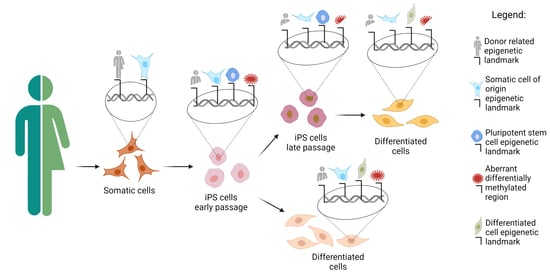
iPSC Preparation and Epigenetic Memory: Does the Tissue Origin Matter?
Abstract
1. iPSCs Production
1.1. Viral Reprogramming Methods
1.1.1. Sendai Virus
1.1.2. Adenovirus
1.1.3. Adeno-Associated Virus
1.2. Non-Viral Reprogramming Methods
1.2.1. mRNA Transfection
1.2.2. miRNA Infection/Transfection
1.2.3. Transposons: PiggyBac and Sleeping Beauty
1.2.4. Episomal Plasmids
1.2.5. Minicircle Vectors
1.2.6. Liposomal Magnetofection
1.2.7. Proteins
1.2.8. Chemical Induction
2. Comparison between Different Methods of Reprogramming
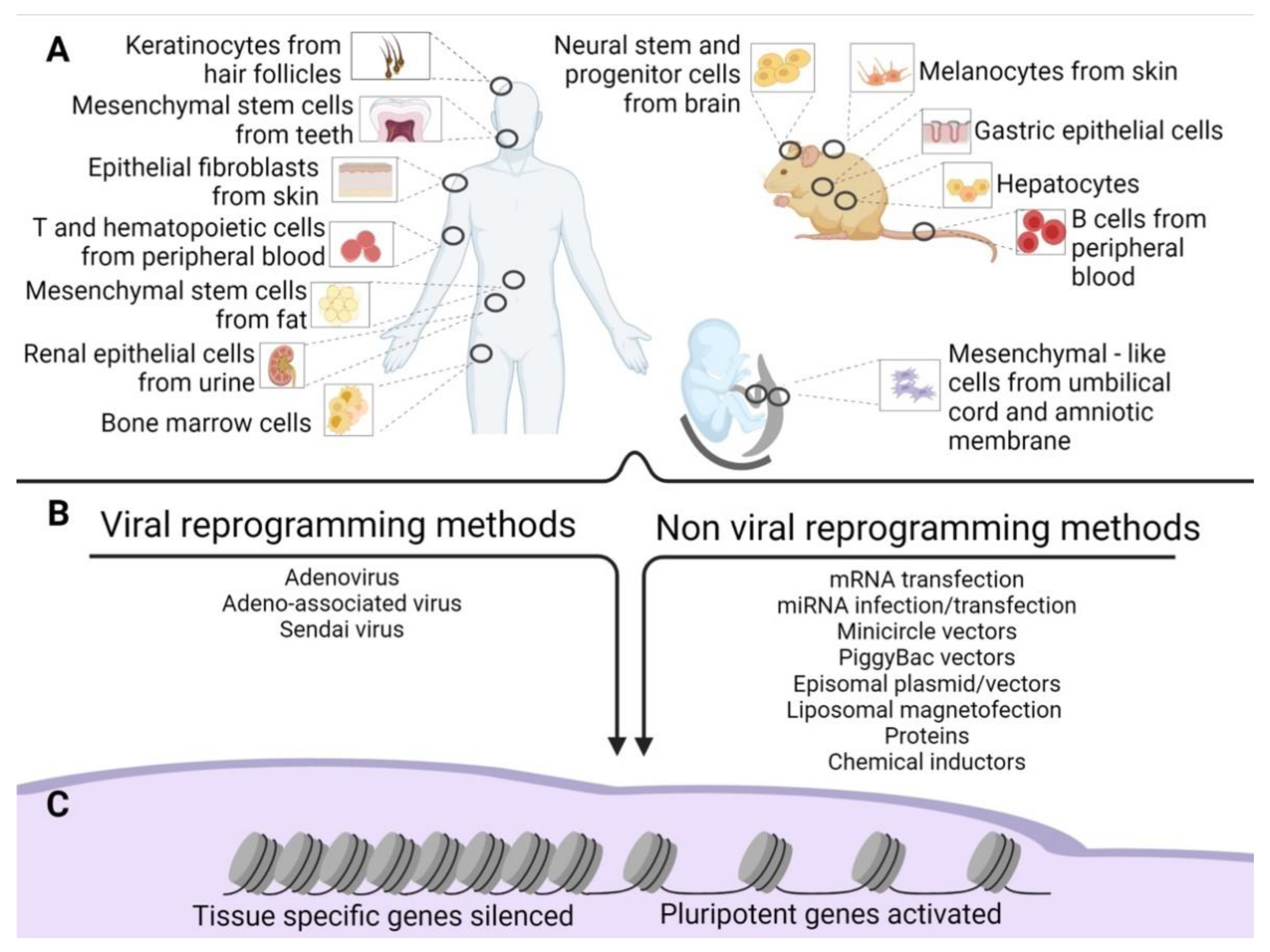
3. Source Cells for Reprogramming
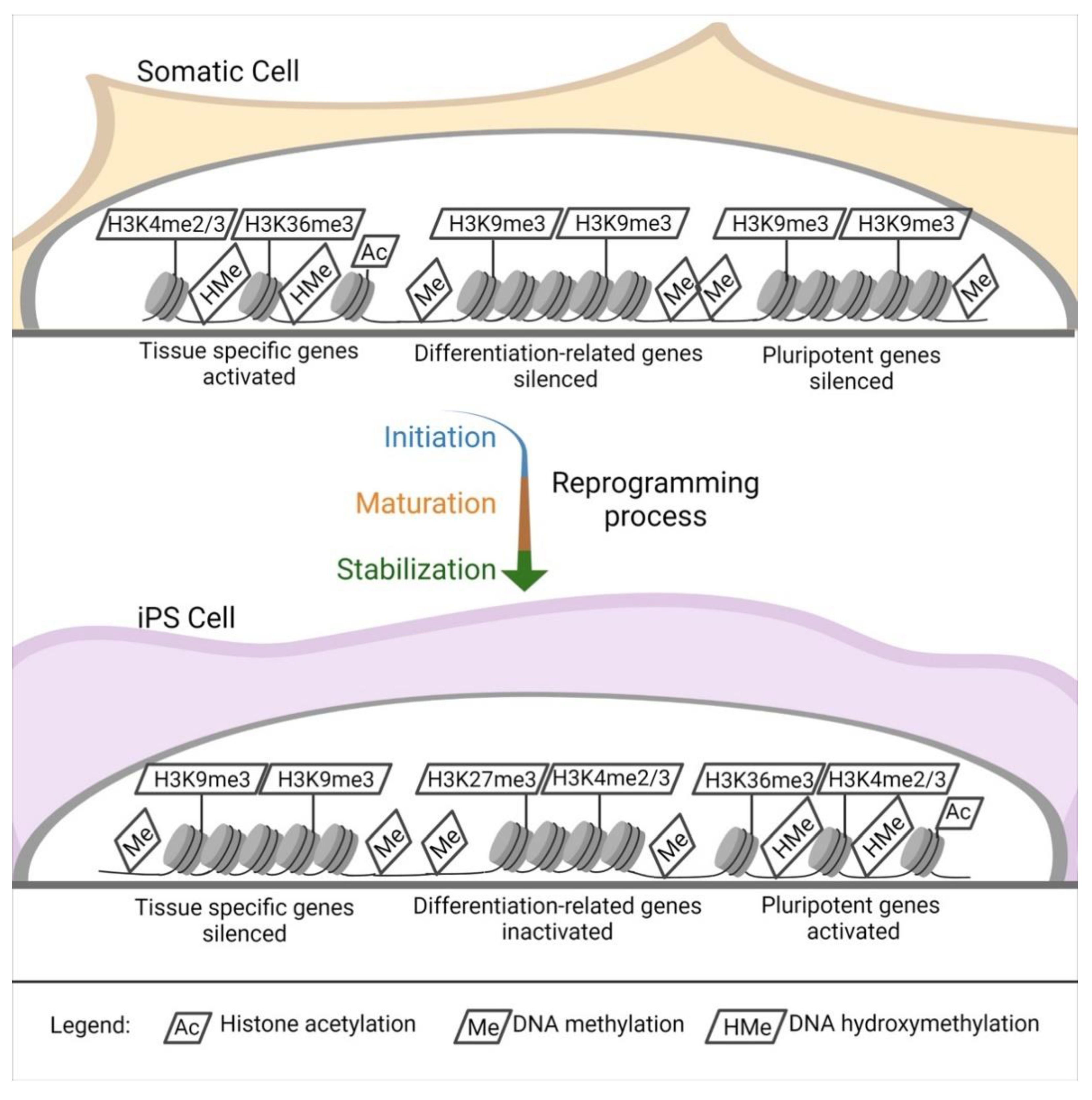
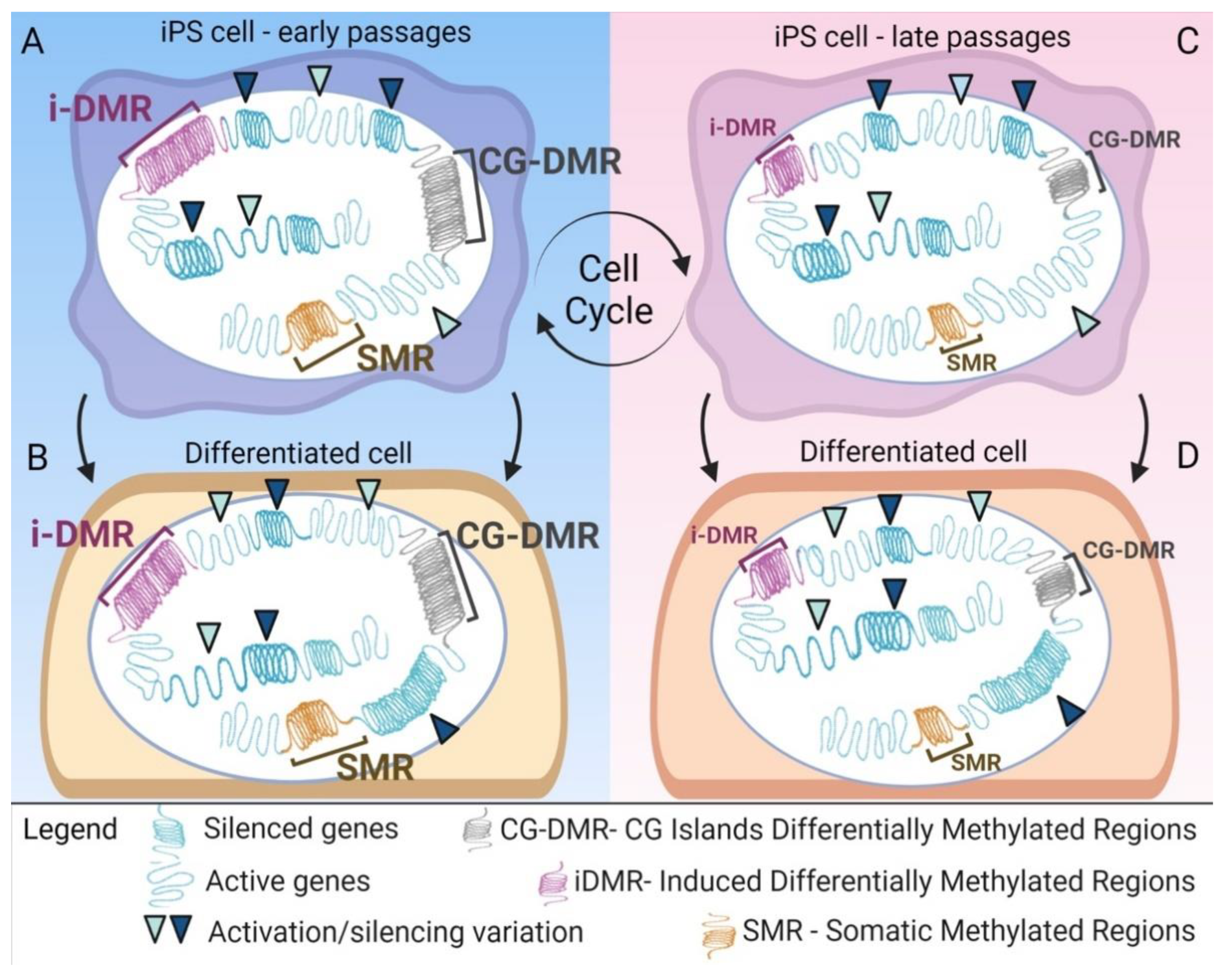
4. Epigenetic Alteration after Reprogramming
5. Similarities and Differences between iPSCs and ESCs
6. Epigenetic Memory Biases iPSCs Differentiation towards the Cell of Origin
7. Genetic Differences Drive Phenotypical Differences in iPSCs vs. ESCs
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Wernig, M.; Meissner, A.; Foreman, R.; Brambrink, T.; Ku, M.; Hochedlinger, K.; Bernstein, B.E.; Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Bayart, E.; Cohen-Haguenauer, O. Technological overview of iPS induction from human adult somatic cells. Curr. Gene Ther. 2013, 13, 73–92. [Google Scholar] [CrossRef]
- Eguchi, T.; Kuboki, T. Cellular Reprogramming Using Defined Factors and MicroRNAs. Stem Cells Int. 2016, 2016, 7530942. [Google Scholar] [CrossRef]
- El-Sayed, A.K.; Zhang, Z.; Zhang, L.; Liu, Z.; Abbott, L.C.; Zhang, Y.; Li, B. Pluripotent state induction in mouse embryonic fibroblast using mRNAs of reprogramming factors. Int. J. Mol. Sci. 2014, 15, 21840–21864. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Li, Y.; Zhang, X.; Liu, C.; Guan, J.; Li, H.; Zhao, T.; Ye, J.; Yang, W.; Liu, K.; et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef]
- Kim, D.; Kim, C.H.; Moon, J.I.; Chung, Y.G.; Chang, M.Y.; Han, B.S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 2009, 4, 472–476. [Google Scholar] [CrossRef]
- Lin, S.L.; Chang, D.C.; Chang-Lin, S.; Lin, C.H.; Wu, D.T.; Chen, D.T.; Ying, S.Y. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 2008, 14, 2115–2124. [Google Scholar] [CrossRef]
- Malik, N.; Rao, M.S. A review of the methods for human iPSC derivation. Methods Mol. Biol. 2013, 997, 23–33. [Google Scholar] [CrossRef]
- Rivera, T.; Zhao, Y.; Ni, Y.; Wang, J. Human-Induced Pluripotent Stem Cell Culture Methods Under cGMP Conditions. Curr. Protoc. Stem Cell Biol. 2020, 54, e117. [Google Scholar] [CrossRef]
- Stadtfeld, M.; Nagaya, M.; Utikal, J.; Weir, G.; Hochedlinger, K. Induced pluripotent stem cells generated without viral integration. Science 2008, 322, 945–949. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Freed, C.R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 2009, 27, 2667–2674. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Nakagawa, M.; Hyenjong, H.; Ichisaka, T.; Yamanaka, S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008, 322, 949–953. [Google Scholar] [CrossRef]
- Haridhasapavalan, K.K.; Borgohain, M.P.; Dey, C.; Saha, B.; Narayan, G.; Kumar, S.; Thummer, R.P. An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene 2019, 686, 146–159. [Google Scholar] [CrossRef]
- Nagai, Y. Paramyxovirus replication and pathogenesis. Reverse genetics transforms understanding. Rev. Med. Virol. 1999, 9, 83–99. [Google Scholar] [CrossRef]
- Li, H.O.; Zhu, Y.F.; Asakawa, M.; Kuma, H.; Hirata, T.; Ueda, Y.; Lee, Y.S.; Fukumura, M.; Iida, A.; Kato, A.; et al. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J. Virol. 2000, 74, 6564–6569. [Google Scholar] [CrossRef]
- Yonemitsu, Y.; Kitson, C.; Ferrari, S.; Farley, R.; Griesenbach, U.; Judd, D.; Steel, R.; Scheid, P.; Zhu, J.; Jeffery, P.K.; et al. Efficient gene transfer to airway epithelium using recombinant Sendai virus. Nat. Biotechnol. 2000, 18, 970–973. [Google Scholar] [CrossRef]
- Ban, H.; Nishishita, N.; Fusaki, N.; Tabata, T.; Saeki, K.; Shikamura, M.; Takada, N.; Inoue, M.; Hasegawa, M.; Kawamata, S.; et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. USA 2011, 108, 14234–14239. [Google Scholar] [CrossRef]
- Isomi, M.; Sadahiro, T.; Fujita, R.; Abe, Y.; Yamada, Y.; Akiyama, T.; Mizukami, H.; Shu, T.; Fukuda, K.; Ieda, M. Direct reprogramming with Sendai virus vectors repaired infarct hearts at the chronic stage. Biochem. Biophys. Res. Commun. 2021, 560, 87–92. [Google Scholar] [CrossRef]
- Honda, T.; Ando, M.; Ando, J.; Ishii, M.; Sakiyama, Y.; Ohara, K.; Toyota, T.; Ohtaka, M.; Masuda, A.; Terao, Y.; et al. Sustainable Tumor-Suppressive Effect of iPSC-Derived Rejuvenated T Cells Targeting Cervical Cancers. Mol. Ther. 2020, 28, 2394–2405. [Google Scholar] [CrossRef]
- Okumura, T.; Horie, Y.; Lai, C.Y.; Lin, H.T.; Shoda, H.; Natsumoto, B.; Fujio, K.; Kumaki, E.; Okano, T.; Ono, S.; et al. Robust and highly efficient hiPSC generation from patient non-mobilized peripheral blood-derived CD34. Stem Cell Res. Ther. 2019, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 348–362. [Google Scholar] [CrossRef]
- Seki, T.; Yuasa, S.; Oda, M.; Egashira, T.; Yae, K.; Kusumoto, D.; Nakata, H.; Tohyama, S.; Hashimoto, H.; Kodaira, M.; et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell 2010, 7, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Seki, T.; Yuasa, S.; Fukuda, K. Derivation of induced pluripotent stem cells from human peripheral circulating T cells. Curr. Protoc. Stem Cell Biol. 2011, 18, 4A-3. [Google Scholar] [CrossRef]
- Adami, R.; Bottai, D. Spinal Muscular Atrophy Modeling and Treatment Advances by Induced Pluripotent Stem Cells Studies. Stem Cell Rev. Rep. 2019, 15, 795–813. [Google Scholar] [CrossRef] [PubMed]
- Beers, J.; Linask, K.L.; Chen, J.A.; Siniscalchi, L.I.; Lin, Y.; Zheng, W.; Rao, M.; Chen, G. A cost-effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci. Rep. 2015, 5, 11319. [Google Scholar] [CrossRef] [PubMed]
- Schlaeger, T.M.; Daheron, L.; Brickler, T.R.; Entwisle, S.; Chan, K.; Cianci, A.; DeVine, A.; Ettenger, A.; Fitzgerald, K.; Godfrey, M.; et al. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015, 33, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Ohtaka, M.; Takada, H.; Kurisaki, A.; Tran, N.V.K.; Tran, Y.T.H.; Hisatake, K.; Sano, M.; Nakanishi, M. Simple and effective generation of transgene-free induced pluripotent stem cells using an auto-erasable Sendai virus vector responding to microRNA-302. Stem Cell Res. 2017, 23, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Al Abbar, A.; Ngai, S.C.; Nograles, N.; Alhaji, S.Y.; Abdullah, S. Induced Pluripotent Stem Cells: Reprogramming Platforms and Applications in Cell Replacement Therapy. Biores. Open Access 2020, 9, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Wu, W.S. Gene-delivery systems for iPS cell generation. Expert Opin. Biol. Ther. 2010, 10, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Martino, A.T.; Markusic, D.M. Immune Response Mechanisms against AAV Vectors in Animal Models. Mol. Ther. Methods Clin. Dev. 2020, 17, 198–208. [Google Scholar] [CrossRef]
- Li, C.; Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef]
- Hirsch, M.L.; Wolf, S.J.; Samulski, R.J. Delivering Transgenic DNA Exceeding the Carrying Capacity of AAV Vectors. Methods Mol. Biol. 2016, 1382, 21–39. [Google Scholar] [CrossRef]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef]
- Subramanyam, D.; Lamouille, S.; Judson, R.L.; Liu, J.Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 443–448. [Google Scholar] [CrossRef]
- Anokye-Danso, F.; Trivedi, C.M.; Juhr, D.; Gupta, M.; Cui, Z.; Tian, Y.; Zhang, Y.; Yang, W.; Gruber, P.J.; Epstein, J.A.; et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011, 8, 376–388. [Google Scholar] [CrossRef]
- Hu, K. Vectorology and factor delivery in induced pluripotent stem cell reprogramming. Stem Cells Dev. 2014, 23, 1301–1315. [Google Scholar] [CrossRef]
- Kaji, K.; Norrby, K.; Paca, A.; Mileikovsky, M.; Mohseni, P.; Woltjen, K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 2009, 458, 771–775. [Google Scholar] [CrossRef]
- Woltjen, K.; Michael, I.P.; Mohseni, P.; Desai, R.; Mileikovsky, M.; Hämäläinen, R.; Cowling, R.; Wang, W.; Liu, P.; Gertsenstein, M.; et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 2009, 458, 766–770. [Google Scholar] [CrossRef]
- Mali, P.; Chou, B.K.; Yen, J.; Ye, Z.; Zou, J.; Dowey, S.; Brodsky, R.A.; Ohm, J.E.; Yu, W.; Baylin, S.B.; et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 2010, 28, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Mátés, L.; Chuah, M.K.; Belay, E.; Jerchow, B.; Manoj, N.; Acosta-Sanchez, A.; Grzela, D.P.; Schmitt, A.; Becker, K.; Matrai, J.; et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009, 41, 753–761. [Google Scholar] [CrossRef]
- Nanbo, A.; Sugden, A.; Sugden, B. The coupling of synthesis and partitioning of EBV’s plasmid replicon is revealed in live cells. EMBO J. 2007, 26, 4252–4262. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Yu, J.; Suknuntha, K.; Tian, S.; Montgomery, K.; Choi, K.D.; Stewart, R.; Thomson, J.A.; Slukvin, I.I. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood 2011, 117, e109–e119. [Google Scholar] [CrossRef]
- Gonzalez, F.; Barragan Monasterio, M.; Tiscornia, G.; Montserrat Pulido, N.; Vassena, R.; Batlle Morera, L.; Rodriguez Piza, I.; Izpisua Belmonte, J.C. Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proc. Natl. Acad. Sci. USA 2009, 106, 8918–8922. [Google Scholar] [CrossRef]
- Luke, G.; Escuin, H.; De Felipe, P.; Ryan, M. 2A to the fore—Research, technology and applications. Biotechnol. Genet. Eng. Rev. 2010, 26, 223–260. [Google Scholar] [CrossRef]
- Wen, W.; Zhang, J.P.; Xu, J.; Su, R.J.; Neises, A.; Ji, G.Z.; Yuan, W.; Cheng, T.; Zhang, X.B. Enhanced Generation of Integration-free iPSCs from Human Adult Peripheral Blood Mononuclear Cells with an Optimal Combination of Episomal Vectors. Stem Cell Rep. 2016, 6, 873–884. [Google Scholar] [CrossRef]
- Narsinh, K.H.; Jia, F.; Robbins, R.C.; Kay, M.A.; Longaker, M.T.; Wu, J.C. Generation of adult human induced pluripotent stem cells using nonviral minicircle DNA vectors. Nat. Protoc. 2011, 6, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, M.; Zhou, H.; Nadif Kasri, N. Choices for Induction of Pluripotency: Recent Developments in Human Induced Pluripotent Stem Cell Reprogramming Strategies. Stem Cell Rev. Rep. 2016, 12, 54–72. [Google Scholar] [CrossRef]
- Chabot, S.; Orio, J.; Schmeer, M.; Schleef, M.; Golzio, M.; Teissié, J. Minicircle DNA electrotransfer for efficient tissue-targeted gene delivery. Gene Ther. 2013, 20, 62–68. [Google Scholar] [CrossRef]
- Ma, X.; Kong, L.; Zhu, S. Reprogramming cell fates by small molecules. Protein Cell 2017, 8, 328–348. [Google Scholar] [CrossRef]
- Yoshida, Y.; Takahashi, K.; Okita, K.; Ichisaka, T.; Yamanaka, S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 2009, 5, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Scherer, F.; Anton, M.; Schillinger, U.; Henke, J.; Bergemann, C.; Krüger, A.; Gänsbacher, B.; Plank, C. Magnetofection: Enhancing and targeting gene delivery by magnetic force in vitro and in vivo. Gene Ther. 2002, 9, 102–109. [Google Scholar] [CrossRef]
- Park, H.Y.; Noh, E.H.; Chung, H.M.; Kang, M.J.; Kim, E.Y.; Park, S.P. Efficient generation of virus-free iPS cells using liposomal magnetofection. PLoS ONE 2012, 7, e45812. [Google Scholar] [CrossRef]
- Nordin, F.; Ahmad, R.N.R.; Farzaneh, F. Transactivator protein: An alternative for delivery of recombinant proteins for safer reprogramming of induced Pluripotent Stem Cell. Virus Res. 2017, 235, 106–114. [Google Scholar] [CrossRef]
- Zhou, H.; Wu, S.; Joo, J.Y.; Zhu, S.; Han, D.W.; Lin, T.; Trauger, S.; Bien, G.; Yao, S.; Zhu, Y.; et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 2009, 4, 381–384. [Google Scholar] [CrossRef]
- Ford, E.; Pearlman, J.; Ruan, T.; Manion, J.; Waller, M.; Neely, G.G.; Caron, L. Human Pluripotent Stem Cells-Based Therapies for Neurodegenerative Diseases: Current Status and Challenges. Cells 2020, 9, 2517. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jeong, J.; Choi, D. Small-molecule-mediated reprogramming: A silver lining for regenerative medicine. Exp. Mol. Med. 2020, 52, 213–226. [Google Scholar] [CrossRef]
- Baranek, M.; Markiewicz, W.T.; Barciszewski, J. Selected small molecules as inducers of pluripotency. Acta Biochim. Pol. 2016, 63, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Fu, Y.; Zhu, J.; Liu, Y.; Zhang, Q.; Yi, Z.; Chen, S.; Jiao, Z.; Xu, X.; Xu, J.; et al. Single-Cell RNA-Seq Reveals Dynamic Early Embryonic-like Programs during Chemical Reprogramming. Cell Stem Cell 2018, 23, 31–45.e37. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Guo, Y.; Li, C.; Li, S.; Wan, X. Small Molecules that Promote Self-Renewal of Stem Cells and Somatic Cell Reprogramming. Stem Cell Rev. Rep. 2020, 16, 511–523. [Google Scholar] [CrossRef]
- Steichen, C.; Hannoun, Z.; Luce, E.; Hauet, T.; Dubart-Kupperschmitt, A. Genomic integrity of human induced pluripotent stem cells: Reprogramming, differentiation and applications. World J. Stem Cells 2019, 11, 729–747. [Google Scholar] [CrossRef]
- Salomonis, N.; Dexheimer, P.J.; Omberg, L.; Schroll, R.; Bush, S.; Huo, J.; Schriml, L.; Ho Sui, S.; Keddache, M.; Mayhew, C.; et al. Integrated Genomic Analysis of Diverse Induced Pluripotent Stem Cells from the Progenitor Cell Biology Consortium. Stem Cell Rep. 2016, 7, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Abyzov, A.; Mariani, J.; Palejev, D.; Zhang, Y.; Haney, M.S.; Tomasini, L.; Ferrandino, A.F.; Rosenberg Belmaker, L.A.; Szekely, A.; Wilson, M.; et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature 2012, 492, 438–442. [Google Scholar] [CrossRef]
- Ma, H.; Morey, R.; O’Neil, R.C.; He, Y.; Daughtry, B.; Schultz, M.D.; Hariharan, M.; Nery, J.R.; Castanon, R.; Sabatini, K.; et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 2014, 511, 177–183. [Google Scholar] [CrossRef]
- Cheng, L.; Hansen, N.F.; Zhao, L.; Du, Y.; Zou, C.; Donovan, F.X.; Chou, B.K.; Zhou, G.; Li, S.; Dowey, S.N.; et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell 2012, 10, 337–344. [Google Scholar] [CrossRef]
- Park, H.; Kim, D.; Kim, C.H.; Mills, R.E.; Chang, M.Y.; Iskow, R.C.; Ko, S.; Moon, J.I.; Choi, H.W.; Man Yoo, P.S.; et al. Increased genomic integrity of an improved protein-based mouse induced pluripotent stem cell method compared with current viral-induced strategies. Stem Cells Transl. Med 2014, 3, 599–609. [Google Scholar] [CrossRef]
- Bhutani, K.; Nazor, K.L.; Williams, R.; Tran, H.; Dai, H.; Džakula, Ž.; Cho, E.H.; Pang, A.W.; Rao, M.; Cao, H.; et al. Whole-genome mutational burden analysis of three pluripotency induction methods. Nat. Commun. 2016, 7, 10536. [Google Scholar] [CrossRef]
- Hussein, S.M.; Batada, N.N.; Vuoristo, S.; Ching, R.W.; Autio, R.; Närvä, E.; Ng, S.; Sourour, M.; Hämäläinen, R.; Olsson, C.; et al. Copy number variation and selection during reprogramming to pluripotency. Nature 2011, 471, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Markoulaki, S.; Schorderet, P.; Carey, B.W.; Beard, C.; Wernig, M.; Creyghton, M.P.; Steine, E.J.; Cassady, J.P.; Foreman, R.; et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 2008, 133, 250–264. [Google Scholar] [CrossRef]
- Loh, Y.H.; Agarwal, S.; Park, I.H.; Urbach, A.; Huo, H.; Heffner, G.C.; Kim, K.; Miller, J.D.; Ng, K.; Daley, G.Q. Generation of induced pluripotent stem cells from human blood. Blood 2009, 113, 5476–5479. [Google Scholar] [CrossRef]
- Maherali, N.; Ahfeldt, T.; Rigamonti, A.; Utikal, J.; Cowan, C.; Hochedlinger, K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 2008, 3, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Zahumenska, R.; Nosal, V.; Smolar, M.; Okajcekova, T.; Skovierova, H.; Strnadel, J.; Halasova, E. Induced Pluripotency: A Powerful Tool for In Vitro Modeling. Int. J. Mol. Sci. 2020, 21, 8910. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Benda, C.; Duzinger, S.; Huang, Y.; Li, X.; Li, Y.; Guo, X.; Cao, G.; Chen, S.; Hao, L.; et al. Generation of induced pluripotent stem cells from urine. J. Am. Soc. Nephrol. 2011, 22, 1221–1228. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Panetta, N.J.; Gupta, D.M.; Wilson, K.D.; Lee, A.; Jia, F.; Hu, S.; Cherry, A.M.; Robbins, R.C.; Longaker, M.T.; et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc. Natl. Acad. Sci. USA 2009, 106, 15720–15725. [Google Scholar] [CrossRef]
- Aoi, T.; Yae, K.; Nakagawa, M.; Ichisaka, T.; Okita, K.; Takahashi, K.; Chiba, T.; Yamanaka, S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 2008, 321, 699–702. [Google Scholar] [CrossRef]
- Utikal, J.; Maherali, N.; Kulalert, W.; Hochedlinger, K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 2009, 122, 3502–3510. [Google Scholar] [CrossRef]
- Eminli, S.; Utikal, J.; Arnold, K.; Jaenisch, R.; Hochedlinger, K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells 2008, 26, 2467–2474. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Li, W.; Su, H.; Qin, D.; Yang, J.; Zhu, F.; Xu, J.; He, W.; Guo, X.; Labuda, K.; et al. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J. Biol. Chem. 2010, 285, 11227–11234. [Google Scholar] [CrossRef]
- Lapasset, L.; Milhavet, O.; Prieur, A.; Besnard, E.; Babled, A.; Aït-Hamou, N.; Leschik, J.; Pellestor, F.; Ramirez, J.M.; De Vos, J.; et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011, 25, 2248–2253. [Google Scholar] [CrossRef]
- Alle, Q.; Le Borgne, E.; Milhavet, O.; Lemaitre, J.M. Reprogramming: Emerging Strategies to Rejuvenate Aging Cells and Tissues. Int. J. Mol. Sci. 2021, 22, 3990. [Google Scholar] [CrossRef]
- Adami, R.; Scesa, G.; Bottai, D. Stem cell transplantation in neurological diseases: Improving effectiveness in animal models. Front. Cell Dev. Biol. 2014, 2, 17. [Google Scholar] [CrossRef]
- Weick, J.P.; Held, D.L.; Bonadurer, G.F.; Doers, M.E.; Liu, Y.; Maguire, C.; Clark, A.; Knackert, J.A.; Molinarolo, K.; Musser, M.; et al. Deficits in human trisomy 21 iPSCs and neurons. Proc. Natl. Acad. Sci. USA 2013, 110, 9962–9967. [Google Scholar] [CrossRef]
- Kang, L.; Gao, S. Generation of Viable Mice from Induced Pluripotent Stem Cells (iPSCs) Through Tetraploid Complementation. Methods Mol. Biol. 2015, 1330, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Gładych, M.; Andrzejewska, A.; Oleksiewicz, U.; Estécio, M.R. Epigenetic mechanisms of induced pluripotency. Contemp. Oncol. 2015, 19, A30–A38. [Google Scholar] [CrossRef] [PubMed]
- Van den Hurk, M.; Kenis, G.; Bardy, C.; van den Hove, D.L.; Gage, F.H.; Steinbusch, H.W.; Rutten, B.P. Transcriptional and epigenetic mechanisms of cellular reprogramming to induced pluripotency. Epigenomics 2016, 8, 1131–1149. [Google Scholar] [CrossRef]
- Apostolou, E.; Hochedlinger, K. Chromatin dynamics during cellular reprogramming. Nature 2013, 502, 462–471. [Google Scholar] [CrossRef]
- Orkin, S.H.; Hochedlinger, K. Chromatin connections to pluripotency and cellular reprogramming. Cell 2011, 145, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, G.J.; Bai, Y.; Fletcher, J.; Wilmut, I. Induced pluripotent stem cells: Epigenetic memories and practical implications. Mol. Hum. Reprod. 2010, 16, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Cevallos, R.R.; Hossain, M.E.; Zhang, R.; Hu, K. Evaluating Reprogramming Efficiency and Pluripotency of the Established Human iPSCS by Pluripotency Markers. Methods Mol. Biol. 2021, 2239, 235–249. [Google Scholar] [CrossRef]
- Marchetto, M.C.; Yeo, G.W.; Kainohana, O.; Marsala, M.; Gage, F.H.; Muotri, A.R. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE 2009, 4, e7076. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Kida, Y.S.; Hawkins, R.D.; Nery, J.R.; Hon, G.; Antosiewicz-Bourget, J.; O’Malley, R.; Castanon, R.; Klugman, S.; et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011, 471, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Saha, K.; Pando, B.; van Zon, J.; Lengner, C.J.; Creyghton, M.P.; van Oudenaarden, A.; Jaenisch, R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 2009, 462, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Wernig, M.; Jaenisch, R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 2007, 25, 1177–1181. [Google Scholar] [CrossRef] [PubMed]
- Ohi, Y.; Qin, H.; Hong, C.; Blouin, L.; Polo, J.M.; Guo, T.; Qi, Z.; Downey, S.L.; Manos, P.D.; Rossi, D.J.; et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell. Biol. 2011, 13, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Stadtfeld, M.; Apostolou, E.; Akutsu, H.; Fukuda, A.; Follett, P.; Natesan, S.; Kono, T.; Shioda, T.; Hochedlinger, K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 2010, 465, 175–181. [Google Scholar] [CrossRef]
- Lindoso, R.S.; Kasai-Brunswick, T.H.; Monnerat Cahli, G.; Collino, F.; Bastos Carvalho, A.; Campos de Carvalho, A.C.; Vieyra, A. Proteomics in the World of Induced Pluripotent Stem Cells. Cells 2019, 8, 703. [Google Scholar] [CrossRef]
- Enterina, J.R.; Enfield, K.S.S.; Anderson, C.; Marshall, E.A.; Ng, K.W.; Lam, W.L. DLK1-DIO3 imprinted locus deregulation in development, respiratory disease, and cancer. Expert Rev. Respir. Med. 2017, 11, 749–761. [Google Scholar] [CrossRef]
- Doi, A.; Park, I.H.; Wen, B.; Murakami, P.; Aryee, M.J.; Irizarry, R.; Herb, B.; Ladd-Acosta, C.; Rho, J.; Loewer, S.; et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009, 41, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Chin, M.H.; Mason, M.J.; Xie, W.; Volinia, S.; Singer, M.; Peterson, C.; Ambartsumyan, G.; Aimiuwu, O.; Richter, L.; Zhang, J.; et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 2009, 5, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.J.; Amano, T.; Zhang, J.; Chen, Y.E.; Tian, X.C. Acceptance of embryonic stem cells by a wide developmental range of mouse tetraploid embryos. Biol. Reprod. 2010, 83, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Eakin, G.S.; Hadjantonakis, A.K.; Papaioannou, V.E.; Behringer, R.R. Developmental potential and behavior of tetraploid cells in the mouse embryo. Dev. Biol. 2005, 288, 150–159. [Google Scholar] [CrossRef]
- Wen, D.; Saiz, N.; Rosenwaks, Z.; Hadjantonakis, A.K.; Rafii, S. Completely ES cell-derived mice produced by tetraploid complementation using inner cell mass (ICM) deficient blastocysts. PLoS ONE 2014, 9, e94730. [Google Scholar] [CrossRef]
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolou, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855. [Google Scholar] [CrossRef]
- Rizzi, R.; Di Pasquale, E.; Portararo, P.; Papait, R.; Cattaneo, P.; Latronico, M.V.; Altomare, C.; Sala, L.; Zaza, A.; Hirsch, E.; et al. Post-natal cardiomyocytes can generate iPS cells with an enhanced capacity toward cardiomyogenic re-differentation. Cell Death Differ. 2012, 19, 1162–1174. [Google Scholar] [CrossRef]
- Hu, S.; Zhao, M.T.; Jahanbani, F.; Shao, N.Y.; Lee, W.H.; Chen, H.; Snyder, M.P.; Wu, J.C. Effects of cellular origin on differentiation of human induced pluripotent stem cell-derived endothelial cells. JCI Insight 2016, 1. [Google Scholar] [CrossRef]
- Streckfuss-Bömeke, K.; Wolf, F.; Azizian, A.; Stauske, M.; Tiburcy, M.; Wagner, S.; Hübscher, D.; Dressel, R.; Chen, S.; Jende, J.; et al. Comparative study of human-induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes, and skin fibroblasts. Eur. Heart J. 2013, 34, 2618–2629. [Google Scholar] [CrossRef]
- Hargus, G.; Ehrlich, M.; Hallmann, A.L.; Kuhlmann, T. Human stem cell models of neurodegeneration: A novel approach to study mechanisms of disease development. Acta Neuropathol. 2014, 127, 151–173. [Google Scholar] [CrossRef]
- Hargus, G.; Ehrlich, M.; Araúzo-Bravo, M.J.; Hemmer, K.; Hallmann, A.L.; Reinhardt, P.; Kim, K.P.; Adachi, K.; Santourlidis, S.; Ghanjati, F.; et al. Origin-dependent neural cell identities in differentiated human iPSCs in vitro and after transplantation into the mouse brain. Cell Rep. 2014, 8, 1697–1703. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hiler, D.; Xu, B.; AlDiri, I.; Chen, X.; Zhou, X.; Griffiths, L.; Valentine, M.; Shirinifard, A.; Sablauer, A.; et al. Retinal Cell Type DNA Methylation and Histone Modifications Predict Reprogramming Efficiency and Retinogenesis in 3D Organoid Cultures. Cell Rep. 2018, 22, 2601–2614. [Google Scholar] [CrossRef] [PubMed]
- Hiler, D.; Chen, X.; Hazen, J.; Kupriyanov, S.; Carroll, P.A.; Qu, C.; Xu, B.; Johnson, D.; Griffiths, L.; Frase, S.; et al. Quantification of Retinogenesis in 3D Cultures Reveals Epigenetic Memory and Higher Efficiency in iPSCs Derived from Rod Photoreceptors. Cell Stem Cell 2015, 17, 101–115. [Google Scholar] [CrossRef]
- Choompoo, N.; Bartley, O.J.M.; Precious, S.V.; Vinh, N.N.; Schnell, C.; Garcia, A.; Roberton, V.H.; Williams, N.M.; Kemp, P.J.; Kelly, C.M.; et al. Induced pluripotent stem cells derived from the developing striatum as a potential donor source for cell replacement therapy for Huntington disease. Cytotherapy 2021, 23, 111–118. [Google Scholar] [CrossRef]
- Chlebanowska, P.; Sułkowski, M.; Skrzypek, K.; Tejchman, A.; Muszyńska, A.; Noroozi, R.; Majka, M. Origin of the Induced Pluripotent Stem Cells Affects Their Differentiation into Dopaminergic Neurons. Int. J. Mol. Sci. 2020, 21, 5705. [Google Scholar] [CrossRef] [PubMed]
- Pianezzi, E.; Altomare, C.; Bolis, S.; Balbi, C.; Torre, T.; Rinaldi, A.; Camici, G.G.; Barile, L.; Vassalli, G. Role of somatic cell sources in the maturation degree of human induced pluripotent stem cell-derived cardiomyocytes. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118538. [Google Scholar] [CrossRef] [PubMed]
- Karamboulas, C.; Swedani, A.; Ward, C.; Al-Madhoun, A.S.; Wilton, S.; Boisvenue, S.; Ridgeway, A.G.; Skerjanc, I.S. HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J. Cell Sci. 2006, 119, 4305–4314. [Google Scholar] [CrossRef]
- Osafune, K.; Caron, L.; Borowiak, M.; Martinez, R.J.; Fitz-Gerald, C.S.; Sato, Y.; Cowan, C.A.; Chien, K.R.; Melton, D.A. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008, 26, 313–315. [Google Scholar] [CrossRef]
- Bock, C.; Kiskinis, E.; Verstappen, G.; Gu, H.; Boulting, G.; Smith, Z.D.; Ziller, M.; Croft, G.F.; Amoroso, M.W.; Oakley, D.H.; et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011, 144, 439–452. [Google Scholar] [CrossRef]
- Rouhani, F.; Kumasaka, N.; de Brito, M.C.; Bradley, A.; Vallier, L.; Gaffney, D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014, 10, e1004432. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Lee, S.; Mallard, W.; Clement, K.; Tagliazucchi, G.M.; Lim, H.; Choi, I.Y.; Ferrari, F.; Tsankov, A.M.; Pop, R.; et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 2015, 33, 1173–1181. [Google Scholar] [CrossRef]
- Burrows, C.K.; Banovich, N.E.; Pavlovic, B.J.; Patterson, K.; Gallego Romero, I.; Pritchard, J.K.; Gilad, Y. Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs. PLoS Genet. 2016, 12, e1005793. [Google Scholar] [CrossRef] [PubMed]
- Kyttälä, A.; Moraghebi, R.; Valensisi, C.; Kettunen, J.; Andrus, C.; Pasumarthy, K.K.; Nakanishi, M.; Nishimura, K.; Ohtaka, M.; Weltner, J.; et al. Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem Cell Rep. 2016, 6, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Féraud, O.; Valogne, Y.; Melkus, M.W.; Zhang, Y.; Oudrhiri, N.; Haddad, R.; Daury, A.; Rocher, C.; Larbi, A.; Duquesnoy, P.; et al. Donor Dependent Variations in Hematopoietic Differentiation among Embryonic and Induced Pluripotent Stem Cell Lines. PLoS ONE 2016, 11, e0149291. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.R.; Mallon, B.S.; Fann, Y.C.; Chen, K.G. Multivariate meta-analysis reveals global transcriptomic signatures underlying distinct human naive-like pluripotent states. PLoS ONE 2021, 16, e0251461. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and primed pluripotent states. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scesa, G.; Adami, R.; Bottai, D. iPSC Preparation and Epigenetic Memory: Does the Tissue Origin Matter? Cells 2021, 10, 1470. https://doi.org/10.3390/cells10061470
Scesa G, Adami R, Bottai D. iPSC Preparation and Epigenetic Memory: Does the Tissue Origin Matter? Cells. 2021; 10(6):1470. https://doi.org/10.3390/cells10061470
Chicago/Turabian StyleScesa, Giuseppe, Raffaella Adami, and Daniele Bottai. 2021. "iPSC Preparation and Epigenetic Memory: Does the Tissue Origin Matter?" Cells 10, no. 6: 1470. https://doi.org/10.3390/cells10061470
APA StyleScesa, G., Adami, R., & Bottai, D. (2021). iPSC Preparation and Epigenetic Memory: Does the Tissue Origin Matter? Cells, 10(6), 1470. https://doi.org/10.3390/cells10061470