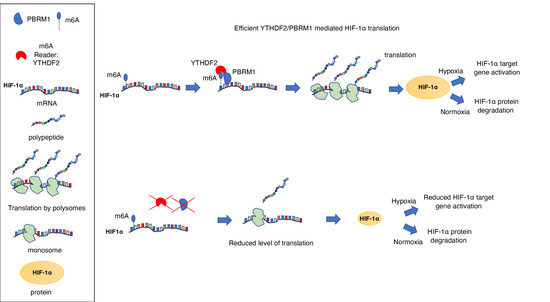
PBRM1 Cooperates with YTHDF2 to Control HIF-1α Protein Translation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment
2.2. Transfection of siRNA and DNA
2.3. Immunoblotting
2.4. Luciferase Assay
2.5. Quantitative PCR (qPCR)
2.6. Polysome Profiling
2.7. Coimmunoprecipitation
2.8. RNA Immunoprecipitation
2.9. In Vitro RNA Binding Assay
2.10. Recombinant Protein Expression and In Vitro Pulldown Assay
3. Results
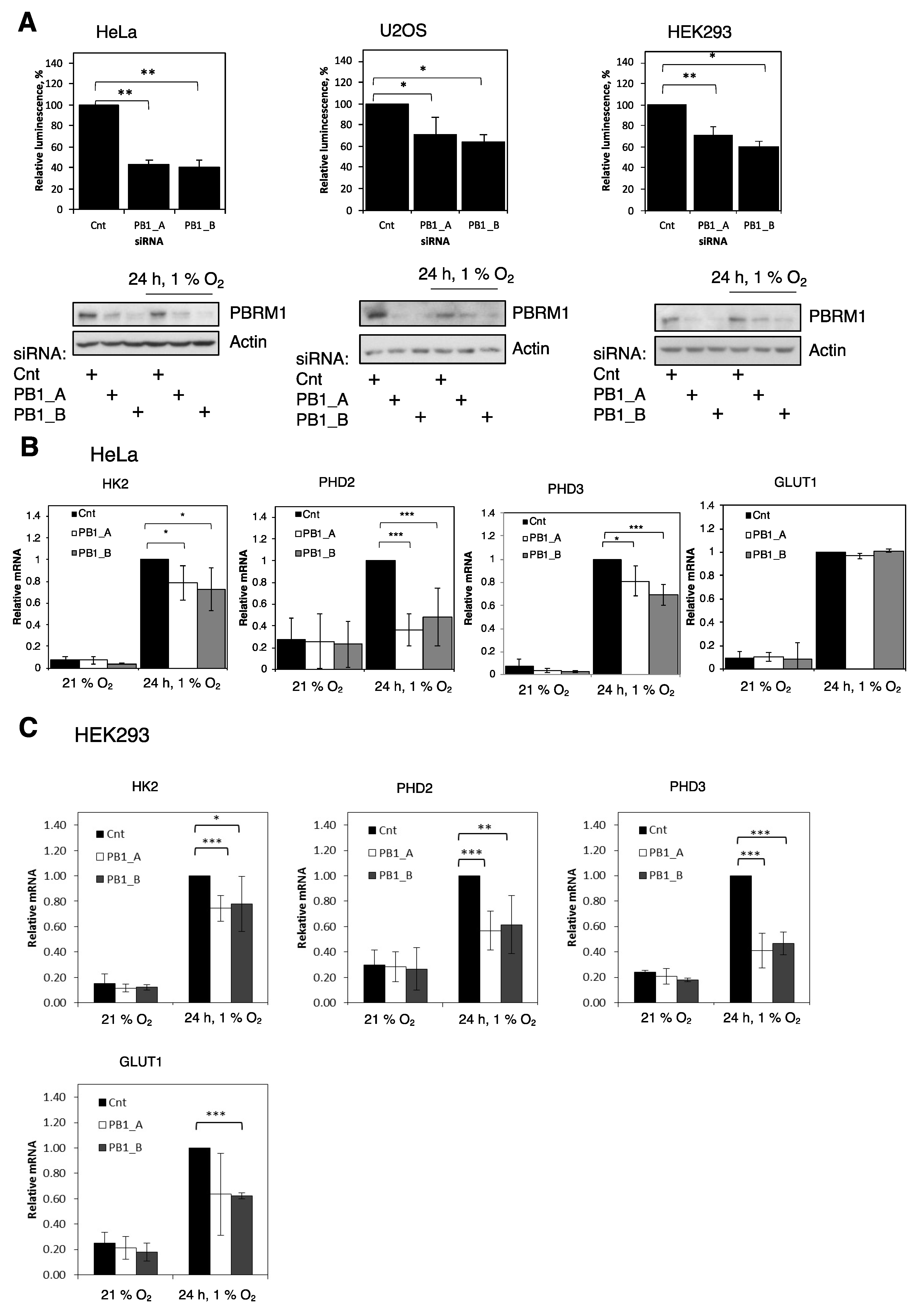
3.1. PBRM1 Is Required for HIF Transcriptional Activity
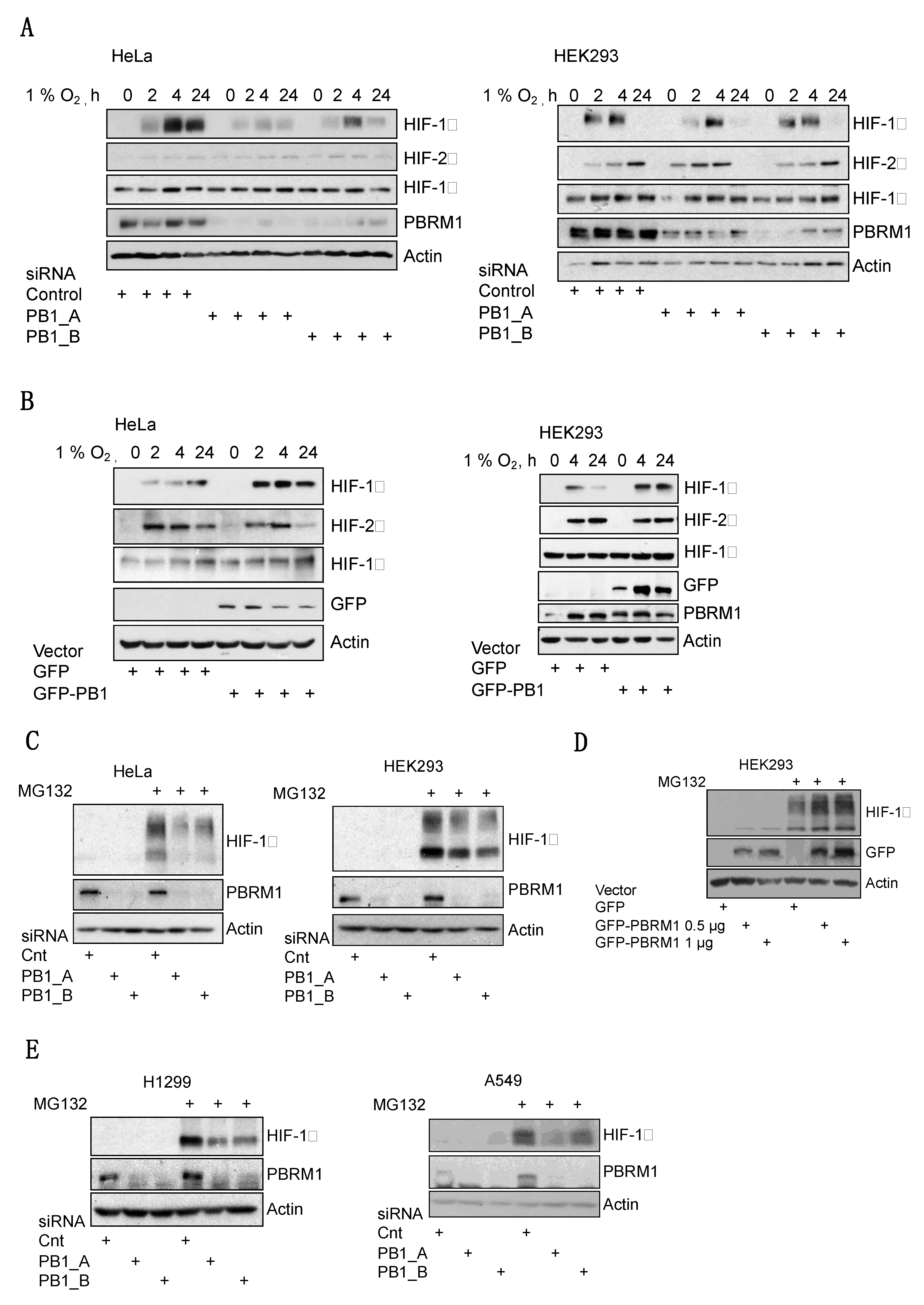
3.2. PBRM1 Is Required for HIF-1α Protein Expression in Normoxia and Hypoxia
3.3. PBRM1 Binds Selectively to HIF-1α mRNA and Promotes Polysome Processing
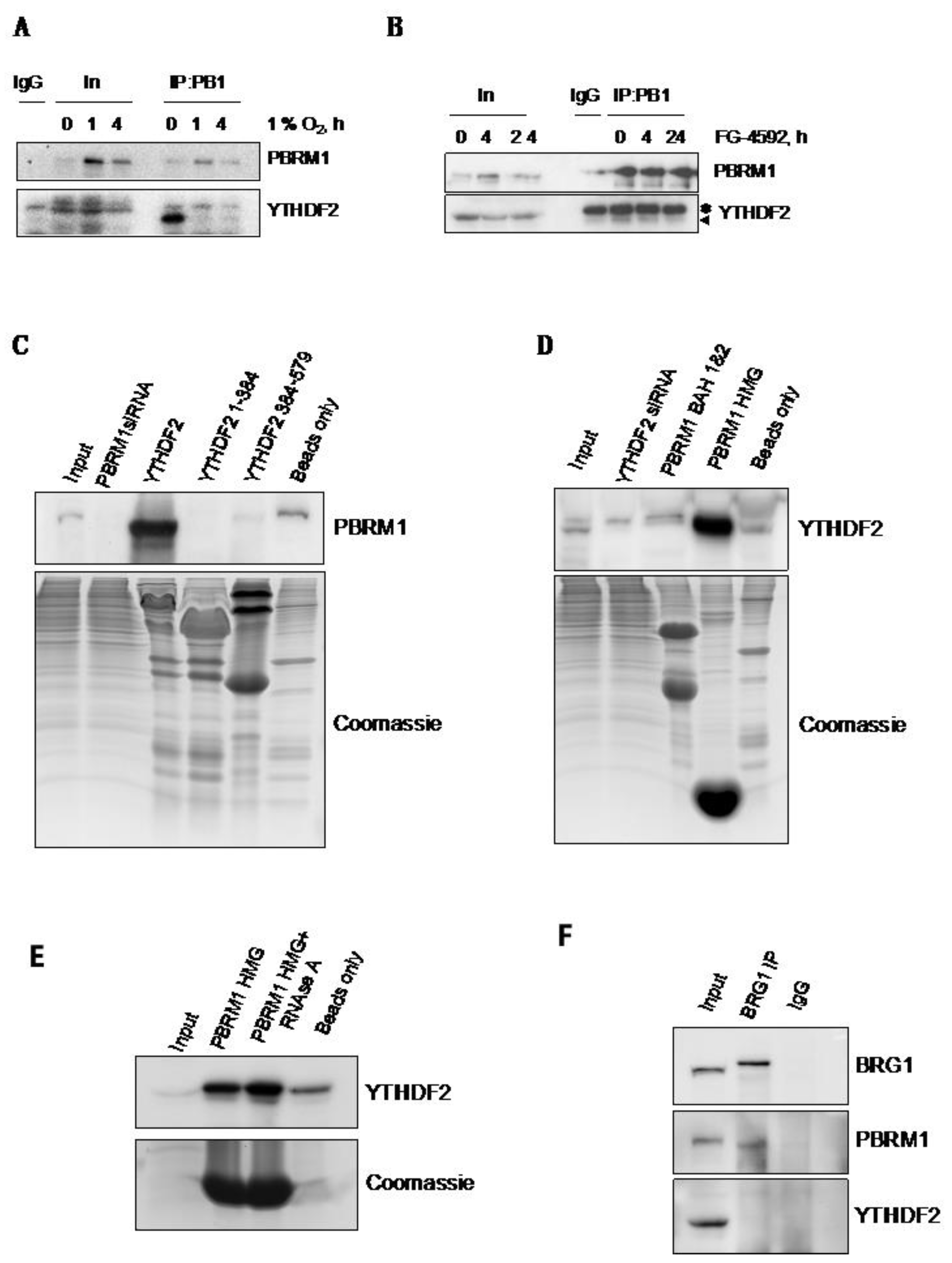
3.4. PBRM1 but Not BRG1 Can Bind to the m6A-Binding Protein YTHDF2
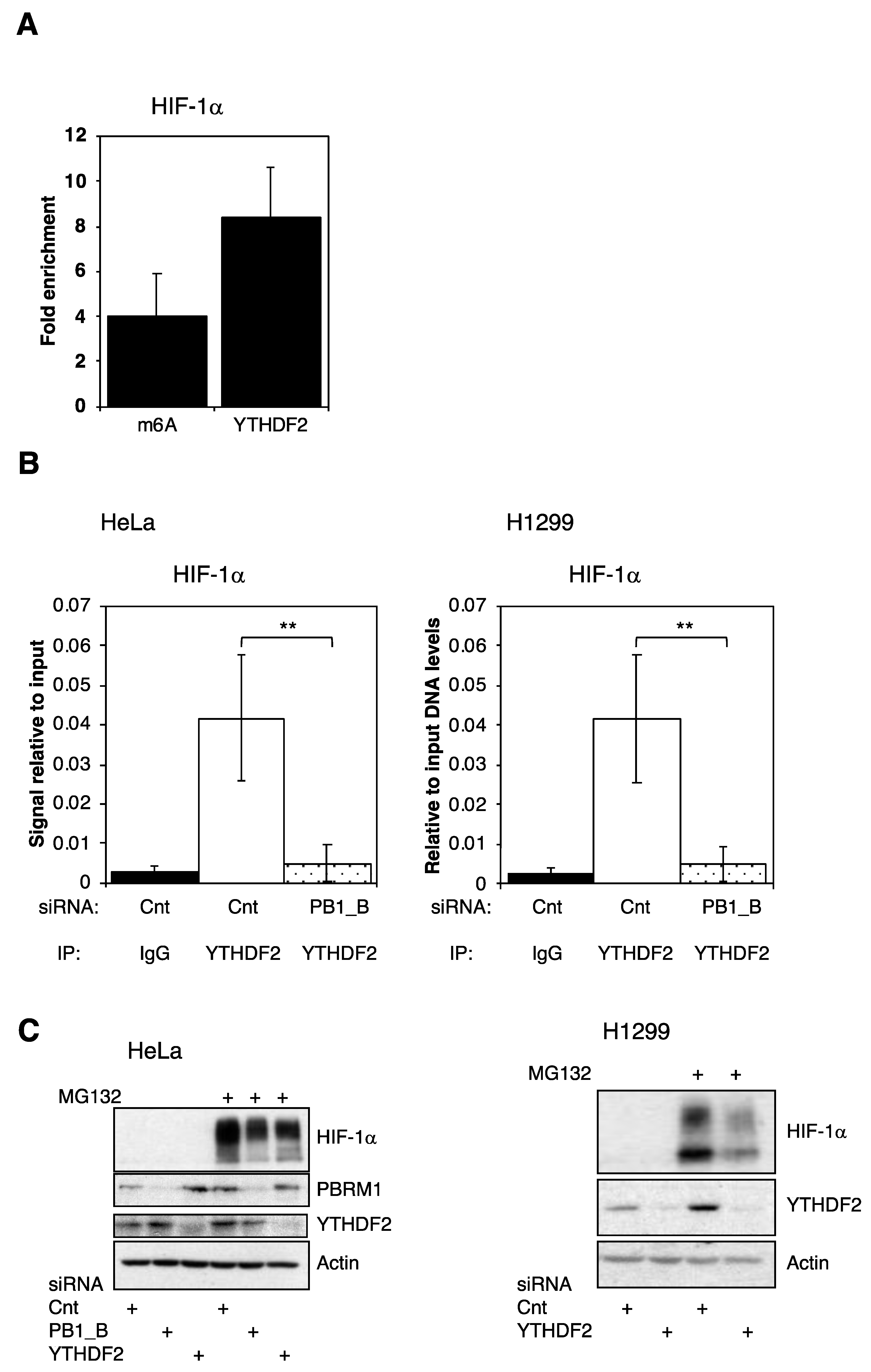
3.5. PBRM1 Is Required for YTHDF2 Binding to HIF-1α mRNA and for HIF-1α Protein Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gossage, L.; Eisen, T.; Maher, E.R. VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer 2015, 15, 55–64. [Google Scholar] [CrossRef]
- Rocha, S. Gene regulation under low oxygen: Holding your breath for transcription. Trends Biochem. Sci. 2007, 32, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and Functions of ATP-Dependent Chromatin-Remodeling Enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melvin, A.; Rocha, S. Chromatin as an oxygen sensor and active player in the hypoxia response. Cell. Signal. 2012, 24, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Shmakova, A.; Batie, M.; Druker, J.; Rocha, S. Chromatin and oxygen sensing in the context of JmjC histone demethylases. Biochem. J. 2014, 462, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Centore, R.C.; Sandoval, G.J.; Soares, L.M.M.; Kadoch, C.; Chan, H.M. Mammalian SWI/SNF Chromatin Remodeling Complexes: Emerging Mechanisms and Therapeutic Strategies. Trends Genet. 2020, 36, 936–950. [Google Scholar] [CrossRef]
- Kenneth, N.; Mudie, S.; van Uden, P.; Rocha, S. SWI/SNF Regulates the Cellular Response to Hypoxia*. J. Biol. Chem. 2009, 284, 4123–4131. [Google Scholar] [CrossRef] [Green Version]
- Sena, J.A.; Wang, L.; Hu, C.-J.; Kim, S.-G.; Lee, B.; Kim, D.-H.; Kim, J.; Lee, S.; Lee, S.-K.; Lee, J.W. BRG1 and BRM Chromatin-Remodeling Complexes Regulate the Hypoxia Response by Acting as Coactivators for a Subset of Hypoxia-Inducible Transcription Factor Target Genes. Mol. Cell. Biol. 2013, 33, 3849–3863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, B.; Porter, E.G.; Stewart, J.C.; Ferreira, C.R.; Schipma, M.J.; Dykhuizen, E.C. PBRM1 Regulates the Expression of Genes Involved in Metabolism and Cell Adhesion in Renal Clear Cell Carcinoma. PLoS ONE 2016, 11, e0153718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linehan, W.M.; Ricketts, C.J. The Cancer Genome Atlas of renal cell carcinoma: Findings and clinical implications. Nat. Rev. Urol. 2019, 16, 539–552. [Google Scholar] [CrossRef]
- Thompson, M. Polybromo-1: The chromatin targeting subunit of the PBAF complex. Biochimie 2009, 91, 309–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Canman, J.C.; Lee, C.S.; Nie, Z.; Yang, D.; Moreno, G.T.; Young, M.K.; Salmon, E.D.; Wang, W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast Rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13015–13020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brownlee, P.M.; Chambers, A.; Cloney, R.; Bianchi, A.; Downs, J.A. BAF180 Promotes Cohesion and Prevents Genome Instability and Aneuploidy. Cell Rep. 2014, 6, 973–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakarougkas, A.; Downs, J.A.; Jeggo, P.A. The PBAF chromatin remodeling complex represses transcription and promotes rapid repair at DNA double-strand breaks. Mol. Cell. Oncol. 2014, 2, e970072. [Google Scholar] [CrossRef]
- Yan, Z.; Cui, K.; Murray, D.M.; Ling, C.; Xue, Y.; Gerstein, A.; Parsons, R.; Zhao, K.; Wang, W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005, 19, 1662–1667. [Google Scholar] [CrossRef] [Green Version]
- Wurster, A.L.; Precht, P.; Becker, K.G.; Wood, W.H.; Zhang, Y.; Wang, Z.; Pazin, M.J. IL-10 transcription is negatively regulated by BAF180, a component of the SWI/SNF chromatin remodeling enzyme. BMC Immunol. 2012, 13, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Orvis, T.; Hepperla, A.; Walter, V.; Song, S.; Simon, J.; Parker, J.; Wilkerson, M.D.; Desai, N.; Major, M.B.; Hayes, D.N.; et al. BRG1/SMARCA4 Inactivation Promotes Non–Small Cell Lung Cancer Aggressiveness by Altering Chromatin Organization. Cancer Res. 2014, 74, 6486–6498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5′ Sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nat. Cell Biol. 2015, 526, 591–594. [Google Scholar] [CrossRef] [Green Version]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m 6 A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierre, R.S.; Kadoch, C. Mammalian SWI/SNF complexes in cancer: Emerging therapeutic opportunities. Curr. Opin. Genet. Dev. 2017, 42, 56–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cajigas, I.; Leib, D.; Cochrane, J.; Luo, H.; Swyter, K.R.; Chen, S.; Clark, B.S.; Thompson, J.; Yates, J.R.; Kingston, R.E.; et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent chromatin remodeling inhibition. Development 2015, 142, 2641–2652. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, A.; Ryme, J.; Brodin, D.; Farrants, A.K.; Östlund, V.N. SWI/SNF Associates with Nascent Pre-mRNPs and Regulates Alternative Pre-mRNA Processing. PLoS Genet. 2009, 5, e1000470. [Google Scholar] [CrossRef] [PubMed]
- Bentley, D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014, 15, 163–175. [Google Scholar] [CrossRef] [Green Version]
- Phatnani, H.P.; Greenleaf, A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006, 20, 2922–2936. [Google Scholar] [CrossRef] [Green Version]
- Xiao, T.; Hall, H.; Kizer, K.O.; Shibata, Y.; Hall, M.C.; Borchers, C.H.; Strahl, B.D. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003, 17, 654–663. [Google Scholar] [CrossRef] [Green Version]
- Perales, R.; Bentley, D. “Cotranscriptionality”: The Transcription Elongation Complex as a Nexus for Nuclear Transactions. Mol. Cell 2009, 36, 178–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batsché, E.; Yaniv, M.; Muchardt, C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2005, 13, 22–29. [Google Scholar] [CrossRef]
- Genzor, P.; Bortvin, A. A Unique HMG-Box Domain of Mouse Maelstrom Binds Structured RNA but Not Double Stranded DNA. PLoS ONE 2015, 10, e0120268. [Google Scholar] [CrossRef]
- Holmes, Z.E.; Hamilton, D.J.; Hwang, T.; Parsonnet, N.V.; Rinn, J.L.; Wuttke, D.S.; Batey, R.T. The Sox2 transcription factor binds RNA. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, G.R.; Rahal, R.; Buxton, F.; Xiang, K.; McAllister, G.; Frias, E.; Bagdasarian, L.; Huber, J.; Lindeman, A.; Chen, D.; et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 3128–3133. [Google Scholar] [CrossRef] [Green Version]
- Karki, M.; Jangid, R.K.; Anish, R.; Seervai, R.N.H.; Bertocchio, J.-P.; Hotta, T.; Msaouel, P.; Jung, S.Y.; Grimm, S.L.; Coarfa, C.; et al. A cytoskeletal function for PBRM1 reading methylated microtubules. Sci. Adv. 2021, 7, eabf2866. [Google Scholar] [CrossRef]
- Liu, L.; Cash, T.P.; Jones, R.G.; Keith, B.; Thompson, C.B.; Simon, M.C. Hypoxia-Induced Energy Stress Regulates mRNA Translation and Cell Growth. Mol. Cell 2006, 21, 521–531. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, I.G.; Park, C.V.; Kenneth, N.S. Translating the Hypoxic Response—the Role of HIF Protein Translation in the Cellular Response to Low Oxygen. Cells 2019, 8, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P.; Baginski, B.; Wirecki, T.K.; De Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nat. Cell Biol. 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nat. Cell Biol. 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Meyer, K.; Patil, D.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m6A Modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Frost, J.; Frost, M.; Batie, M.; Jiang, H.; Rocha, S. Roles of HIF and 2-Oxoglutarate-Dependent Dioxygenases in Controlling Gene Expression in Hypoxia. Cancers 2021, 13, 350. [Google Scholar] [CrossRef]
- Thalhammer, A.; Bencokova, Z.; Poole, R.; Loenarz, C.; Adam, J.; O’Flaherty, L.; Schödel, J.; Mole, D.; Giaslakiotis, K.; Schofield, C.J.; et al. Human AlkB Homologue 5 Is a Nuclear 2-Oxoglutarate Dependent Oxygenase and a Direct Target of Hypoxia-Inducible Factor 1α (HIF-1α). PLoS ONE 2011, 6, e16210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, J.; Zhang, H.; Liu, J.; Zhao, Z.; Wang, J.; Lu, Z.; Hu, B.; Zhou, J.; Zhao, Z.; Feng, M.; et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 2019, 18, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Y.; Qiao, Y.; Li, L.; Luo, E.; Wang, D.; Yao, Y.; Tang, C.; Yan, G. The m6A methyltransferase METTL3 promotes hypoxic pulmonary arterial hypertension. Life Sci. 2021, 274, 119366. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shmakova, A.; Frost, M.; Batie, M.; Kenneth, N.S.; Rocha, S. PBRM1 Cooperates with YTHDF2 to Control HIF-1α Protein Translation. Cells 2021, 10, 1425. https://doi.org/10.3390/cells10061425
Shmakova A, Frost M, Batie M, Kenneth NS, Rocha S. PBRM1 Cooperates with YTHDF2 to Control HIF-1α Protein Translation. Cells. 2021; 10(6):1425. https://doi.org/10.3390/cells10061425
Chicago/Turabian StyleShmakova, Alena, Mark Frost, Michael Batie, Niall S. Kenneth, and Sonia Rocha. 2021. "PBRM1 Cooperates with YTHDF2 to Control HIF-1α Protein Translation" Cells 10, no. 6: 1425. https://doi.org/10.3390/cells10061425
APA StyleShmakova, A., Frost, M., Batie, M., Kenneth, N. S., & Rocha, S. (2021). PBRM1 Cooperates with YTHDF2 to Control HIF-1α Protein Translation. Cells, 10(6), 1425. https://doi.org/10.3390/cells10061425