Epigenetically Mediated Ciliogenesis and Cell Cycle Regulation, and Their Translational Potential
Abstract
1. Introduction
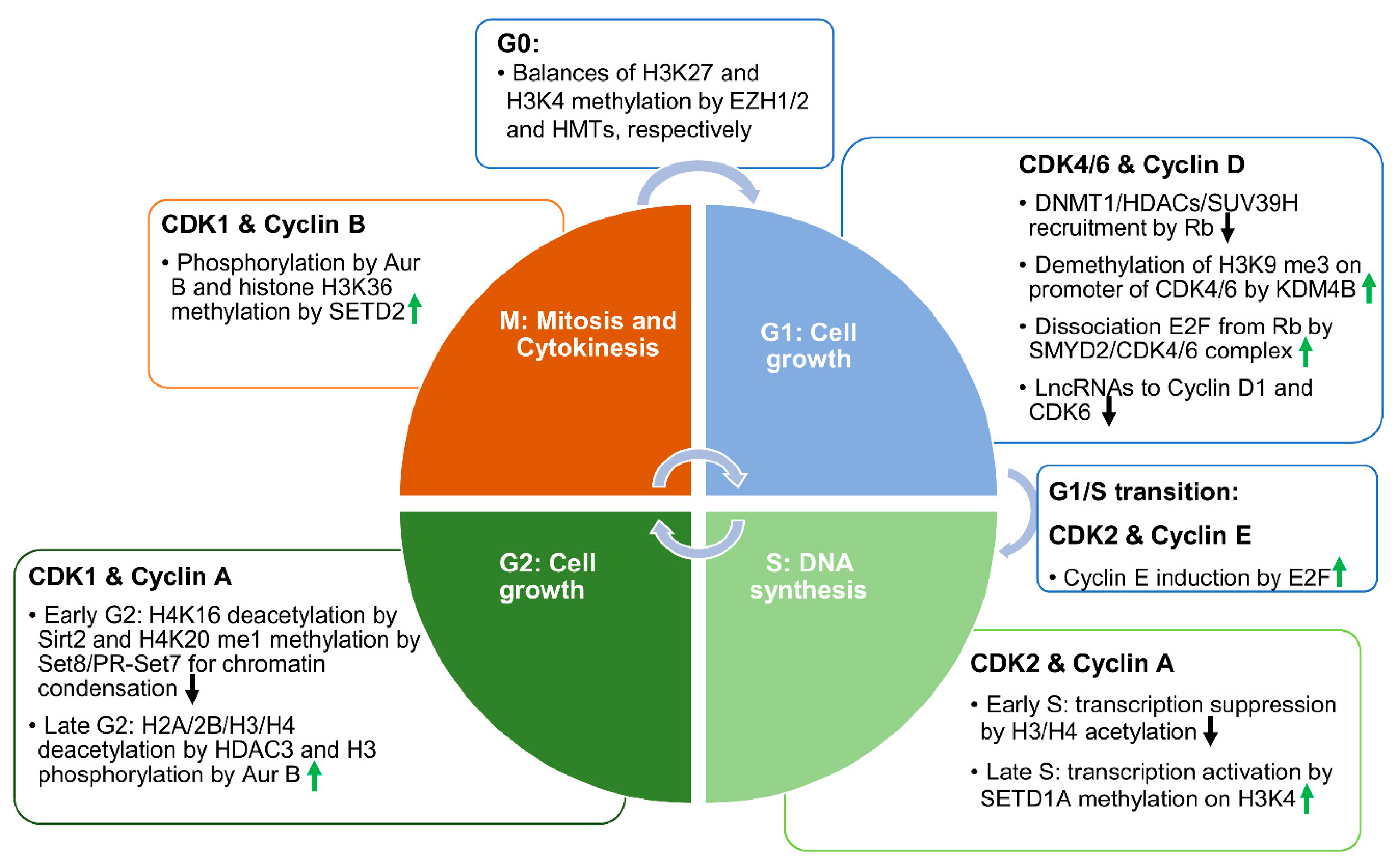
2. Epigenetic Regulation on Cell Cycle
2.1. Epigenetic Mechanisms in the Regulation of CDKs and Cyclins at Different Stages of the Cell Cycle
2.2. Epigenetic Mechanisms in the Regulation of Cell-Cycle-Associated Transcriptional Factors and Chromatin
2.3. Epigenetic Mechanisms in the Regulation of the Cell-Cycle-Associated Cytoskeleton
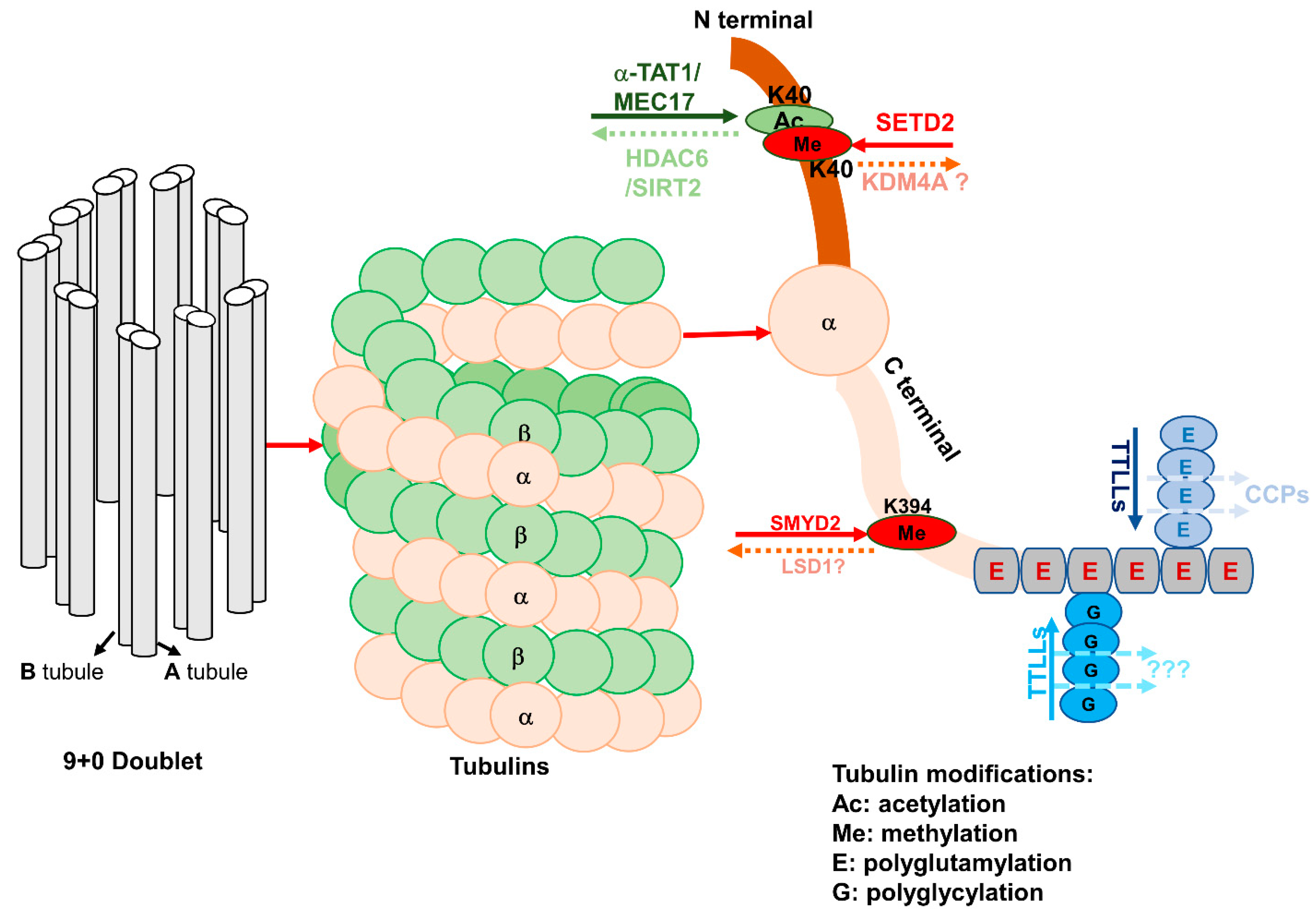
3. Epigenetic Regulation on Ciliogenesis
3.1. Epigenetic Mechanisms Mediate the Modification of α-Tubulin in Ciliogenesis
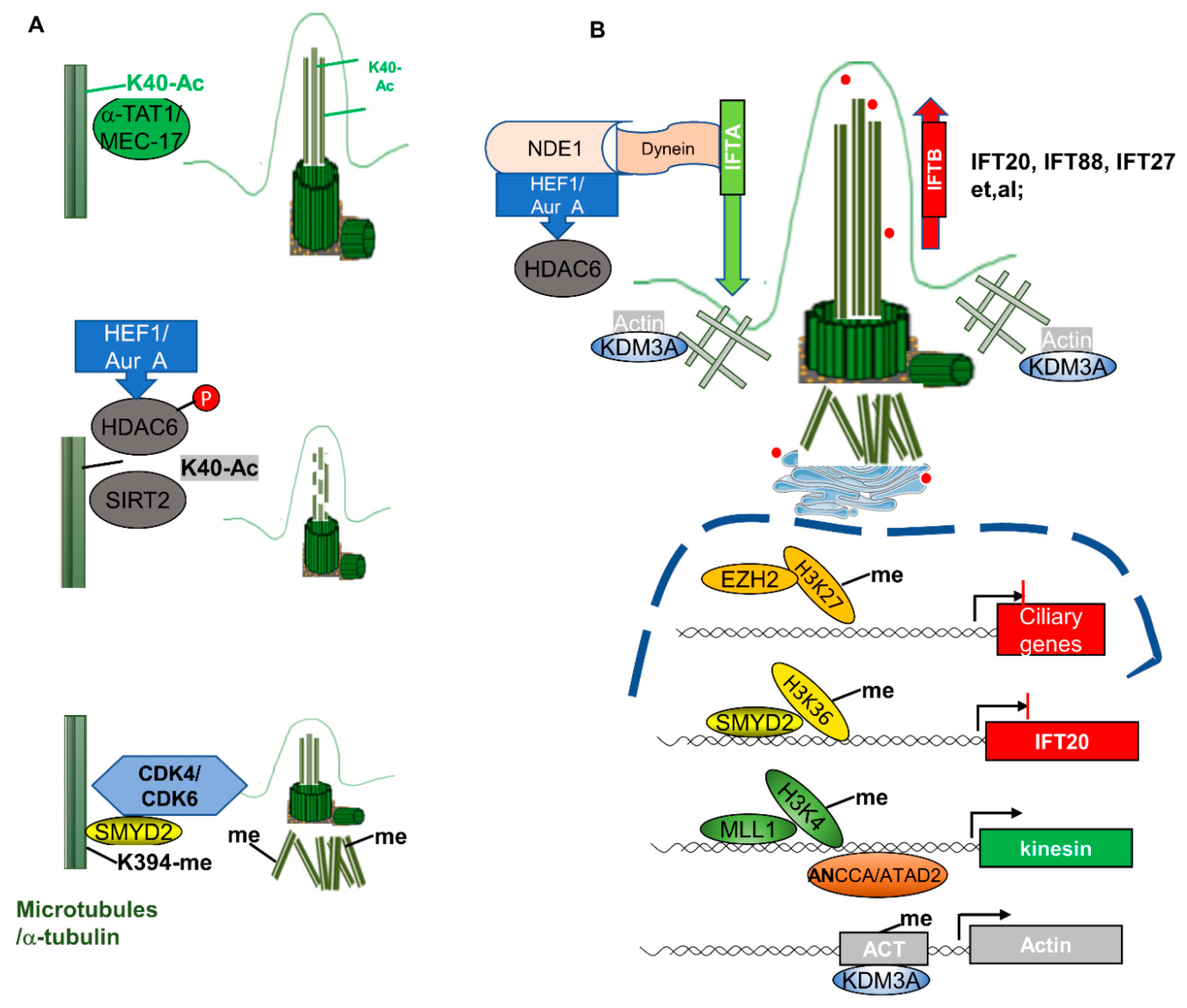
3.2. Epigenetic Mechanisms Mediate Ciliary Gene Expression in Ciliogenesis
3.3. Epigenetic Mechanisms Mediate the Modification of Other Cytoskeleton Proteins in Ciliogenesis
4. Crosstalk between the Cell Cycle and Ciliogenesis
4.1. Canonical View of Ciliogenesis and the Cell Cycle
4.2. Cilia Play a Predominant Role in Cell Cycle and Cell Proliferation Regulation
4.3. Epigenetic Mechanisms Regulate Crosstalk between the Cell Cycle and Cilia Biogenesis
5. Cilia-Dependent Signaling Pathways in Cell Cycle Regulation, and the Association of Epigenetic Mechanisms with These Signaling Pathways
5.1. Cilia-Dependent Signaling Pathways in Cell Cycle Regulation
5.2. The Association of Epigenetic Mechanisms with Cilia-Dependent Signaling Pathways
6. Therapeutic Targets of Cell-Cycle- and Ciliogenesis-Associated Epigenetic Regulators
7. Conclusions and Perspectives
Funding
Conflicts of Interest
References
- Schafer, K.A. The cell cycle: A review. Vet. Pathol. 1998, 35, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.X.; Fakhreddin, R.I.; Shimerov, H.K.; Kedziora, K.M.; Kumar, R.J.; Perez, J.; Limas, J.C.; Grant, G.D.; Cook, J.G.; Gupta, G.P.; et al. Evidence that the human cell cycle is a series of uncoupled, memoryless phases. Mol. Syst. Biol. 2019, 15, e8604. [Google Scholar] [CrossRef]
- Matson, J.P.; Cook, J.G. Cell cycle proliferation decisions: The impact of single cell analyses. FEBS J. 2017, 284, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, T.; Xu, G.; Liu, H.; Ren, C.; Xie, W.; Wang, M. Cyclin-Dependent Kinase 2 Promotes Tumor Proliferation and Induces Radio Resistance in Glioblastoma. Transl. Oncol. 2016, 9, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Sviderskiy, V.O.; Blumenberg, L.; Gorodetsky, E.; Karakousi, T.R.; Hirsh, N.; Alvarez, S.W.; Terzi, E.M.; Kaparos, E.; Whiten, G.C.; Ssebyala, S.; et al. Hyperactive CDK2 Activity in Basal-like Breast Cancer Imposes a Genome Integrity Liability that Can Be Exploited by Targeting DNA Polymerase epsilon. Mol. Cell 2020, 80, 682–698 e687. [Google Scholar] [CrossRef] [PubMed]
- Onesti, C.E.; Jerusalem, G. CDK4/6 inhibitors in breast cancer: Differences in toxicity profiles and impact on agent choice. A systematic review and meta-analysis. Expert Rev. Anticancer 2021, 21, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Servetto, A.; Napolitano, F.; De Angelis, C.; De Placido, P.; Giuliano, M.; Arpino, G.; De Placido, S.; Bianco, R.; Formisano, L. A review of the use of next generation sequencing methodologies to identify biomarkers of resistance to CDK4/6 inhibitors in ER+/HER2- breast cancer. Crit. Rev. Oncol. Hematol. 2021, 157, 103191. [Google Scholar] [CrossRef] [PubMed]
- Duronio, R.J.; Xiong, Y. Signaling pathways that control cell proliferation. Cold Spring Harb. Perspect. Biol. 2013, 5, a008904. [Google Scholar] [CrossRef]
- Besson, A.; Dowdy, S.F.; Roberts, J.M. CDK inhibitors: Cell cycle regulators and beyond. Dev. Cell 2008, 14, 159–169. [Google Scholar] [CrossRef]
- Pan, J.; Snell, W. The primary cilium: Keeper of the key to cell division. Cell 2007, 129, 1255–1257. [Google Scholar] [CrossRef]
- Plotnikova, O.V.; Pugacheva, E.N.; Golemis, E.A. Primary cilia and the cell cycle. Methods Cell Biol. 2009, 94, 137–160. [Google Scholar] [CrossRef] [PubMed]
- Li, L.X.; Zhou, J.X.; Wang, X.; Zhang, H.; Harris, P.C.; Calvet, J.P.; Li, X. Cross-talk between CDK4/6 and SMYD2 regulates gene transcription, tubulin methylation, and ciliogenesis. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef]
- Hu, S.; Danilov, A.V.; Godek, K.; Orr, B.; Tafe, L.J.; Rodriguez-Canales, J.; Behrens, C.; Mino, B.; Moran, C.A.; Memoli, V.A.; et al. CDK2 Inhibition Causes Anaphase Catastrophe in Lung Cancer through the Centrosomal Protein CP110. Cancer Res. 2015, 75, 2029–2038. [Google Scholar] [CrossRef] [PubMed]
- Pugacheva, E.N.; Jablonski, S.A.; Hartman, T.R.; Henske, E.P.; Golemis, E.A. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007, 129, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Devaskar, S.U.; Raychaudhuri, S. Epigenetics--a science of heritable biological adaptation. Pediatr. Res. 2007, 61, 1R–4R. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, X. Epigenetics and cell cycle regulation in cystogenesis. Cell. Signal. 2020, 68, 109509. [Google Scholar] [CrossRef] [PubMed]
- McGill, C.J.; Brooks, G. Cell cycle control mechanisms and their role in cardiac growth. Cardiovasc. Res. 1995, 30, 557–569. [Google Scholar] [CrossRef]
- Stein, G.S.; Stein, J.L.; van Wijnen, A.; Lian, J.B.; Quesenberry, P.J. Molecular mechanisms mediating control of cell cycle and cell growth. Exp. Hematol. 1995, 23, 1053–1061. [Google Scholar]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef]
- Vidal-Laliena, M.; Gallastegui, E.; Mateo, F.; Martinez-Balbas, M.; Pujol, M.J.; Bachs, O. Histone deacetylase 3 regulates cyclin A stability. J. Biol. Chem. 2013, 288, 21096–21104. [Google Scholar] [CrossRef]
- Li, Y.; Peng, L.; Seto, E. Histone Deacetylase 10 Regulates the Cell Cycle G2/M Phase Transition via a Novel Let-7-HMGA2-Cyclin A2 Pathway. Mol. Cell Biol. 2015, 35, 3547–3565. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Cubizolles, F.; Zhang, Y.; Reichert, N.; Kohler, H.; Seiser, C.; Matthias, P. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010, 24, 455–469. [Google Scholar] [CrossRef]
- Li, S.; Liu, X.; Chen, X.; Zhang, L.; Wang, X. Histone deacetylase 6 promotes growth of glioblastoma through inhibition of SMAD2 signaling. Tumour Biol. 2015, 36, 9661–9665. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Sun, L.; Li, Q.; Liang, J.; Yu, W.; Yi, X.; Yang, X.; Li, Y.; Han, X.; Zhang, Y.; et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 7541–7546. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jubb, A.M.; Pike, L.; Buffa, F.M.; Turley, H.; Baban, D.; Leek, R.; Gatter, K.C.; Ragoussis, J.; Harris, A.L. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010, 70, 6456–6466. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.Y.; Xuan, F.; Liu, W.; Cui, H.J. MINA controls proliferation and tumorigenesis of glioblastoma by epigenetically regulating cyclins and CDKs via H3K9me3 demethylation. Oncogene 2017, 36, 387–396. [Google Scholar] [CrossRef]
- Zhou, J.X.; Li, X. Non-Coding RNAs in Hereditary Kidney Disorders. Int. J. Mol. Sci. 2021, 22, 3014. [Google Scholar] [CrossRef]
- Wang, X.; Arai, S.; Song, X.; Reichart, D.; Du, K.; Pascual, G.; Tempst, P.; Rosenfeld, M.G.; Glass, C.K.; Kurokawa, R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008, 454, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, D.; Zhang, W.; Guo, M.; Zhan, Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012, 31, 4415–4427. [Google Scholar] [CrossRef]
- Arshi, A.; Sharifi, F.S.; Khorramian Ghahfarokhi, M.; Faghih, Z.; Doosti, A.; Ostovari, S.; Mahmoudi Maymand, E.; Ghahramani Seno, M.M. Expression Analysis of MALAT1, GAS5, SRA, and NEAT1 lncRNAs in Breast Cancer Tissues from Young Women and Women over 45 Years of Age. Mol. Nucleic Acids 2018, 12, 751–757. [Google Scholar] [CrossRef]
- Zhang, H.S.; Gavin, M.; Dahiya, A.; Postigo, A.A.; Ma, D.; Luo, R.X.; Harbour, J.W.; Dean, D.C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 2000, 101, 79–89. [Google Scholar] [CrossRef]
- Li, L.X.; Fan, L.X.; Zhou, J.X.; Grantham, J.J.; Calvet, J.P.; Sage, J.; Li, X. Lysine methyltransferase SMYD2 promotes cyst growth in autosomal dominant polycystic kidney disease. J. Clin. Investig. 2017, 127, 2751–2764. [Google Scholar] [CrossRef]
- Akiyama, T.; Ohuchi, T.; Sumida, S.; Matsumoto, K.; Toyoshima, K. Phosphorylation of the retinoblastoma protein by cdk2. Proc. Natl. Acad. Sci. USA 1992, 89, 7900–7904. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.D.; Levin, P.A. Metabolism, cell growth and the bacterial cell cycle. Nat. Rev. Microbiol. 2009, 7, 822–827. [Google Scholar] [CrossRef]
- Rhind, N.; Gilbert, D.M. DNA replication timing. Cold Spring Harb. Perspect. Biol. 2013, 5, a010132. [Google Scholar] [CrossRef] [PubMed]
- Voichek, Y.; Mittelman, K.; Gordon, Y.; Bar-Ziv, R.; Lifshitz Smit, D.; Shenhav, R.; Barkai, N. Epigenetic Control of Expression Homeostasis during Replication Is Stabilized by the Replication Checkpoint. Mol. Cell 2018, 70, 1121–1133, e1129. [Google Scholar] [CrossRef] [PubMed]
- Hendzel, M.J.; Wei, Y.; Mancini, M.A.; Van Hooser, A.; Ranalli, T.; Brinkley, B.R.; Bazett-Jones, D.P.; Allis, C.D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 1997, 106, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kao, G.D.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Qin, J.; Phelan, C.; Lazar, M.A. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006, 20, 2566–2579. [Google Scholar] [CrossRef]
- Bonenfant, D.; Towbin, H.; Coulot, M.; Schindler, P.; Mueller, D.R.; van Oostrum, J. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell Proteom. 2007, 6, 1917–1932. [Google Scholar] [CrossRef]
- Vaquero, A.; Scher, M.B.; Lee, D.H.; Sutton, A.; Cheng, H.L.; Alt, F.W.; Serrano, L.; Sternglanz, R.; Reinberg, D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006, 20, 1256–1261. [Google Scholar] [CrossRef]
- Jorgensen, S.; Schotta, G.; Sorensen, C.S. Histone H4 lysine 20 methylation: Key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013, 41, 2797–2806. [Google Scholar] [CrossRef] [PubMed]
- Cheung, T.H.; Rando, T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol 2013, 14, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Berry, W.L.; Janknecht, R. KDM4/JMJD2 histone demethylases: Epigenetic regulators in cancer cells. Cancer Res. 2013, 73, 2936–2942. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wang, Z.; Zhang, H.; Gan, H.; Zhang, Z. H3K9me3 demethylase Kdm4d facilitates the formation of pre-initiative complex and regulates DNA replication. Nucleic Acids Res. 2017, 45, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Nair, V.D.; Ge, Y.; Balasubramaniyan, N.; Kim, J.; Okawa, Y.; Chikina, M.; Troyanskaya, O.; Sealfon, S.C. Involvement of histone demethylase LSD1 in short-time-scale gene expression changes during cell cycle progression in embryonic stem cells. Mol. Cell Biol. 2012, 32, 4861–4876. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gachet, Y.; Tournier, S.; Millar, J.B.; Hyams, J.S. A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature 2001, 412, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Koenning, M.; Wang, X.; Karki, M.; Jangid, R.K.; Kearns, S.; Tripathi, D.N.; Cianfrocco, M.; Verhey, K.J.; Jung, S.Y.; Coarfa, C.; et al. Neuronal SETD2 activity links microtubule methylation to an anxiety-like phenotype in mice. Brain 2021. [Google Scholar] [CrossRef]
- Li, F.; Mao, G.; Tong, D.; Huang, J.; Gu, L.; Yang, W.; Li, G.M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013, 153, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Powell, R.T.; Tripathi, D.N.; Dere, R.; Ho, T.H.; Blasius, T.L.; Chiang, Y.C.; Davis, I.J.; Fahey, C.C.; Hacker, K.E.; et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell 2016, 166, 950–962. [Google Scholar] [CrossRef]
- Yeyati, P.L.; Schiller, R.; Mali, G.; Kasioulis, I.; Kawamura, A.; Adams, I.R.; Playfoot, C.; Gilbert, N.; van Heyningen, V.; Wills, J.; et al. KDM3A coordinates actin dynamics with intraflagellar transport to regulate cilia stability. J. Cell Biol. 2017, 216, 999–1013. [Google Scholar] [CrossRef]
- Zacharopoulou, N.; Tsapara, A.; Kallergi, G.; Schmid, E.; Tsichlis, P.N.; Kampranis, S.C.; Stournaras, C. The epigenetic factor KDM2B regulates cell adhesion, small rho GTPases, actin cytoskeleton and migration in prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 587–597. [Google Scholar] [CrossRef]
- Pazour, G.J.; Witman, G.B. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 2003, 15, 105–110. [Google Scholar] [CrossRef]
- Jain, R.; Javidan-Nejad, C.; Alexander-Brett, J.; Horani, A.; Cabellon, M.C.; Walter, M.J.; Brody, S.L. Sensory functions of motile cilia and implication for bronchiectasis. Front. Biosci. (Sch. Ed.) 2012, 4, 1088–1098. [Google Scholar] [CrossRef][Green Version]
- Ringers, C.; Olstad, E.W.; Jurisch-Yaksi, N. The role of motile cilia in the development and physiology of the nervous system. Philos Trans. R Soc. Lond B Biol. Sci. 2020, 375, 20190156. [Google Scholar] [CrossRef]
- Singh, M.; Chaudhry, P.; Merchant, A.A. Primary cilia are present on human blood and bone marrow cells and mediate Hedgehog signaling. Exp. Hematol. 2016, 44, 1181–1187 e1182. [Google Scholar] [CrossRef]
- Hua, K.; Ferland, R.J. Primary cilia proteins: Ciliary and extraciliary sites and functions. Cell Mol. Life Sci. 2018, 75, 1521–1540. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, J.L.; Witman, G.B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002, 3, 813–825. [Google Scholar] [CrossRef]
- Breslow, D.K.; Holland, A.J. Mechanism and Regulation of Centriole and Cilium Biogenesis. Annu. Rev. Biochem. 2019, 88, 691–724. [Google Scholar] [CrossRef]
- Garcin, C.; Straube, A. Microtubules in cell migration. Essays Biochem. 2019, 63, 509–520. [Google Scholar] [CrossRef]
- Pitaval, A.; Senger, F.; Letort, G.; Gidrol, X.; Guyon, L.; Sillibourne, J.; Thery, M. Microtubule stabilization drives 3D centrosome migration to initiate primary ciliogenesis. J. Cell Biol. 2017, 216, 3713–3728. [Google Scholar] [CrossRef]
- Shida, T.; Cueva, J.G.; Xu, Z.; Goodman, M.B.; Nachury, M.V. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. USA 2010, 107, 21517–21522. [Google Scholar] [CrossRef] [PubMed]
- Wloga, D.; Joachimiak, E.; Fabczak, H. Tubulin Post-Translational Modifications and Microtubule Dynamics. Int. J. Mol. Sci. 2017, 18, 2207. [Google Scholar] [CrossRef]
- Akella, J.S.; Wloga, D.; Kim, J.; Starostina, N.G.; Lyons-Abbott, S.; Morrissette, N.S.; Dougan, S.T.; Kipreos, E.T.; Gaertig, J. MEC-17 is an alpha-tubulin acetyltransferase. Nature 2010, 467, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Kalebic, N.; Sorrentino, S.; Perlas, E.; Bolasco, G.; Martinez, C.; Heppenstall, P.A. alphaTAT1 is the major alpha-tubulin acetyltransferase in mice. Nat. Commun. 2013, 4, 1962. [Google Scholar] [CrossRef]
- Zhou, X.; Fan, L.X.; Li, K.; Ramchandran, R.; Calvet, J.P.; Li, X. SIRT2 regulates ciliogenesis and contributes to abnormal centrosome amplification caused by loss of polycystin-1. Hum. Mol. Genet. 2014, 23, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Eshun-Wilson, L.; Zhang, R.; Portran, D.; Nachury, M.V.; Toso, D.B.; Lohr, T.; Vendruscolo, M.; Bonomi, M.; Fraser, J.S.; Nogales, E. Effects of alpha-tubulin acetylation on microtubule structure and stability. Proc. Natl. Acad. Sci. USA 2019, 116, 10366–10371. [Google Scholar] [CrossRef]
- Aguilar, A.; Becker, L.; Tedeschi, T.; Heller, S.; Iomini, C.; Nachury, M.V. Alpha-tubulin K40 acetylation is required for contact inhibition of proliferation and cell-substrate adhesion. Mol. Biol. Cell 2014, 25, 1854–1866. [Google Scholar] [CrossRef]
- Mirvis, M.; Stearns, T.; James Nelson, W. Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem. J. 2018, 475, 2329–2353. [Google Scholar] [CrossRef]
- Edde, B.; Rossier, J.; Le Caer, J.P.; Desbruyeres, E.; Gros, F.; Denoulet, P. Posttranslational glutamylation of alpha-tubulin. Science 1990, 247, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.R. Polyglutamylation: The GLU that makes microtubules sticky. Curr. Biol. 2010, 20, R234–R236. [Google Scholar] [CrossRef] [PubMed]
- Bosch Grau, M.; Masson, C.; Gadadhar, S.; Rocha, C.; Tort, O.; Marques Sousa, P.; Vacher, S.; Bieche, I.; Janke, C. Alterations in the balance of tubulin glycylation and glutamylation in photoreceptors leads to retinal degeneration. J. Cell Sci. 2017, 130, 938–949. [Google Scholar] [CrossRef]
- Rogowski, K.; Juge, F.; van Dijk, J.; Wloga, D.; Strub, J.M.; Levilliers, N.; Thomas, D.; Bre, M.H.; Van Dorsselaer, A.; Gaertig, J.; et al. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell 2009, 137, 1076–1087. [Google Scholar] [CrossRef]
- Gadadhar, S.; Dadi, H.; Bodakuntla, S.; Schnitzler, A.; Bieche, I.; Rusconi, F.; Janke, C. Tubulin glycylation controls primary cilia length. J. Cell Biol. 2017, 216, 2701–2713. [Google Scholar] [CrossRef]
- Gadadhar, S.; Alvarez Viar, G.; Hansen, J.N.; Gong, A.; Kostarev, A.; Ialy-Radio, C.; Leboucher, S.; Whitfield, M.; Ziyyat, A.; Toure, A.; et al. Tubulin glycylation controls axonemal dynein activity, flagellar beat, and male fertility. Science 2021, 371. [Google Scholar] [CrossRef] [PubMed]
- Zingg, D.; Debbache, J.; Pena-Hernandez, R.; Antunes, A.T.; Schaefer, S.M.; Cheng, P.F.; Zimmerli, D.; Haeusel, J.; Calcada, R.R.; Tuncer, E.; et al. EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation. Cancer Cell 2018, 34, 69–84 e14. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.X.; Duan, Z.; Wang, J.; Sokolov, A.; Xu, J.; Chen, C.Z.; Li, J.J.; Chen, H.W. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol. Cancer Res. 2014, 12, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012, 2, 419–431. [Google Scholar] [CrossRef]
- Copeland, S.J.; McRae, A.; Guarguaglini, G.; Trinkle-Mulcahy, L.; Copeland, J.W. Actin-dependent regulation of cilia length by the inverted formin FHDC1. Mol. Biol Cell 2018, 29, 1611–1627. [Google Scholar] [CrossRef] [PubMed]
- Avasthi, P.; Marshall, W.F. Stages of ciliogenesis and regulation of ciliary length. Differentiation 2012, 83, S30–S42. [Google Scholar] [CrossRef] [PubMed]
- Malicki, J.J.; Johnson, C.A. The Cilium: Cellular Antenna and Central Processing Unit. Trends Cell Biol. 2017, 27, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Inaba, H.; Inagaki, M. Mechanisms of ciliogenesis suppression in dividing cells. Cell Mol. Life Sci. 2017, 74, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Inoko, A.; Matsuyama, M.; Goto, H.; Ohmuro-Matsuyama, Y.; Hayashi, Y.; Enomoto, M.; Ibi, M.; Urano, T.; Yonemura, S.; Kiyono, T.; et al. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J. Cell Biol. 2012, 197, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, O.V.; Golemis, E.A.; Pugacheva, E.N. Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 2008, 68, 2058–2061. [Google Scholar] [CrossRef] [PubMed]
- Higgins, M.; Obaidi, I.; McMorrow, T. Primary cilia and their role in cancer. Oncol. Lett. 2019, 17, 3041–3047. [Google Scholar] [CrossRef]
- Thoma, C.R.; Frew, I.J.; Hoerner, C.R.; Montani, M.; Moch, H.; Krek, W. pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nat. Cell Biol. 2007, 9, 588–595. [Google Scholar] [CrossRef]
- Pazour, G.J.; Dickert, B.L.; Vucica, Y.; Seeley, E.S.; Rosenbaum, J.L.; Witman, G.B.; Cole, D.G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000, 151, 709–718. [Google Scholar] [CrossRef]
- Robert, A.; Margall-Ducos, G.; Guidotti, J.E.; Bregerie, O.; Celati, C.; Brechot, C.; Desdouets, C. The intraflagellar transport component IFT88/polaris is a centrosomal protein regulating G1-S transition in non-ciliated cells. J. Cell Sci. 2007, 120, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Wang, Z.; Diener, D.; Rosenbaum, J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 2007, 17, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Zaghloul, N.A.; Bubenshchikova, E.; Oh, E.C.; Rankin, S.; Katsanis, N.; Obara, T.; Tsiokas, L. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat. Cell Biol. 2011, 13, 351–360. [Google Scholar] [CrossRef]
- Li, L.X.; Zhou, J.X.; Calvet, J.P.; Godwin, A.K.; Jensen, R.A.; Li, X. Lysine methyltransferase SMYD2 promotes triple negative breast cancer progression. Cell Death Dis. 2018, 9, 326. [Google Scholar] [CrossRef]
- Kaneko, S.; Li, G.; Son, J.; Xu, C.F.; Margueron, R.; Neubert, T.A.; Reinberg, D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010, 24, 2615–2620. [Google Scholar] [CrossRef]
- Jiang, J.; Hui, C.C. Hedgehog signaling in development and cancer. Dev. Cell 2008, 15, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, J.; Therond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Raleigh, D.R.; Reiter, J.F. Misactivation of Hedgehog signaling causes inherited and sporadic cancers. J. Clin. Investig. 2019, 129, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Tukachinsky, H.; Lopez, L.V.; Salic, A. A mechanism for vertebrate Hedgehog signaling: Recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 2010, 191, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Corbit, K.C.; Aanstad, P.; Singla, V.; Norman, A.R.; Stainier, D.Y.; Reiter, J.F. Vertebrate Smoothened functions at the primary cilium. Nature 2005, 437, 1018–1021. [Google Scholar] [CrossRef]
- Duman-Scheel, M.; Weng, L.; Xin, S.; Du, W. Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature 2002, 417, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Ingham, P.W. Hedgehogs tryst with the cell cycle. J. Cell Sci. 2002, 115, 4393–4397. [Google Scholar] [CrossRef]
- Chiarini, F.; Paganelli, F.; Martelli, A.M.; Evangelisti, C. The Role Played by Wnt/beta-Catenin Signaling Pathway in Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2020, 21, 1098. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Johmura, Y.; Yu, L.R.; Park, J.E.; Gao, Y.; Bang, J.K.; Zhou, M.; Veenstra, T.D.; Yeon Kim, B.; Lee, K.S. Identification of a novel Wnt5a-CK1varepsilon-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J. 2012, 31, 3104–3117. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, M.; Sancho, E.; Verweij, C.; de Lau, W.; Oving, I.; Hurlstone, A.; van der Horn, K.; Batlle, E.; Coudreuse, D.; Haramis, A.P.; et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002, 111, 241–250. [Google Scholar] [CrossRef]
- Lokker, N.A.; Sullivan, C.M.; Hollenbach, S.J.; Israel, M.A.; Giese, N.A. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: Evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res. 2002, 62, 3729–3735. [Google Scholar]
- Heldin, C.H.; Lennartsson, J. Structural and functional properties of platelet-derived growth factor and stem cell factor receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a009100. [Google Scholar] [CrossRef]
- Schneider, L.; Clement, C.A.; Teilmann, S.C.; Pazour, G.J.; Hoffmann, E.K.; Satir, P.; Christensen, S.T. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr. Biol. 2005, 15, 1861–1866. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Liu, X.W.; Kim, H.R. Platelet-derived growth factor (PDGF) receptor-alpha-activated c-Jun NH2-terminal kinase-1 is critical for PDGF-induced p21WAF1/CIP1 promoter activity independent of p53. J. Biol. Chem. 2003, 278, 49582–49588. [Google Scholar] [CrossRef]
- Schmid, F.M.; Schou, K.B.; Vilhelm, M.J.; Holm, M.S.; Breslin, L.; Farinelli, P.; Larsen, L.A.; Andersen, J.S.; Pedersen, L.B.; Christensen, S.T. IFT20 modulates ciliary PDGFRalpha signaling by regulating the stability of Cbl E3 ubiquitin ligases. J. Cell Biol. 2018, 217, 151–161. [Google Scholar] [CrossRef]
- Wheway, G.; Nazlamova, L.; Hancock, J.T. Signaling through the Primary Cilium. Front. Cell Dev. Biol. 2018, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Leitch, C.C.; Lodh, S.; Prieto-Echague, V.; Badano, J.L.; Zaghloul, N.A. Basal body proteins regulate Notch signaling through endosomal trafficking. J. Cell Sci. 2014, 127, 2407–2419. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.H.; Yang, L.; Dessaud, E.; Chuang, K.; Moore, D.M.; Rohatgi, R.; Briscoe, J.; Novitch, B.G. Notch activity modulates the responsiveness of neural progenitors to sonic hedgehog signaling. Dev. Cell 2015, 33, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.T.; Morthorst, S.K.; Mogensen, J.B.; Pedersen, L.B. Primary Cilia and Coordination of Receptor Tyrosine Kinase (RTK) and Transforming Growth Factor beta (TGF-beta) Signaling. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Wils, L.J.; Bijlsma, M.F. Epigenetic regulation of the Hedgehog and Wnt pathways in cancer. Crit. Rev. Oncol. Hematol. 2018, 121, 23–44. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, E.; De Smaele, E.; Miele, E.; Laneve, P.; Po, A.; Pelloni, M.; Paganelli, A.; Di Marcotullio, L.; Caffarelli, E.; Screpanti, I.; et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008, 27, 2616–2627. [Google Scholar] [CrossRef]
- Cai, J.; Guan, H.; Fang, L.; Yang, Y.; Zhu, X.; Yuan, J.; Wu, J.; Li, M. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J. Clin. Investig. 2013, 123, 566–579. [Google Scholar] [CrossRef]
- Drinane, M.C.; Yaqoob, U.; Yu, H.; Luo, F.; Greuter, T.; Arab, J.P.; Kostallari, E.; Verma, V.K.; Maiers, J.; De Assuncao, T.M.; et al. Synectin promotes fibrogenesis by regulating PDGFR isoforms through distinct mechanisms. JCI Insight 2017, 2. [Google Scholar] [CrossRef]
- Bhagat, T.D.; Zou, Y.; Huang, S.; Park, J.; Palmer, M.B.; Hu, C.; Li, W.; Shenoy, N.; Giricz, O.; Choudhary, G.; et al. Notch Pathway Is Activated via Genetic and Epigenetic Alterations and Is a Therapeutic Target in Clear Cell Renal Cancer. J. Biol. Chem. 2017, 292, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Guarani, V.; Deflorian, G.; Franco, C.A.; Kruger, M.; Phng, L.K.; Bentley, K.; Toussaint, L.; Dequiedt, F.; Mostoslavsky, R.; Schmidt, M.H.H.; et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature 2011, 473, 234–238. [Google Scholar] [CrossRef]
- Bae, Y.; Yang, T.; Zeng, H.C.; Campeau, P.M.; Chen, Y.; Bertin, T.; Dawson, B.C.; Munivez, E.; Tao, J.; Lee, B.H. miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 2012, 21, 2991–3000. [Google Scholar] [CrossRef] [PubMed]
- Suriyamurthy, S.; Baker, D.; Ten Dijke, P.; Iyengar, P.V. Epigenetic Reprogramming of TGF-beta Signaling in Breast Cancer. Cancers (Basel) 2019, 11, 726. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Xi, Q. Crosstalk between TGF-beta signaling and epigenome. Acta Biochim. Biophys. Sin. 2018, 50, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Kim, M. EZH2 as a Potential Target for NAFLD Therapy. Int. J. Mol. Sci. 2020, 21, 8617. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Jiang, X.J.; Fang, Z.M. Histone methyltransferase SMYD2: Ubiquitous regulator of disease. Clin. Epigenet. 2019, 11, 112. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.N.; Sharma, N. Epigenetic Modulators as Potential Multi-targeted Drugs Against Hedgehog Pathway for Treatment of Cancer. Protein J. 2019, 38, 537–550. [Google Scholar] [CrossRef] [PubMed]
| Epigenetic Regulators | Cell-Cycle-Related Target(s) | Ciliary Target(s) | References |
|---|---|---|---|
| SMYD2 | CDK4/6, H3K4 me3 and H3K36 me3 | IFT20, α-tubulin | [12,33,92] |
| SETD2 | H3K36me3 | α-tubulin | [48,49,50] |
| EZH2 | H3K27me3 | WDR19, IFT81, FUZ, etc. | [76,93] |
| KDM3A | Actin | Actin at basal body | [51] |
| HDAC6 | p21 | α-tubulin | [14,24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.X.; Li, X. Epigenetically Mediated Ciliogenesis and Cell Cycle Regulation, and Their Translational Potential. Cells 2021, 10, 1662. https://doi.org/10.3390/cells10071662
Li LX, Li X. Epigenetically Mediated Ciliogenesis and Cell Cycle Regulation, and Their Translational Potential. Cells. 2021; 10(7):1662. https://doi.org/10.3390/cells10071662
Chicago/Turabian StyleLi, Linda Xiaoyan, and Xiaogang Li. 2021. "Epigenetically Mediated Ciliogenesis and Cell Cycle Regulation, and Their Translational Potential" Cells 10, no. 7: 1662. https://doi.org/10.3390/cells10071662
APA StyleLi, L. X., & Li, X. (2021). Epigenetically Mediated Ciliogenesis and Cell Cycle Regulation, and Their Translational Potential. Cells, 10(7), 1662. https://doi.org/10.3390/cells10071662





