Patterns of Gene Expression in Cutaneous T-Cell Lymphoma: Systematic Review of Transcriptomic Studies in Mycosis Fungoides
Abstract
1. Introduction
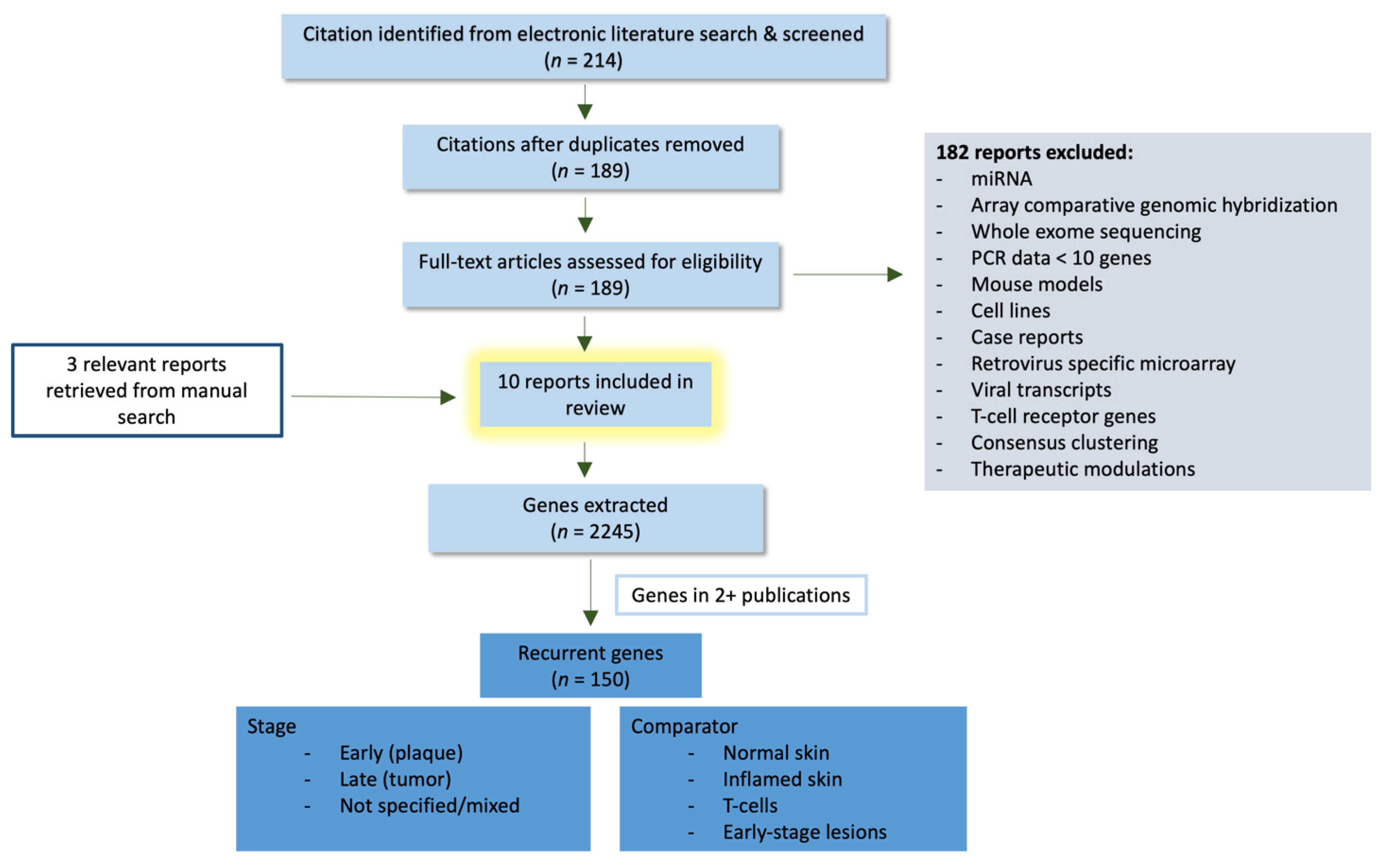
2. Methods
3. Results
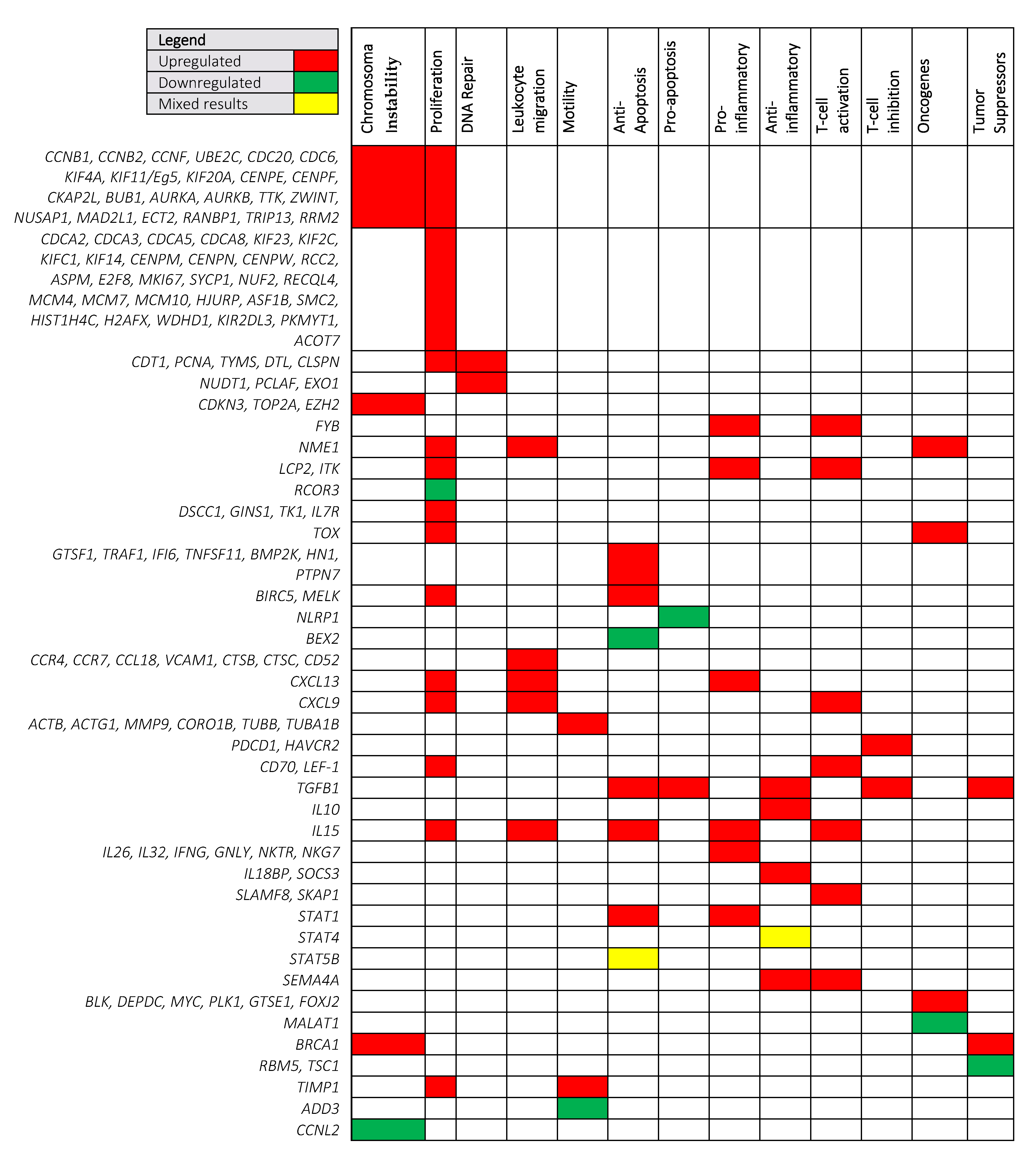
3.1. Shared Cancer Pathways Upregulated in MF
3.1.1. Chromosomal Instability and Proliferation
3.1.2. Leukocyte Migration and Motility
3.1.3. Pro-Tumorigenic Cytokines
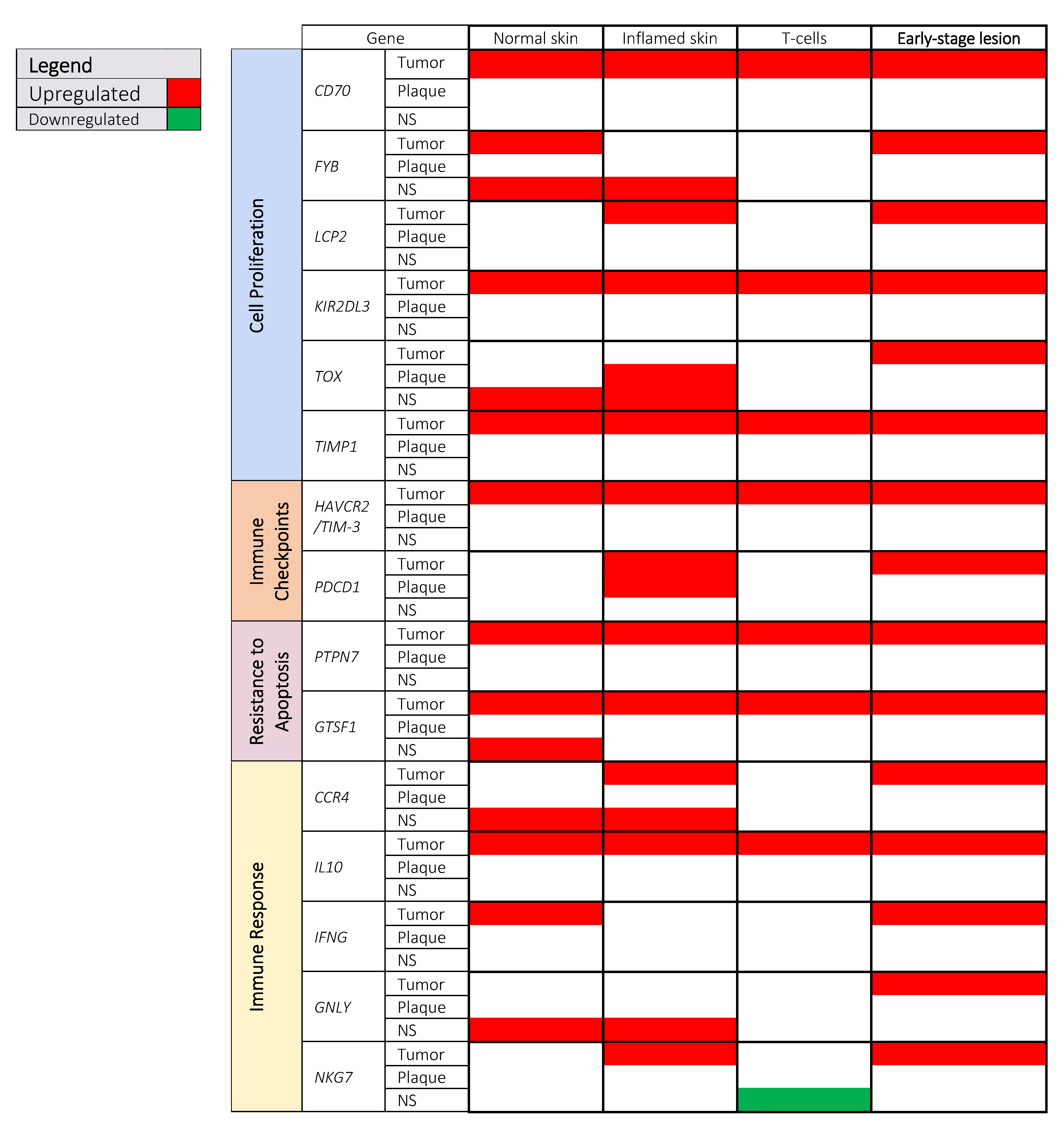
3.2. Prognostic Cancer Pathways in MF
3.2.1. Cell Proliferation
3.2.2. Immune Checkpoints
3.2.3. Resistance to Apoptosis
3.2.4. Immune Response
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willemze, R.; Cerroni, L.; Kempf, W.; Berti, E.; Facchetti, F.; Swerdlow, S.H.; Jaffe, E.S. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood 2019, 133, 1703–1714. [Google Scholar] [CrossRef] [PubMed]
- Talpur, R.; Singh, L.; Daulat, S.; Liu, P.; Seyfer, S.; Trynosky, T.; Wei, W.; Duvic, M. Long-term Outcomes of 1263 Patients with Mycosis Fungoides and Sézary Syndrome from 1982 to 2009. Clin. Cancer Res. 2012, 18, 5051–5060. [Google Scholar] [CrossRef]
- Scarisbrick, J.J.; Prince, H.M.; Vermeer, M.H.; Quaglino, P.; Horwitz, S.; Porcu, P.; Stadler, R.; Wood, G.S.; Beylot-Barry, M.; Pham-Ledard, A.; et al. Cutaneous Lymphoma International Consortium Study of Outcome in Advanced Stages of Mycosis Fungoides and Sézary Syndrome: Effect of Specific Prognostic Markers on Survival and Development of a Prognostic Model. J. Clin. Oncol. 2015, 33, 3766–3773. [Google Scholar] [CrossRef] [PubMed]
- Agar, N.S.; Wedgeworth, E.; Crichton, S.; Mitchell, T.J.; Cox, M.; Ferreira, S.; Robson, A.; Calonje, E.; Stefanato, C.M.; Wain, E.M.; et al. Survival Outcomes and Prognostic Factors in Mycosis fungoides/Sézary Syndrome: Validation of the Revised In-ternational Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer Staging Proposal. J. Clin. Oncol. 2010, 28, 4730–4739. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research, N.; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Iyer, A.; Hennessey, D.; O’Keefe, S.; Patterson, J.; Wang, W.; Wong, G.K.-S.; Gniadecki, R. Branched evolution and genomic intratumor heterogeneity in the pathogenesis of cutaneous T-cell lymphoma. Blood Adv. 2020, 4, 2489–2500. [Google Scholar] [CrossRef]
- Chang, L.-W.; Patrone, C.C.; Yang, W.; Rabionet, R.; Gallardo, F.; Espinet, B.; Sharma, M.K.; Girardi, M.; Tensen, C.P.; Vermeer, M.; et al. An Integrated Data Resource for Genomic Analysis of Cutaneous T-Cell Lymphoma. J. Investig. Dermatol. 2018, 138, 2681–2683. [Google Scholar] [CrossRef]
- Choi, J.; Goh, G.; Walradt, T.; Hong, B.S.; Bunick, C.G.; Chen, K.; Bjornson, R.D.; Maman, Y.; Wang, T.; Tordoff, J.; et al. Ge-nomic Landscape of Cutaneous T Cell Lymphoma. Nat. Genet. 2015, 47, 1011–1019. [Google Scholar] [CrossRef]
- Almeida, A.C.D.S.; Abate, F.; Khiabanian, H.; Martinez-Escala, E.; Guitart, J.; Tensen, C.P.; Vermeer, M.H.; Rabadan, R.; Ferrando, A.; Palomero, T. The mutational landscape of cutaneous T cell lymphoma and Sézary syndrome. Nat. Genet. 2015, 47, 1465–1470. [Google Scholar] [CrossRef]
- Ungewickell, A.; Bhaduri, A.; Rios, E.J.; Reuter, J.A.; Lee, C.S.; Mah, A.; Zehnder, A.M.; Ohgami, R.S.; Kulkarni, S.; Armstrong, R.; et al. Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR. Nat. Genet. 2015, 47, 1056–1060. [Google Scholar] [CrossRef]
- Campbell, J.; Clark, R.A.; Watanabe, R.; Kupper, T.S. Sézary syndrome and mycosis fungoides arise from distinct T-cell subsets: A biologic rationale for their distinct clinical behaviors. Blood 2010, 116, 767–771. [Google Scholar] [CrossRef]
- Van Doorn, R.; Van Kester, M.S.; Dijkman, R.; Vermeer, M.H.; Mulder, A.A.; Szuhai, K.; Knijnenburg, J.; Boer, J.M.; Willemze, R.; Tensen, C.P. Oncogenomic analysis of mycosis fungoides reveals major differences with Sézary syndrome. Blood 2009, 113, 127–136. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Yu, R.; Huang, Y.; Su, M.; Xiao, C.; Martinka, M.; Dutz, J.P.; Zhang, X.; Zheng, Z.; et al. Molecular Markers of Early-Stage Mycosis Fungoides. J. Investig. Dermatol. 2012, 132, 1698–1706. [Google Scholar] [CrossRef] [PubMed]
- Querfeld, C.; Leung, S.; Myskowski, P.L.; Curran, S.A.; Goldman, D.A.; Heller, G.; Wu, X.; Kil, S.H.; Sharma, S.; Finn, K.J.; et al. Primary T Cells from Cutaneous T-cell Lymphoma Skin Explants Display an Exhausted Immune Checkpoint Profile. Cancer Immunol. Res. 2018, 6, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Lefrançois, P.; Xie, P.; Wang, L.; Tetzlaff, M.T.; Moreau, L.; Watters, A.K.; Netchiporouk, E.; Provost, N.; Gilbert, M.; Ni, X.; et al. Gene expression profiling and immune cell-type deconvolution highlight robust disease progression and survival markers in multiple cohorts of CTCL patients. Oncoimmunology 2018, 7, e1467856. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, I.V.; Tetzlaff, M.T.; Thibault, P.; Gangar, P.; Moreau, L.; Watters, A.K.; Netchiporouk, E.; Pehr, K.; Prieto, V.G.; Rahme, E.; et al. Gene expression analysis in Cutaneous T-Cell Lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology 2017, 6, e1306618. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, I.V.; Cordeiro, B.; Huang, Y.; Zargham, H.; Pehr, K.; Doré, M.-A.; Gilbert, M.; Zhou, Y.; Kupper, T.S.; Sasseville, D. Ectopic Expression of Cancer–Testis Antigens in Cutaneous T-cell Lymphoma Patients. Clin. Cancer Res. 2014, 20, 3799–3808. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, I.V.; Netchiporouk, E.; Cordeiro, B.; Doré, M.-A.; Moreau, L.; Pehr, K.; Gilbert, M.; Zhou, Y.; Sasseville, D.; Kupper, T.S. The Use of Transcriptional Profiling to Improve Personalized Diagnosis and Management of Cutaneous T-Cell Lympho-ma (CTCL). Clin. Cancer Res. 2015, 21, 2820–2829. [Google Scholar] [CrossRef]
- Gaydosik, A.M.; Tabib, T.; Geskin, L.J.; Bayan, C.-A.; Conway, J.F.; Lafyatis, R.; Fuschiotti, P. Single-Cell Lymphocyte Heter-ogeneity in Advanced Cutaneous T-Cell Lymphoma Skin Tumors. Clin. Cancer Res. 2019, 25, 4443–4454. [Google Scholar] [CrossRef] [PubMed]
- Gaydosik, A.M.; Queen, D.S.; Trager, M.H.; Akilov, O.E.; Geskin, L.J.; Fuschiotti, P. Genome-wide transcriptome analysis of the STAT6-regulated genes in advanced-stage cutaneous T-cell lymphoma. Blood 2020, 136, 1748–1759. [Google Scholar] [CrossRef]
- Hahtola, S.; Tuomela, S.; Elo, L.; Häkkinen, T.; Karenko, L.; Nedoszytko, B.; Heikkilä, H.; Saarialho-Kere, U.; Roszkiewicz, J.; Aittokallio, T.; et al. Th1 Response and Cytotoxicity Genes Are down-Regulated in Cutaneous T-Cell Lymphoma. Clin. Cancer Res. 2006, 12, 4812–4821. [Google Scholar] [CrossRef]
- Bastidas Torres, A.N.; Cats, D.; Mei, H.; Szuhai, K.; Willemze, R.; Vermeer, M.H.; Tensen, C.P. Genomic Analysis Reveals Recurrent Deletion of JAK-STAT Signaling Inhibitors HNRNPK and SOCS1 in Mycosis Fungoides. Genes Chromosomes Cancer 2018, 57, 653–664. [Google Scholar] [CrossRef]
- van Kester, M.S.; Borg, M.K.; Zoutman, W.H.; Out-Luiting, J.J.; Jansen, P.M.; Dreef, E.J.; Vermeer, M.H.; van Doorn, R.; Wil-lemze, R.; Tensen, C.P. A Meta-Analysis of Gene Expression Data Identifies a Molecular Signature Characteristic for Tu-mor-Stage Mycosis Fungoides. J. Investig. Dermatol. 2012, 132, 2050–2059. [Google Scholar] [CrossRef] [PubMed]
- Dulmage, B.; Geskin, L. Lessons learned from gene expression profiling of cutaneous T-cell lymphoma. Br. J. Dermatol. 2013, 169, 1188–1197. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Monti, S.; Aires, D.J.; Duvic, M.; Golub, T.; Jones, D.A.; Kupper, T.S. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood 2007, 110, 3015–3027. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, I.V.; Jones, D.A.; Sasseville, D.; Kupper, T.S. Transcriptional Profiles Predict Disease Outcome in Patients with Cu-taneous T-Cell Lymphoma. Clin. Cancer Res. 2010, 16, 2106–2114. [Google Scholar] [CrossRef] [PubMed]
- Alhothali, G.I. Review of the Treatment of Mycosis Fungoides and SéZary Syndrome: A Stage-Based Approach. Int. J. Health Sci. 2013, 7, 220–239. [Google Scholar] [CrossRef]
- Tracey, L.; Villuendas, R.; Dotor, A.M.; Spiteri, I.; Ortiz, P.; Garcia, J.F.; Peralto, J.L.R.; Lawler, M.; Piris, M.A. Mycosis Fun-goides Shows Concurrent Deregulation of Multiple Genes Involved in the TNF Signaling Pathway: An Expression Profile Study. Blood 2003, 102, 1042–1050. [Google Scholar] [CrossRef]
- Chae, S.W.; Sohn, J.H.; Kim, D.-H.; Choi, Y.J.; Park, Y.L.; Kim, K.; Cho, Y.H.; Pyo, J.-S.; Kim, J.H. Overexpressions of Cyclin B1, cdc2, p16 and p53 in Human Breast Cancer: The Clinicopathologic Correlations and Prognostic Implications. Yonsei Med. J. 2011, 52, 445–453. [Google Scholar] [CrossRef]
- Murakami, H.; Furihata, M.; Ohtsuki, Y.; Ogoshi, S. Determination of the Prognostic Significance of Cyclin B1 Overexpres-sion in Patients with Esophageal Squamous Cell Carcinoma. Virchows Arch. 1999, 434, 153–158. [Google Scholar] [CrossRef]
- Banerjee, S.K.; Weston, A.P.; Zoubine, M.N.; Campbell, D.R.; Cherian, R. Expression of Cdc2 and Cyclin B1 in Helicobacter pylori-Associated Gastric MALT and MALT Lymphoma: Relationship to Cell Death, Proliferation, and Transformation. Am. J. Pathol. 2000, 156, 217–225. [Google Scholar] [CrossRef]
- Soria, J.C.; Jang, S.J.; Khuri, F.R.; Hassan, K.; Liu, D.; Hong, W.K.; Mao, L. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000, 60, 4000–4004. [Google Scholar]
- Androic, I.; Krämer, A.; Yan, R.; Rödel, F.; Gätje, R.; Kaufmann, M.; Strebhardt, K.; Yuan, J. Targeting Cyclin B1 Inhibits Pro-liferation and Sensitizes Breast Cancer Cells to Taxol. BMC Cancer 2008, 8, 391. [Google Scholar] [CrossRef]
- Savorani, C.; Manfé, V.; Biskup, E.; Gniadecki, R. Ellipticine induces apoptosis in T-cell lymphoma via oxidative DNA damage. Leuk. Lymphoma 2014, 56, 739–747. [Google Scholar] [CrossRef]
- Biskup, E.; Naym, D.G.; Gniadecki, R. Small-molecule inhibitors of Ataxia Telangiectasia and Rad3 related kinase (ATR) sensitize lymphoma cells to UVA radiation. J. Dermatol. Sci. 2016, 84, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Biskup, E.; Manfé, V.; Kamstrup, M.R.; Gniadecki, R. Growth Dynamics and Cyclin Expression in Cutaneous T-Cell Lym-phoma Cell Lines. Dermatol. Rep. 2010, 2, e8. [Google Scholar] [CrossRef][Green Version]
- Nam, H.-J.; van Deursen, J.M. Cyclin B2 and p53 Control Proper Timing of Centrosome Separation. Nat. Cell Biol. 2014, 16, 538–549. [Google Scholar] [CrossRef]
- Yang, L.; Li, N.; Wang, C.; Yu, Y.; Yuan, L.; Zhang, M.; Cao, X. Cyclin L2, a Novel RNA Polymerase II-Associated Cyclin, Is Involved in Pre-mRNA Splicing and Induces Apoptosis of Human Hepatocellular Carcinoma Cells. J. Biol. Chem. 2004, 279, 11639–11648. [Google Scholar] [CrossRef] [PubMed]
- Dastsooz, H.; Cereda, M.; Donna, D.; Oliviero, S. A Comprehensive Bioinformatics Analysis of UBE2C in Cancers. Int. J. Mol. Sci. 2019, 20, 2228. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Cheng, Y.; Wang, B. Chapter 27-Cancer and Genomic Instability. In Genome Stability; Kovalchuk, I., Kovalchuk, O., Eds.; Academic Press: Boston, MA, USA, 2016; pp. 463–486. [Google Scholar]
- Bakhoum, S.F.; Cantley, L.C. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 2018, 174, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.B.; Horne, S.D.; Abdallah, B.Y.; Ye, C.J.; Heng, H.H. Chromosomal instability and transcriptome dynamics in cancer. Cancer Metastasis Rev. 2013, 32, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Thangavelu, M.; Finn, W.G.; Yelavarthi, K.K.; Roenigk, H.H., Jr.; Samuelson, E.; Peterson, L.; Kuzel, T.M.; Rosen, S.T. Recur-ring Structural Chromosome Abnormalities in Peripheral Blood Lymphocytes of Patients with Mycosis fungoides/Sézary Syndrome. Blood 1997, 89, 3371–3377. [Google Scholar] [CrossRef]
- Karenko, L.; Hyytinen, E.; Sarna, S.; Ranki, A. Chromosomal Abnormalities in Cutaneous T-Cell Lymphoma and in Its Premalignant Conditions as Detected by G-Banding and Interphase Cytogenetic Methods. J. Investig. Dermatol. 1997, 108, 22–29. [Google Scholar] [CrossRef]
- Prochazkova, M.; Chevret, E.; Mainhaguiet, G.; Sobotka, J.; Vergier, B.; Belaud-Rotureau, M.-A.; Beylot-Barry, M.; Merlio, J.-P. Common chromosomal abnormalities in mycosis fungoides transformation. Genes Chromosom. Cancer 2007, 46, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.L.; Bakhoum, S.; Compton, D.A. Mechanisms of Chromosomal Instability. Curr. Biol. 2010, 20, R285–R295. [Google Scholar] [CrossRef]
- Gollin, S.M. Mechanisms leading to chromosomal instability. Semin. Cancer Biol. 2005, 15, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Marumoto, T.; Zhang, D.; Saya, H. Aurora-A—A Guardian of Poles. Nat. Rev. Cancer 2005, 5, 42–50. [Google Scholar] [CrossRef]
- Lentini, L.; Amato, A.; Schillaci, T.; Di Leonardo, A. Simultaneous Aurora-A/STK15 Overexpression and Centrosome Ampli-fication Induce Chromosomal Instability in Tumour Cells with a MIN Phenotype. BMC Cancer 2007, 7, 212. [Google Scholar] [CrossRef]
- Nishida, N.; Nagasaka, T.; Kashiwagi, K.; Boland, C.R.; Goel, A. High Copy Amplification of the Aurora-A Gene Is Associ-ated with Chromosomal Instability Phenotype in Human Colorectal Cancers. Cancer Biol. Ther. 2007, 6, 525–533. [Google Scholar] [CrossRef][Green Version]
- Siddiqi, T.; Frankel, P.; Beumer, J.H.; Kiesel, B.F.; Christner, S.; Ruel, C.; Song, J.Y.; Chen, R.; Kelly, K.R.; Ailawadhi, S.; et al. Phase 1 Study of the Aurora Kinase A Inhibitor Alisertib (MLN8237) Combined with the Histone Deacetylase Inhibitor Vo-rinostat in Lymphoid Malignancies. Leuk. Lymphoma 2020, 61, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Hauf, S.; Cole, R.W.; LaTerra, S.; Zimmer, C.; Schnapp, G.; Walter, R.; Heckel, A.; van Meel, J.; Rieder, C.L.; Peters, J.-M. The Small Molecule Hesperadin Reveals a Role for Aurora B in Correcting Kinetochore–microtubule Attachment and in Main-taining the Spindle Assembly Checkpoint. J. Cell Biol. 2003, 161, 281–294. [Google Scholar] [CrossRef]
- Ditchfield, C.; Johnson, V.L.; Tighe, A.; Ellston, R.; Haworth, C.; Johnson, T.; Mortlock, A.; Keen, N.; Taylor, S.S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003, 161, 267–280. [Google Scholar] [CrossRef]
- Gruneberg, U.; Neef, R.; Honda, R.; Nigg, E.A.; Barr, F.A. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 2004, 166, 167–172. [Google Scholar] [CrossRef]
- Acquaviva, C.; Herzog, F.; Kraft, C.; Pines, J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat. Cell Biol. 2004, 6, 892–898. [Google Scholar] [CrossRef]
- Pablo, L.-G.; Westhorpe, F.G.; Taylor, S.S. The Spindle Assembly Checkpoint. Curr. Biol. 2012, 22, R966–R980. [Google Scholar]
- Marks, D.H.; Thomas, R.; Chin, Y.; Shah, R.; Khoo, C.; Benezra, R. Mad2 Overexpression Uncovers a Critical Role for TRIP13 in Mitotic Exit. Cell Rep. 2017, 19, 1832–1845. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Yang, G.; Yang, H.; Song, D.; Hu, L.; Xie, B.; Wang, H.; Gao, L.; Gao, M.; Xu, H.; et al. TRIP13 impairs mitotic checkpoint surveillance and is associated with poor prognosis in multiple myeloma. Oncotarget 2017, 8, 26718–26731. [Google Scholar] [CrossRef] [PubMed]
- Eytan, E.; Wang, K.; Miniowitz-Shemtov, S.; Sitry-Shevah, D.; Kaisari, S.; Yen, T.J.; Liu, S.-T.; Hershko, A. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31comet. Proc. Natl. Acad. Sci. USA 2014, 111, 12019–12024. [Google Scholar] [CrossRef]
- Saito, S.; Tatsumoto, T.; Lorenzi, M.V.; Chedid, M.; Kapoor, V.; Sakata, H.; Rubin, J.; Miki, T. Rho Exchange Factor ECT2 Is Induced by Growth Factors and Regulates Cytokinesis through the N-Terminal Cell Cycle Regulator-Related Do-mains. J. Cell. Biochem. 2003, 90, 819–836. [Google Scholar] [CrossRef]
- Lee, C.-H.; Hwang, S.T.-Y. Pathophysiology of Chemokines and Chemokine Receptors in Dermatological Science: A Focus on Psoriasis and Cutaneous T-Cell Lymphoma. Dermatol. Sin. 2012, 30, 128–135. [Google Scholar] [CrossRef]
- Krejsgaard, T.; Lindahl, L.M.; Mongan, N.P.; Wasik, M.A.; Litvinov, I.V.; Iversen, L.; Langhoff, E.; Woetmann, A.; Odum, N. Malignant Inflammation in Cutaneous T-Cell Lymphoma-a Hostile Takeover. Semin. Immunopathol. 2017, 39, 269–282. [Google Scholar] [CrossRef]
- Stolearenco, V.; Namini, M.R.J.; Hasselager, S.S.; Gluud, M.; Buus, T.B.; Willerslev-Olsen, A.; Ødum, N.; Krejsgaard, T. Cel-lular Interactions and Inflammation in the Pathogenesis of Cutaneous T-Cell Lymphoma. Front Cell Dev. Biol. 2020, 8, 851. [Google Scholar] [CrossRef] [PubMed]
- Günther, C.; Zimmermann, N.; Berndt, N.; Großer, M.; Stein, A.; Koch, A.; Meurer, M. Up-Regulation of the Chemokine CCL18 by Macrophages Is a Potential Immunomodulatory Pathway in Cutaneous T-Cell Lymphoma. Am. J. Pathol. 2011, 179, 1434–1442. [Google Scholar] [CrossRef]
- Miyagaki, T.; Sugaya, M.; Suga, H.; Ohmatsu, H.; Fujita, H.; Asano, Y.; Tada, Y.; Kadono, T.; Sato, S. Increased CCL18 Ex-pression in Patients with Cutaneous T-Cell Lymphoma: Association with Disease Severity and Prognosis. J. Eur. Acad. Dermatol. Venereol. 2013, 27, e60–e67. [Google Scholar] [CrossRef]
- Kallinich, T.; Muche, J.M.; Qin, S.; Sterry, W.; Audring, H.; Kroczek, R.A. Chemokine Receptor Expression on Neoplastic and Reactive T Cells in the Skin at Different Stages of Mycosis Fungoides. J. Investig. Dermatol. 2003, 121, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.C.-S.; Lin, C.-L.; Hong, C.-H.; Yu, H.-S.; Chen, G.-S.; Lee, C.-H. CCR7 Expression Correlates with Subcutaneous In-volvement in Mycosis Fungoides Skin Lesions and Promotes Migration of Mycosis Fungoides Cells (MyLa) through mTOR Activation. J. Dermatol. Sci. 2014, 74, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Vacca, A.; Ribatti, D.; Ria, R.; Pellegrino, A.; Bruno, M.; Merchionne, F.; Dammacco, F. Proteolytic Activity of Human Lym-phoid Tumor Cells. Correlation with Tumor Progression. Dev. Immunol. 2000, 7, 77–88. [Google Scholar] [CrossRef]
- Vacca, A.; Moretti, S.; Ribatti, D.; Pellegrino, A.; Pimpinelli, N.; Bianchi, B.; Bonifazi, E.; Ria, R.; Serio, G.; Dammacco, F. Pro-gression of Mycosis Fungoides Is Associated with Changes in Angiogenesis and Expression of the Matrix Metalloproteinases 2 and 9. Eur. J. Cancer 1997, 33, 1685–1692. [Google Scholar]
- Guenova, E.; Watanabe, R.; Teague, J.E.; Desimone, J.A.; Jiang, Y.; Dowlatshahi, M.; Schlapbach, C.; Schaekel, K.; Rook, A.H.; Tawa, M.; et al. TH2 Cytokines from Malignant Cells Suppress TH1 Responses and Enforce a Global TH2 Bias in Leukemic Cutaneous T-Cell Lymphoma. Clin. Cancer Res. 2013, 19, 3755–3763. [Google Scholar] [CrossRef]
- Wu, X.; Hsu, D.K.; Wang, K.-H.; Huang, Y.; Mendoza, L.; Zhou, Y.; Hwang, S.T. IL-10 Is Overexpressed in Human Cutane-ous T-Cell Lymphoma and Is Required for Maximal Tumor Growth in a Mouse Model. Leuk. Lymphoma 2019, 60, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Asadullah, K.; Haeussler-Quade, A.; Gellrich, S.; Hanneken, S.; Hansen-Hagge, T.E.; Döcke, W.D.; Volk, H.D.; Sterry, W. IL-15 and IL-16 Overexpression in Cutaneous T-Cell Lymphomas: Stage-Dependent Increase in Mycosis Fungoides Progression. Exp. Dermatol. 2000, 9, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Ohmatsu, H.; Humme, D.; Gulati, N.; Gonzalez, J.; Möbs, M.; Suárez-Fariñas, M.; Cardinale, I.; Mitsui, H.; Guttman-Yassky, E.; Sterry, W.; et al. IL32 Is Progressively Expressed in Mycosis Fungoides Independent of Helper T-cell 2 and Helper T-cell 9 Polarization. Cancer Immunol. Res. 2014, 2, 890–900. [Google Scholar] [CrossRef]
- Willerslev-Olsen, A.; Litvinov, I.V.; Fredholm, S.M.; Petersen, D.L.; Sibbesen, N.A.; Gniadecki, R.; Zhang, Q.; Bonefeld, C.M.; Wasik, M.A.; Geisler, C.; et al. IL-15 and IL-17F are differentially regulated and expressed in mycosis fungoides (MF). Cell Cycle 2014, 13, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Suga, H.; Sugaya, M.; Miyagaki, T.; Kawaguchi, M.; Fujita, H.; Asano, Y.; Tada, Y.; Kadono, T.; Sato, S. The Role of IL-32 in Cutaneous T-Cell Lymphoma. J. Investig. Dermatol. 2014, 134, 1428–1435. [Google Scholar] [CrossRef] [PubMed]
- Brender, C.; Lovato, P.; Sommer, V.H.; Woetmann, A.; Mathiesen, A.-M.; Geisler, C.; Wasik, M.; Ødum, N. Constitutive SOCS-3 Expression Protects T-Cell Lymphoma against Growth Inhibition by IFNalpha. Leukemia 2005, 19, 209–213. [Google Scholar] [CrossRef]
- Lens, S.M.; Baars, P.A.; Hooibrink, B.; van Oers, M.H.; van Lier, R.A. Antigen-Presenting Cell-Derived Signals Determine Expression Levels of CD70 on Primed T Cells. Immunology 1997, 90, 38–45. [Google Scholar] [CrossRef]
- Jacobs, J.; Deschoolmeester, V.; Zwaenepoel, K.; Rolfo, C.; Silence, K.; Rottey, S.; Lardon, F.; Smits, E.; Pauwels, P. CD70: An emerging target in cancer immunotherapy. Pharmacol. Ther. 2015, 155, 1–10. [Google Scholar] [CrossRef]
- Bagot, M.; Maerevoet, M.; Zinzani, P.L.; Offner, F.; Morschhauser, F.; Michot, J.-M.; Ribrag, V.; Battistella, M.; Moins, H.; Calleri, A.; et al. Argx-110 for Treatment of CD70-Positive Advanced Cutaneous T-Cell Lymphoma in a Phase 1/2 Clinical Trial. Blood 2018, 132, 1627. [Google Scholar] [CrossRef]
- Peterson, E.J.; Woods, M.L.; Dmowski, S.A.; Derimanov, G.; Jordan, M.S.; Wu, J.N.; Myung, P.S.; Liu, Q.-H.; Pribila, J.T.; Freedman, B.D.; et al. Coupling of the TCR to Integrin Activation by SLAP-130/Fyb. Science 2001, 293, 2263–2265. [Google Scholar] [CrossRef]
- Baker, R.G.; Hsu, C.J.; Lee, D.; Jordan, M.S.; Maltzman, J.S.; Hammer, D.A.; Baumgart, T.; Koretzky, G.A. The Adapter Protein SLP-76 Mediates “Outside-in” Integrin Signaling and Function in T Cells. Mol. Cell. Biol. 2009, 29, 5578–5589. [Google Scholar] [CrossRef]
- Judd, B.A.; Koretzky, G.A. The role of the adapter molecule SLP-76 in platelet function. Oncogene 2001, 20, 6291–6299. [Google Scholar] [CrossRef][Green Version]
- Yumeen, S.; Girardi, M. Insights into the Molecular and Cellular Underpinnings of Cutaneous T Cell Lymphoma. Yale J. Biol. Med. 2020, 93, 111–121. [Google Scholar] [PubMed]
- Bagot, M.; Moretta, A.; Sivori, S.; Biassoni, R.; Cantoni, C.; Bottino, C.; Boumsell, L.; Bensussan, A. CD4+ cutaneous T-cell lymphoma cells express the p140–killer cell immunoglobulin-like receptor. Blood 2001, 97, 1388–1391. [Google Scholar] [CrossRef]
- Marie-Cardine, A.; Huet, D.; Ortonne, N.; Remtoula, N.; Le Gouvello, S.; Bagot, M.; Bensussan, A. Killer Cell Ig-like Receptors CD158a and CD158b Display a Coactivatory Function, Involving the c-Jun NH2-Terminal Protein Kinase Signaling Pathway, When Expressed on Malignant CD4+ T Cells from a Patient with Sezary Syndrome. Blood 2007, 109, 5064–5065. [Google Scholar] [CrossRef] [PubMed]
- Long, E.O. Negative signaling by inhibitory receptors: The NK cell paradigm. Immunol. Rev. 2008, 224, 70–84. [Google Scholar] [CrossRef]
- Aliahmad, P.; Seksenyan, A.; Kaye, J. The many roles of TOX in the immune system. Curr. Opin. Immunol. 2012, 24, 173–177. [Google Scholar] [CrossRef]
- Huang, Y.; Litvinov, I.V.; Wang, Y.; Su, M.-W.; Tu, P.; Jiang, X.; Kupper, T.S.; Dutz, J.P.; Sasseville, D.; Zhou, Y. Thymocyte Selection-Associated High Mobility Group Box Gene (TOX) Is Aberrantly over-Expressed in Mycosis Fungoides and Corre-lates with Poor Prognosis. Oncotarget 2014, 5, 4418–4425. [Google Scholar] [CrossRef]
- Yu, X.; Luo, Y.; Liu, J.; Liu, Y.; Sun, Q. TOX Acts an Oncological Role in Mycosis Fungoides. PLoS ONE 2015, 10, e0117479. [Google Scholar] [CrossRef]
- Neinaa, Y.M.E.-H.; Sallam, F.A. TOX as a diagnostic and prognostic marker for mycosis fungoides. J. Egypt. Womens Dermatol. Soc. 2018, 15, 15–22. [Google Scholar] [CrossRef]
- Toricelli, M.; Melo, F.H.M.; Peres, G.B.; Silva, D.C.P.; Jasiulionis, M.G. Erratum: Timp1 interacts with beta-1 integrin and CD63 along melanoma genesis and confers anoikis resistance by activating PI3-K signaling pathway independently of Akt phosphorylation. Mol. Cancer 2015, 14, 161. [Google Scholar] [CrossRef]
- Terpos, E.; Dimopoulos, M.; Shrivastava, V.; Leitzel, K.; Christoulas, D.; Migkou, M.; Gavriatopoulou, M.; Anargyrou, K.; Hamer, P.; Kastritis, E.; et al. High levels of serum TIMP-1 correlate with advanced disease and predict for poor survival in patients with multiple myeloma treated with novel agents. Leuk. Res. 2010, 34, 399–402. [Google Scholar] [CrossRef]
- Dechaphunkul, A.; Phukaoloun, M.; Kanjanapradit, K.; Graham, K.; Ghosh, S.; Santos, C.; Mackey, J.R. Prognostic Signifi-cance of Tissue Inhibitor of Metalloproteinase-1 in Breast Cancer. Int. J. Breast Cancer 2012, 2012, 290854. [Google Scholar] [CrossRef]
- Ries, C. Cytokine functions of TIMP-1. Cell Mol. Life Sci. 2014, 71, 659–672. [Google Scholar] [CrossRef]
- Hayakawa, T.; Yamashita, K.; Tanzawa, K.; Uchijima, E.; Iwata, K. Growth-Promoting Activity of Tissue Inhibitor of Metal-loproteinases-1 (TIMP-1) for a Wide Range of Cells A Possible New Growth Factor in Serum. FEBS Lett. 1992, 298, 29–32. [Google Scholar] [CrossRef]
- Pennanen, H.; Kuittinen, O.; Soini, Y.; Turpeenniemi-Hujanen, T. Clinicopathological correlations of TIMP-1 and TIMP-2 in Hodgkin’s lymphoma. Eur. J. Haematol. 2003, 72, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Hess, S.; Richardson, S.K.; Newton, S.; Showe, L.C.; Benoit, B.M.; Ubriani, R.; Vittorio, C.C.; Junkins-Hopkins, J.M.; Wysocka, M.; et al. Immunopathogenesis and Therapy of Cutaneous T Cell Lymphoma. J. Clin. Investig. 2005, 115, 798–812. [Google Scholar] [CrossRef]
- Murray, D.; McMurray, J.L.; Eldershaw, S.; Pearce, H.; Davies, N.; Scarisbrick, J.J.; Moss, P. Progression of mycosis fungoides occurs through divergence of tumor immunophenotype by differential expression of HLA-DR. Blood Adv. 2019, 3, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Yi, J.S.; Cox, M.; Zajac, A.J. T-cell exhaustion: Characteristics, causes and conversion. Immunology 2010, 129, 474–481. [Google Scholar] [CrossRef]
- Riley, J.L. PD-1 Signaling in Primary T Cells. Immunol. Rev. 2009, 229, 114–125. [Google Scholar] [CrossRef]
- Kantekure, K.; Yang, Y.; Raghunath, P.; Schaffer, A.; Woetmann, A.; Zhang, Q.; Odum, N.; Wasik, M. Expression Patterns of the Immunosuppressive Proteins PD-1/CD279 and PD-L1/CD274 at Different Stages of Cutaneous T-Cell Lympho-ma/mycosis Fungoides. Am. J. Dermatopathol. 2012, 34, 126–128. [Google Scholar] [CrossRef]
- Lesokhin, A.M.; Ansell, S.M.; Armand, P.; Scott, E.C.; Halwani, A.; Gutierrez, M.; Millenson, M.M.; Cohen, A.D.; Schuster, S.J.; Lebovic, D.; et al. Nivolumab in Patients with Relapsed or Refractory Hematologic Malignancy: Preliminary Results of a Phase Ib Study. J. Clin. Oncol. 2016, 34, 2698–2704. [Google Scholar] [CrossRef]
- Khodadoust, M.S.; Rook, A.H.; Porcu, P.; Foss, F.; Moskowitz, A.J.; Shustov, A.; Shanbhag, S.; Sokol, L.; Fling, S.P.; Ramchurren, N.; et al. Pembrolizumab in Relapsed and Refractory Mycosis Fungoides and Sézary Syndrome: A Multicenter Phase II Study. J. Clin. Oncol. 2020, 38, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.; Anderson, A.C.; Kuchroo, V.K. TIM3 Comes of Age as an Inhibitory Receptor. Nat. Rev. Immunol. 2020, 20, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Rousseaux, S.; Debernardi, A.; Jacquiau, B.; Vitte, A.-L.; Vesin, A.; Nagy-Mignotte, H.; Moro-Sibilot, D.; Brichon, P.-Y.; Lantuejoul, S.; Hainaut, P.; et al. Ectopic Activation of Germline and Placental Genes Identifies Aggressive Metastasis-Prone Lung Cancers. Sci. Transl. Med. 2013, 5, 186ra66. [Google Scholar] [CrossRef]
- Gantchev, J.; Martínez Villarreal, A.; Xie, P.; Lefrançois, P.; Gunn, S.; Netchiporouk, E.; Sasseville, D.; Litvinov, I.V. The Ectopic Expression of Meiosis Regulatory Genes in Cutaneous T-Cell Lymphomas (CTCL). Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- MacKeigan, J.P.; Murphy, L.O.; Blenis, J. Sensitized RNAi Screen of Human Kinases and Phosphatases Identifies New Regulators of Apoptosis and Chemoresistance. Nat. Cell Biol. 2005, 7, 591–600. [Google Scholar] [CrossRef]
- Zanke, B.; Squire, J.; Griesser, H.; Henry, M.; Suzuki, H.; Patterson, B.; Minden, M.; Mak, T.W. A Hematopoietic Protein Tyrosine Phosphatase (HePTP) Gene That Is Amplified and Overexpressed in Myeloid Malignancies Maps to Chromosome 1q32.1. Leukemia 1994, 8, 236–244. [Google Scholar]
- Ferenczi, K.; Fuhlbrigge, R.C.; Pinkus, J.; Pinkus, G.S.; Kupper, T.S. Increased CCR4 Expression in Cutaneous T Cell Lymphoma. J. Invest. Dermatol. 2002, 119, 1405–1410. [Google Scholar] [CrossRef] [PubMed]
- Shono, Y.; Suga, H.; Kamijo, H.; Fujii, H.; Oka, T.; Miyagaki, T.; Shishido-Takahashi, N.; Sugaya, M.; Sato, S. Expression of CCR3 and CCR4 Suggests a Poor Prognosis in Mycosis Fungoides and Sézary Syndrome. Acta Derm. Venereol. 2019, 99, 809–812. [Google Scholar] [CrossRef]
- Gniadecki, R. CCR4-Targeted Therapy in Cutaneous T-Cell Lymphoma. Lancet Oncol. 2018, 19, 1140–1141. [Google Scholar] [CrossRef]
- Kim, Y.H.; Bagot, M.; Pinter-Brown, L.; Rook, A.H.; Porcu, P.; Horwitz, S.M.; Whittaker, S.; Tokura, Y.; Vermeer, M.; Zinzani, P.L.; et al. Mogamulizumab versus Vorinostat in Previously Treated Cutaneous T-Cell Lymphoma (MAVORIC): An International, Open-Label, Randomised, Controlled Phase 3 Trial. Lancet Oncol. 2018, 19, 1192–1204. [Google Scholar] [CrossRef]
- Pérez, C.; Mondéjar, R.; García-Díaz, N.; Cereceda, L.; León, A.; Montes, S.; Durán Vian, C.; Pérez Paredes, M.G.; González-Morán, A.; Alegre de Miguel, V.; et al. Advanced-Stage Mycosis Fungoides: Role of the Signal Transducer and Activator of Transcription 3, Nuclear Factor-κB and Nuclear Factor of Activated T Cells Pathways. Br. J. Dermatol. 2020, 182, 147–155. [Google Scholar] [CrossRef]
- Ralfkiaer, U.; Lindahl, L.M.; Litman, T.; Gjerdrum, L.-M.; Ahler, C.B.; Gniadecki, R.; Marstrand, T.; Fredholm, S.; Iversen, L.; Wasik, M.A.; et al. MicroRNA Expression in Early Mycosis Fungoides Is Distinctly Different from Atopic Dermatitis and Advanced Cutaneous T-Cell Lymphoma. Anticancer Res. 2014, 34, 7207–7217. [Google Scholar]
- Zerdes, I.; Wallerius, M.; Sifakis, E.G.; Wallmann, T.; Betts, S.; Bartish, M.; Tsesmetzis, N.; Tobin, N.P.; Coucoravas, C.; Bergh, J.; et al. STAT3 Activity Promotes Programmed-Death Ligand 1 Expression and Suppresses Immune Responses in Breast Cancer. Cancers 2019, 11, 1479. [Google Scholar] [CrossRef] [PubMed]
- Mojic, M.; Takeda, K.; Hayakawa, Y. The Dark Side of IFN-γ: Its Role in Promoting Cancer Immunoevasion. Int. J. Mol. Sci. 2017, 19, 89. [Google Scholar] [CrossRef]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in Tumor Progression and Regression: A Review. Biomark Res. 2020, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Pai, C.-C.S.; Huang, J.T.; Lu, X.; Simons, D.M.; Park, C.; Chang, A.; Tamaki, W.; Liu, E.; Roybal, K.T.; Seagal, J.; et al. Clonal Deletion of Tumor-Specific T Cells by Interferon-γ Confers Therapeutic Resistance to Combination Immune Checkpoint Blockade. Immunity 2019, 50, 477–492.e8. [Google Scholar] [CrossRef]
- Zaidi, M.R.; Davis, S.; Noonan, F.P.; Graff-Cherry, C.; Hawley, T.S.; Walker, R.L.; Feigenbaum, L.; Fuchs, E.; Lyakh, L.; Young, H.A.; et al. Interferon-γ Links Ultraviolet Radiation to Melanomagenesis in Mice. Nature 2011, 469, 548–553. [Google Scholar] [CrossRef]
- Krensky, A.M.; Clayberger, C. Biology and Clinical Relevance of Granulysin. Tissue Antigens 2009, 73, 193–198. [Google Scholar] [CrossRef]
- Pagès, F.; Berger, A.; Camus, M.; Sanchez-Cabo, F.; Costes, A.; Molidor, R.; Mlecnik, B.; Kirilovsky, A.; Nilsson, M.; Damotte, D.; et al. Effector Memory T Cells, Early Metastasis, and Survival in Colorectal Cancer. N. Engl. J. Med. 2005, 353, 2654–2666. [Google Scholar] [CrossRef]
- Saigusa, S.; Ichikura, T.; Tsujimoto, H.; Sugasawa, H.; Majima, T.; Kawarabayashi, N.; Chochi, K.; Ono, S.; Kinoshita, M.; Seki, S.; et al. Serum Granulysin Level as a Novel Prognostic Marker in Patients with Gastric Carcinoma. J. Gastroenterol. Hepatol. 2007, 22, 1322–1327. [Google Scholar] [CrossRef] [PubMed]
- Kishi, A.; Takamori, Y.; Ogawa, K.; Takano, S.; Tomita, S.; Tanigawa, M.; Niman, M.; Kishida, T.; Fujita, S. Differential Expression of Granulysin and Perforin by NK Cells in Cancer Patients and Correlation of Impaired Granulysin Expression with Progression of Cancer. Cancer Immunol. Immunother. 2002, 50, 604–614. [Google Scholar] [CrossRef]
- Tewary, P.; Yang, D.; de la Rosa, G.; Li, Y.; Finn, M.W.; Krensky, A.M.; Clayberger, C.; Oppenheim, J.J. Granulysin Activates Antigen-Presenting Cells through TLR4 and Acts as an Immune Alarmin. Blood 2010, 116, 3465–3474. [Google Scholar] [CrossRef]
- Wilcox, R.A.; Wada, D.A.; Ziesmer, S.C.; Elsawa, S.F.; Comfere, N.I.; Dietz, A.B.; Novak, A.J.; Witzig, T.E.; Feldman, A.L.; Pittelkow, M.R.; et al. Monocytes Promote Tumor Cell Survival in T-Cell Lymphoproliferative Disorders and Are Impaired in Their Ability to Differentiate into Mature Dendritic Cells. Blood 2009, 114, 2936–2944. [Google Scholar] [CrossRef] [PubMed]
- Berger, C.L.; Hanlon, D.; Kanada, D.; Dhodapkar, M.; Lombillo, V.; Wang, N.; Christensen, I.; Howe, G.; Crouch, J.; El-Fishawy, P.; et al. The Growth of Cutaneous T-Cell Lymphoma Is Stimulated by Immature Dendritic Cells. Blood 2002, 99, 2929–2939. [Google Scholar] [CrossRef] [PubMed]
- Edelson, R.L. Cutaneous T Cell Lymphoma: The Helping Hand of Dendritic Cells. Ann. N. Y. Acad. Sci. 2001, 941, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Malarkannan, S. NKG7 Makes a Better Killer. Nat. Immunol. 2020, 21, 1139–1140. [Google Scholar] [CrossRef]
- Alpdogan, O.; Kartan, S.; Johnson, W.; Sokol, K.; Porcu, P. Systemic Therapy of Cutaneous T-Cell Lymphoma (CTCL). Chin. Clin. Oncol. 2019, 8, 10. [Google Scholar] [CrossRef]
- Sivanand, A.; Surmanowicz, P.; Alhusayen, R.; Hull, P.; Litvinov, I.V.; Zhou, Y.; Gniadecki, R. Immunotherapy for Cutaneous T-Cell Lymphoma: Current Landscape and Future Developments. J. Cutan. Med. Surg. 2019, 23, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Bagot, M.; Porcu, P.; Marie-Cardine, A.; Battistella, M.; William, B.M.; Vermeer, M.; Whittaker, S.; Rotolo, F.; Ram-Wolff, C.; Khodadoust, M.S.; et al. IPH4102, a First-in-Class Anti-KIR3DL2 Monoclonal Antibody, in Patients with Relapsed or Refractory Cutaneous T-Cell Lymphoma: An International, First-in-Human, Open-Label, Phase 1 Trial. Lancet Oncol. 2019, 20, 1160–1170. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Motamedi, M.; Xiao, M.Z.X.; Iyer, A.; Gniadecki, R. Patterns of Gene Expression in Cutaneous T-Cell Lymphoma: Systematic Review of Transcriptomic Studies in Mycosis Fungoides. Cells 2021, 10, 1409. https://doi.org/10.3390/cells10061409
Motamedi M, Xiao MZX, Iyer A, Gniadecki R. Patterns of Gene Expression in Cutaneous T-Cell Lymphoma: Systematic Review of Transcriptomic Studies in Mycosis Fungoides. Cells. 2021; 10(6):1409. https://doi.org/10.3390/cells10061409
Chicago/Turabian StyleMotamedi, Melika, Maggie Z. X. Xiao, Aishwarya Iyer, and Robert Gniadecki. 2021. "Patterns of Gene Expression in Cutaneous T-Cell Lymphoma: Systematic Review of Transcriptomic Studies in Mycosis Fungoides" Cells 10, no. 6: 1409. https://doi.org/10.3390/cells10061409
APA StyleMotamedi, M., Xiao, M. Z. X., Iyer, A., & Gniadecki, R. (2021). Patterns of Gene Expression in Cutaneous T-Cell Lymphoma: Systematic Review of Transcriptomic Studies in Mycosis Fungoides. Cells, 10(6), 1409. https://doi.org/10.3390/cells10061409






