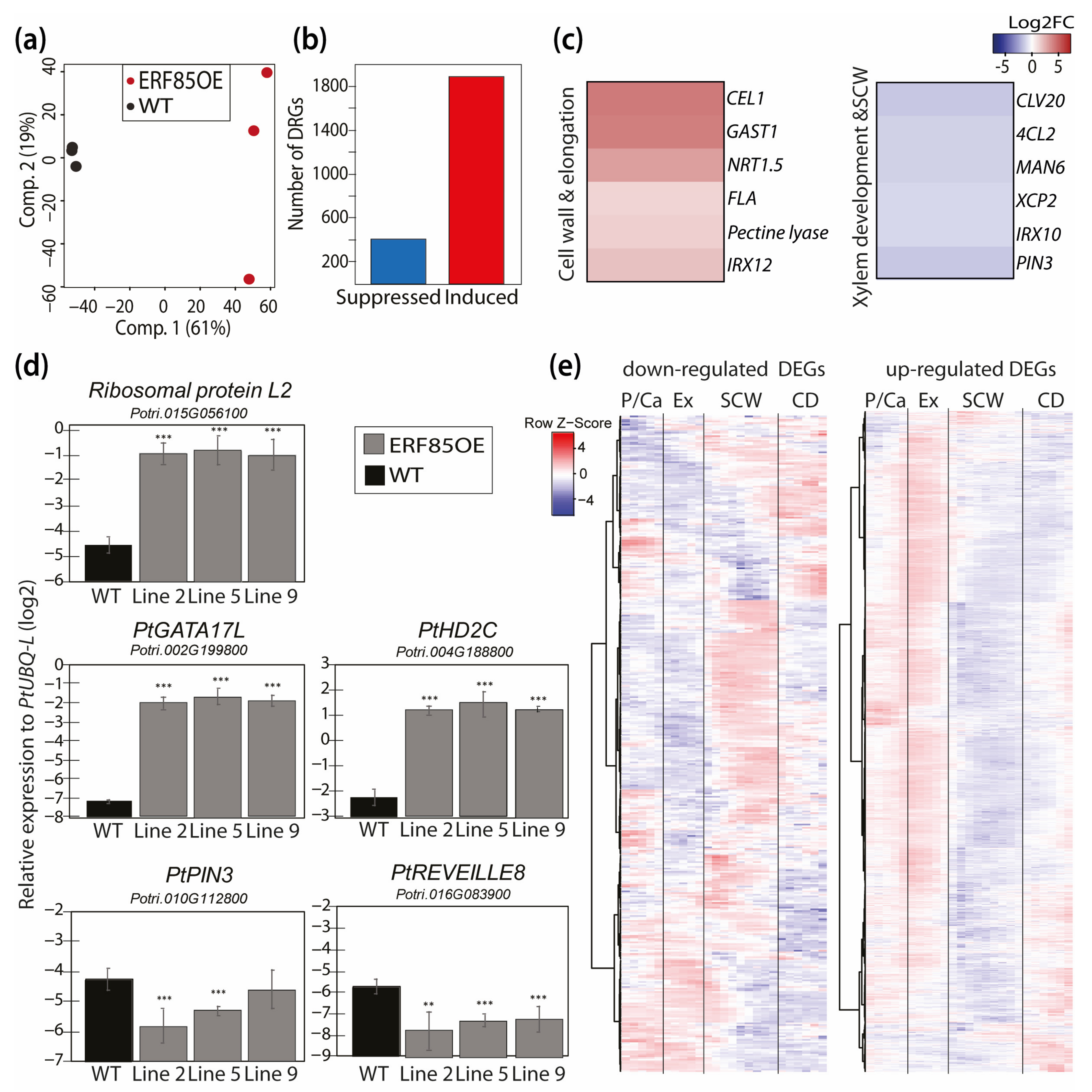
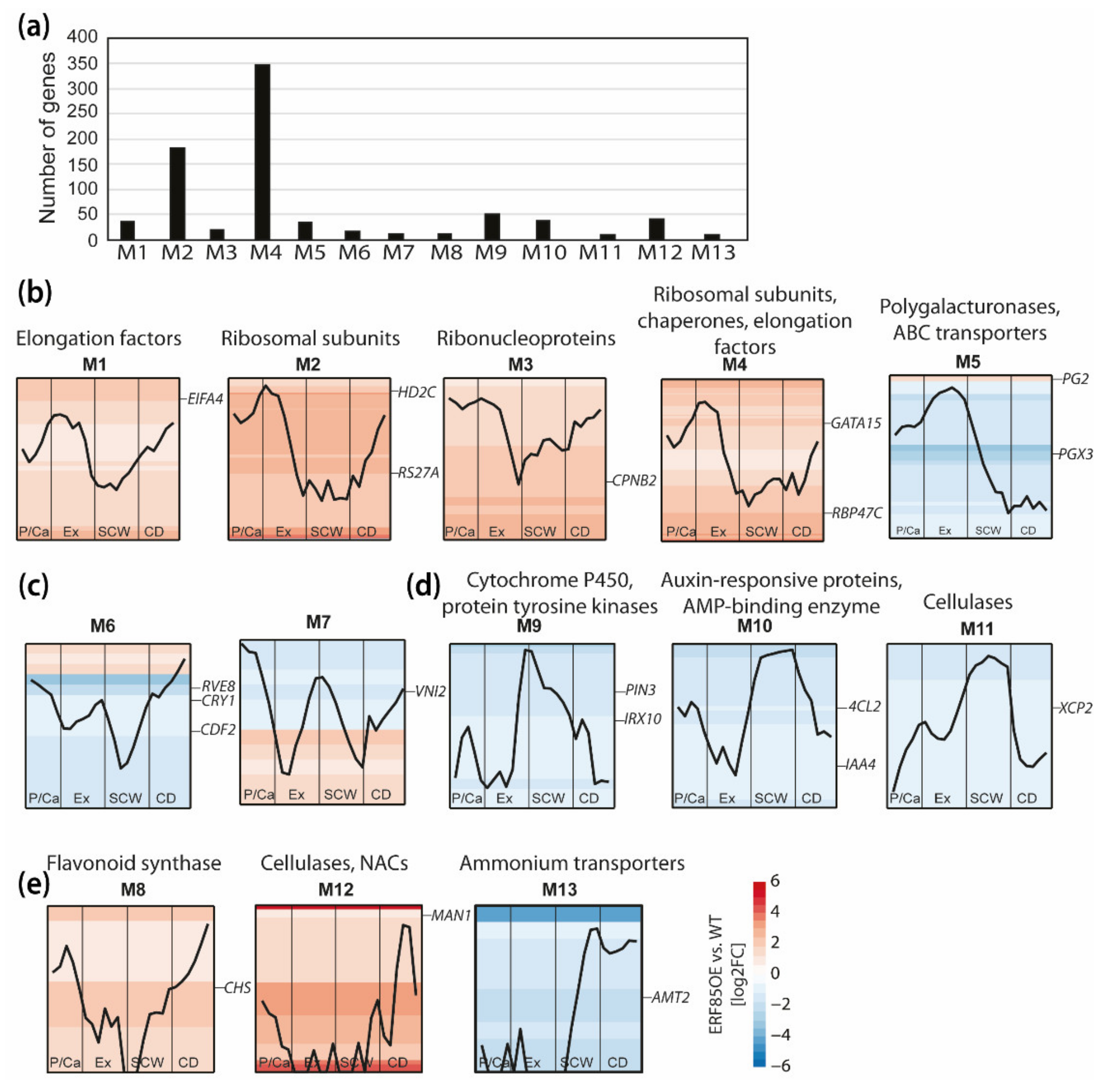
3.4. Ectopic Expression of PtERF85 Activated Expression of Genes Linked to Cell Expansion and Antagonized Induction of Genes Related to SCW Formation
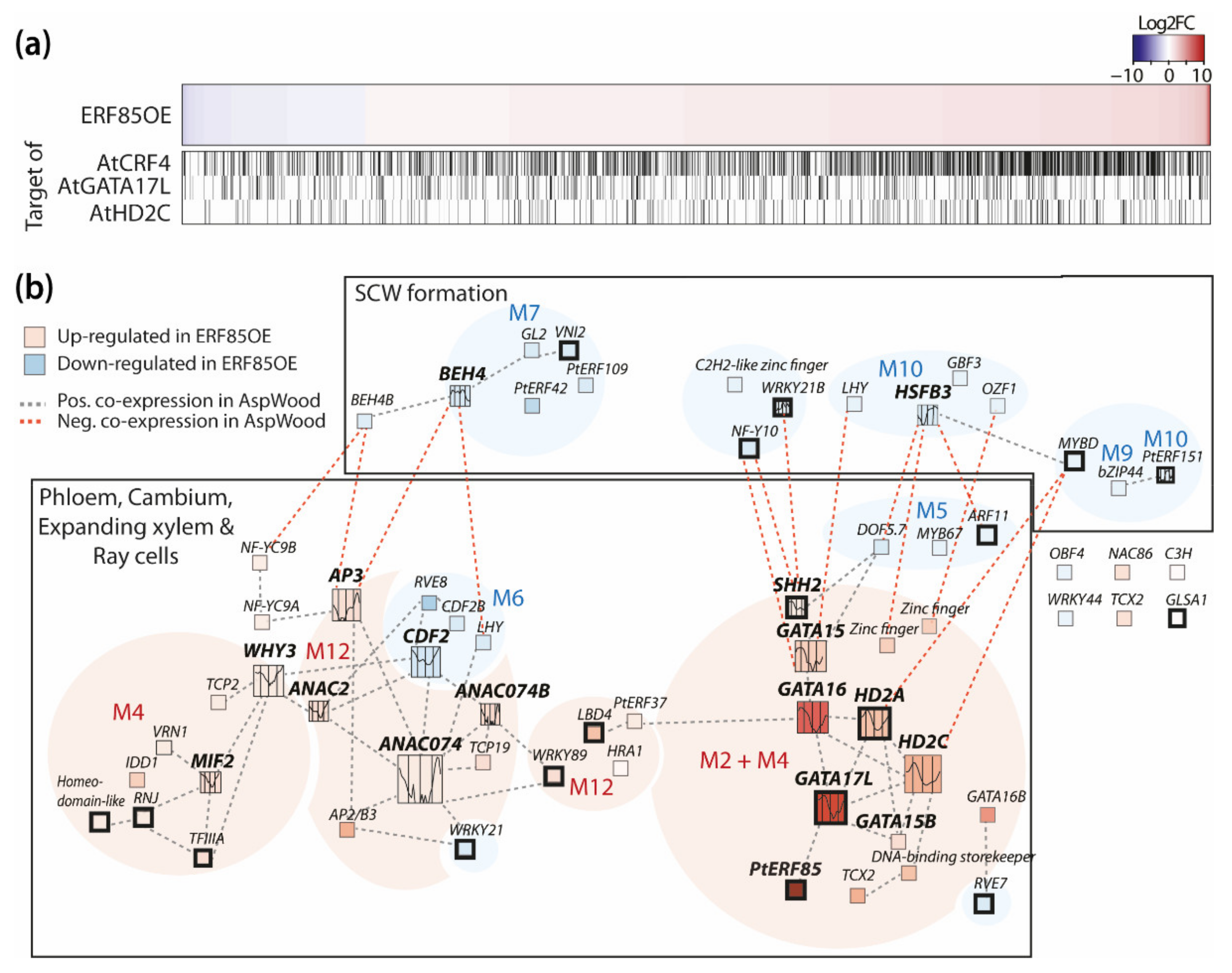
3.5. PtERF85-Mediated Transcriptional Regulation Mechanisms during Xylem Cell Expansion
4. Discussion
4.1. PtERF85 Modulates the Balance between Xylem Cell Expansion and SCW Formation through Transcriptional Regulation
4.2. PtERF85 Might Control Xylem Cell Wall Properties to Allow Cell Expansion
4.3. PtERF85-Mediated Regulation of Translation could Affect the Dynamics of Cell Growth in Expanding Xylem
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531. [Google Scholar] [CrossRef]
- Meents, M.J.; Watanabe, Y.; Samuels, A.L. The cell biology of secondary cell wall biosynthesis. Ann. Bot. 2018, 121, 1107–1125. [Google Scholar] [CrossRef] [Green Version]
- Sundell, D.; Street, N.R.; Kumar, M.; Mellerowicz, E.J.; Kucukoglu, M.; Johnsson, C.; Kumar, V.; Mannapperuma, C.; Delhomme, N.; Nilsson, O.; et al. Aspwood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 2017, 29, 1585–1604. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, R.L.; Zhong, R.; Ye, Z.H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Ye, Z.H. The poplar PtrWNDs are transcriptional activators of secondary cell wall biosynthesis. Plant Signal. Behav. 2010, 5, 469–472. [Google Scholar] [CrossRef] [Green Version]
- Tian, Q.; Li, C.; Lu, W.; Yang, L.; Jiang, Y.; Luo, K. Functional Characterization of the poplar R2R3-MYB transcription factor PtoMYB216 involved in the regulation of lignin biosynthesis during wood formation. PLoS ONE 2013, 8, e76369. [Google Scholar] [CrossRef]
- Zhong, R.; McCarthy, R.L.; Haghighat, M.; Ye, Z.H. The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS ONE 2013, 8, e69219. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, E.; Porth, I.; Chen, J.G.; Mansfield, S.D.; Douglas, C.J. Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis. Sci. Rep. 2014, 4, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wang, X.; Ran, L.; Tian, Q.; Fan, D.; Luo, K. PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in Populus tomentosa. Plant Cell Physiol. 2015, 56, 2436–2446. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Yang, Y.; Zheng, K.; Xie, M.; Feng, K.; Jawdy, S.S.; Gunter, L.E.; Ranjan, P.; Singan, V.R.; Engle, N.; et al. Genome-wide association studies and expression-based quantitative trait loci analyses reveal roles of HCT2 in caffeoylquinic acid biosynthesis and its regulation by defense-responsive transcription factors in Populus. New Phytol. 2018, 220, 502–516. [Google Scholar] [CrossRef] [Green Version]
- Takata, N.; Awano, T.; Nakata, M.T.; Sano, Y.; Sakamoto, S.; Mitsuda, N.; Taniguchi, T. Populus NST/SND orthologs are key regulators of secondary cell wall formation in wood fibers, phloem fibers and xylem ray parenchyma cells. Tree Physiol. 2019, 39, 514–525. [Google Scholar] [CrossRef]
- Love, J.; Björklund, S.; Vahala, J.; Hertzberg, M.; Kangasjärvi, J.; Sundberg, B. Ethylene is an endogenous stimulator of cell division in the cambial meristem of Populus. Proc. Natl. Acad. Sci. USA 2009, 106, 5984–5989. [Google Scholar] [CrossRef] [Green Version]
- Felten, J.; Vahala, J.; Love, J.; Gorzsás, A.; Rüggeberg, M.; Delhomme, N.; Leśniewska, J.; Kangasjärvi, J.; Hvidsten, T.R.; Mellerowicz, E.J.; et al. Ethylene signaling induces gelatinous layers with typical features of tension wood in hybrid aspen. New Phytol. 2018, 218, 999–1014. [Google Scholar] [CrossRef] [Green Version]
- Vahala, J.; Felten, J.; Love, J.; Gorzsás, A.; Gerber, L.; Lamminmäki, A.; Kangasjärvi, J.; Sundberg, B. A genome-wide screen for ethylene-induced Ethylene Response Factors (ERFs) in hybrid aspen stem identifies ERF genes that modify stem growth and wood properties. New Phytol. 2013, 200, 511–522. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, M.; Hou, C.; Lu, T.; Liu, T.; Liu, L.; Wei, H.; Cheng, Y.; Wei, Z. Functional characterization of Populus PsnSHN2 in coordinated regulation of secondary wall components in tobacco. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Seyfferth, C.; Wessels, B.; Jakipii-Lukkari, S.; Sundberg, B.; Delhomme, N.; Felten, J.; Tuominen, H. Ethylene-related gene expression networks in wood formation. Front. Plant Sci. 2018, 9, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wessels, B.; Seyfferth, C.; Escamez, S.; Vain, T.; Antos, K.; Vahala, J.; Delhomme, N.; Kangasjärvi, J.; Eder, M.; Felten, J.; et al. An AP2/ERF transcription factor ERF139 coordinates xylem cell expansion and secondary cell wall deposition. New Phytol. 2019, 224, 1585–1599. [Google Scholar] [CrossRef] [PubMed]
- Zwack, P.J.; Shi, X.; Robinson, B.R.; Gupta, S.; Compton, M.A.; Gerken, D.M.; Goertzen, L.R.; Rashotte, A.M. Vascular expression and C-terminal sequence divergence of cytokinin response factors in flowering plants. Plant Cell Physiol. 2012, 53, 1683–1695. [Google Scholar] [CrossRef]
- Rashotte, A.M.; Mason, M.G.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 11081–11085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutcliffe, J.W.; Hellmann, E.; Heyl, A.; Rashotte, A.M. CRFs form protein-protein interactions with each other and with members of the cytokinin signalling pathway in Arabidopsis via the CRF domain. J. Exp. Bot. 2011, 62, 4995–5002. [Google Scholar] [CrossRef] [Green Version]
- Zwack, P.J.; Compton, M.A.; Adams, C.I.; Rashotte, A.M. Cytokinin response factor 4 (CRF4) is induced by cold and involved in freezing tolerance. Plant Cell Rep. 2016, 35, 573–584. [Google Scholar] [CrossRef]
- Raines, T.; Shanks, C.; Cheng, C.Y.; McPherson, D.; Argueso, C.T.; Kim, H.J.; Franco-Zorrilla, J.M.; Lõpez-Vidriero, I.; Solano, R.; Vaňková, R.; et al. The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 2016, 85, 134–147. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Nilsson, O.; Alden, T.; Sitbon, F.; Little, C.H.A.; Chalupa, V.; Sandberg, G.; Olsson, O. Spatial pattern of cauliflower mosaic virus 35S promoter-luciferase expression in transgenic hybrid aspen trees monitored by enzymatic assay and non-destructive imaging. Transgenic Res. 1992, 1, 209–220. [Google Scholar] [CrossRef]
- Roach, M.; Gerber, L.; Sandquist, D.; Gorzsás, A.; Hedenström, M.; Kumar, M.; Steinhauser, M.C.; Feil, R.; Daniel, G.; Stitt, M.; et al. Fructokinase is required for carbon partitioning to cellulose in aspen wood. Plant J. 2012, 70, 967–977. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Šimášková, M.; O’Brien, J.A.; Khan, M.; Van Noorden, G.; Ötvös, K.; Vieten, A.; De Clercq, I.; Van Haperen, J.M.A.; Cuesta, C.; Hoyerová, K.; et al. Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat. Commun. 2015, 6, 8717. [Google Scholar] [CrossRef] [Green Version]
- Jang, G.; Yoon, Y.; Do Choi, Y. Jasmonic acid modulates xylem development by controlling expression of PIN-FORMED 7. Plant Signal. Behav. 2019, 14, e1637664. [Google Scholar] [CrossRef]
- Shani, Z.; Dekel, M.; Roiz, L.; Horowitz, M.; Kolosovski, N.; Lapidot, S.; Alkan, S.; Koltai, H.; Tsabary, G.; Goren, R.; et al. Expression of endo-1,4-β-glucanase (cel1) in Arabidopsis thaliana is associated with plant growth, xylem development and cell wall thickening. Plant Cell Rep. 2006, 25, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, Y.; Samejima, M.; Shiroishi, M.; Amano, Y.; Kanda, T.; Sakai, F.; Hayashi, T. Evidence that endo-1,4-β-glucanases act on cellulose in suspension-cultured poplar cells. Plant J. 2000, 24, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lake, B.B.; Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 2019, 37, 1452–1457. [Google Scholar] [CrossRef] [PubMed]
- Mellerowicz, E.J.; Sundberg, B. Wood cell walls: Biosynthesis, developmental dynamics and their implications for wood properties. Curr. Opin. Plant Biol. 2008, 11, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Varala, K.; Marshall-Colón, A.; Cirrone, J.; Brooks, M.D.; Pasquino, A.V.; Léran, S.; Mittal, S.; Rock, T.M.; Edwards, M.B.; Kim, G.; et al. Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. USA 2018, 115, 6494–6499. [Google Scholar] [CrossRef] [Green Version]
- Brooks, M.D.; Cirrone, J.; Pasquino, A.V.; Alvarez, J.M.; Swift, J.; Mittal, S.; Juang, C.-L.; Varala, K.; Gutiérrez, R.A.; Krouk, G.; et al. Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Labbe, H.; Sridha, S.; Wang, L.; Tian, L.; Latoszek-Green, M.; Yang, Z.; Brown, D.; Miki, B.; Wu, K. Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 2004, 38, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lim, C.J.; Shen, M.; Park, H.J.; Cha, J.-Y.; Iniesto, E.; Rubio, V.; Mengiste, T.; Zhu, J.-K.; Bressan, R.A.; et al. Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. USA 2018, 115, E5400–E5409. [Google Scholar] [CrossRef] [Green Version]
- Trapalis, M.; Li, S.F.; Parish, R.W. The Arabidopsis GASA10 gene encodes a cell wall protein strongly expressed in developing anthers and seeds. Plant Sci. 2017, 260, 71–79. [Google Scholar] [CrossRef]
- Rui, Y.; Xiao, C.; Yi, H.; Kandemir, B.; Wang, J.Z.; Puri, V.M.; Anderson, C.T. POLYGALACTURONASE INVOLVED IN EXPANSION3 functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell 2017, 29, 2413–2432. [Google Scholar] [CrossRef] [Green Version]
- Majda, M.; Robert, S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef] [Green Version]
- Braybrook, S.A.; Peaucelle, A. Mechano-chemical aspects of organ formation in Arabidopsis thaliana: The relationship between auxin and pectin. PLoS ONE 2013, 8, e57813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerttula, S.; Zinkgraf, M.; Munday, G.K.; Lewis, D.R.; Ibatullin, F.M.; Brumer, H.; Hart, F.; Mansfield, S.D.; Filkov, V.; Groover, A. Transcriptional and hormonal regulation of gravitropism of woody stems in populus. Plant Cell 2015, 27, 2800–2813. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seyfferth, C.; Wessels, B.A.; Vahala, J.; Kangasjärvi, J.; Delhomme, N.; Hvidsten, T.R.; Tuominen, H.; Lundberg-Felten, J. PopulusPtERF85 Balances Xylem Cell Expansion and Secondary Cell Wall Formation in Hybrid Aspen. Cells 2021, 10, 1971. https://doi.org/10.3390/cells10081971
Seyfferth C, Wessels BA, Vahala J, Kangasjärvi J, Delhomme N, Hvidsten TR, Tuominen H, Lundberg-Felten J. PopulusPtERF85 Balances Xylem Cell Expansion and Secondary Cell Wall Formation in Hybrid Aspen. Cells. 2021; 10(8):1971. https://doi.org/10.3390/cells10081971
Chicago/Turabian StyleSeyfferth, Carolin, Bernard A. Wessels, Jorma Vahala, Jaakko Kangasjärvi, Nicolas Delhomme, Torgeir R. Hvidsten, Hannele Tuominen, and Judith Lundberg-Felten. 2021. "PopulusPtERF85 Balances Xylem Cell Expansion and Secondary Cell Wall Formation in Hybrid Aspen" Cells 10, no. 8: 1971. https://doi.org/10.3390/cells10081971
APA StyleSeyfferth, C., Wessels, B. A., Vahala, J., Kangasjärvi, J., Delhomme, N., Hvidsten, T. R., Tuominen, H., & Lundberg-Felten, J. (2021). PopulusPtERF85 Balances Xylem Cell Expansion and Secondary Cell Wall Formation in Hybrid Aspen. Cells, 10(8), 1971. https://doi.org/10.3390/cells10081971






