Deciphering the Long Non-Coding RNAs and MicroRNAs Coregulation Networks in Ovarian Cancer Development: An Overview
Abstract
1. Introduction
2. The Expanding Repertoire of Regulatory LncRNAs and MicroRNAs
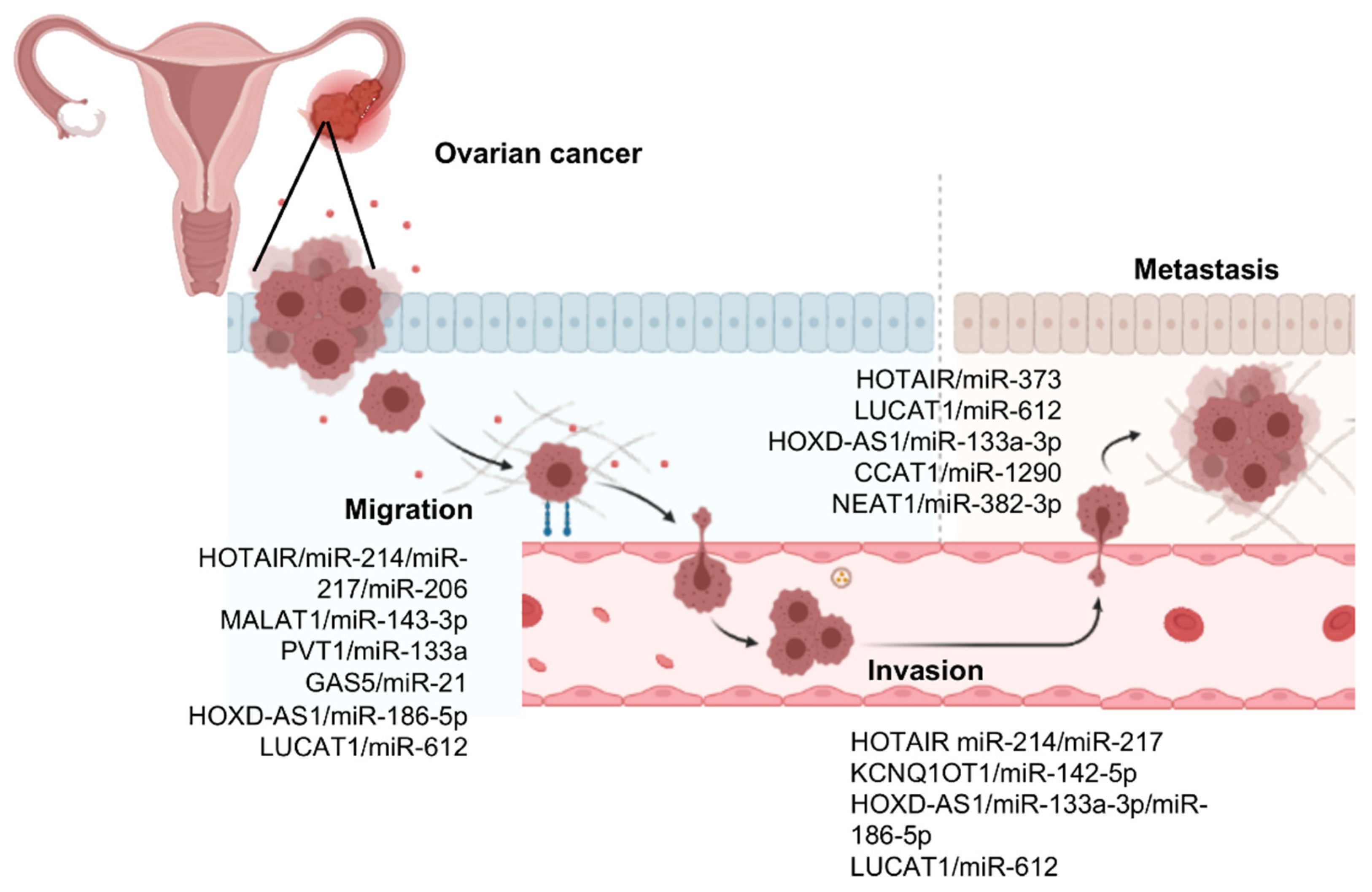
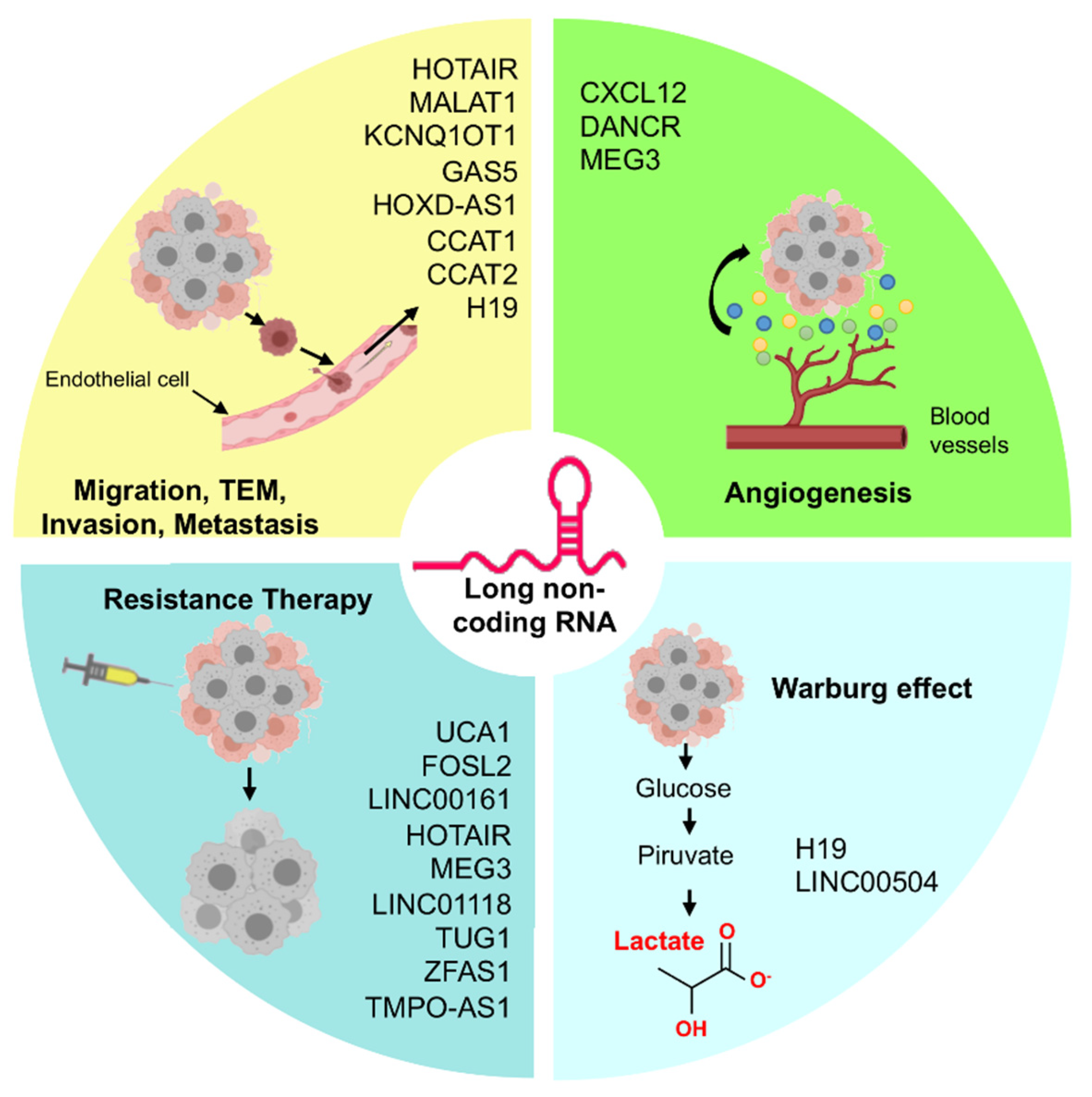
3. LncRNAs/MicroRNAs Pairs Controlling Cell Migration, Invasion, and Metastasis in Ovarian Cancer
3.1. HOX Transcript Antisense RNA (HOTAIR)
3.2. Metastasis-Associated Lung Carcinoma Transcript 1 (MALAT1)
3.3. Plasmacytoma Variant Translocation 1 (PVT1)
3.4. Potassium Voltage-Gated Channel Sub-Family Q Member 1 Opposite Strand Transcript 1 (KCNQ1OT1)
3.5. Growth Arrest-Specific 5 (GAS5)
3.6. HOXD Cluster Antisense RNA 1 (HOXD-AS1)
3.7. Lung Cancer-Associated Transcript 1 (LUCAT1)
3.8. LncRNA Colon Cancer-Associated Transcript 1 (CCAT1)
3.9. Other LncRNAs/MicroRNAs Pairs Involved in Ovarian Cancer Progression
4. LncRNAs and MicroRNAs Regulating Angiogenesis in Ovarian Cancer
5. Role of the LncRNAs/MicroRNAs in Therapy Resistance of Ovarian Cancer
6. Regulation of Warburg Effect by LncRNAs in Ovarian Cancer
7. LncRNAs/MicroRNAs Coregulation Networks
8. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Rojas, V.; Hirshfield, K.; Ganesan, S.; Rodriguez-Rodriguez, L. Molecular characterization of epithelial ovarian cancer: Implications for diagnosis and treatment. IJMS 2016, 17, 2113. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, S.; Coward, J.I.; Bast, R.C.; Berchuck, A.; Berek, J.S.; Brenton, J.D.; Coukos, G.; Crum, C.C.; Drapkin, R.; Etemadmoghadam, D.; et al. Rethinking ovarian cancer: Recommendations for improving outcomes. Nat. Rev. Cancer 2011, 11, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Sandercock, J.; Parmar, M.K.B.; Torri, V.; Qian, W. First-line treatment for advanced ovarian cancer: Paclitaxel, platinum and the evidence. Br. J. Cancer 2002, 87, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef] [PubMed]
- St. Laurent, G.; Wahlestedt, C.; Kapranov, P. The landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef]
- Ribeiro, D.M.; Zanzoni, A.; Cipriano, A.; Delli Ponti, R.; Spinelli, L.; Ballarino, M.; Bozzoni, I.; Tartaglia, G.G.; Brun, C. Protein complex scaffolding predicted as a prevalent function of long non-coding RNAs. Nucleic Acids Res. 2018, 46, 917–928. [Google Scholar] [CrossRef]
- Hou, Y.; Zhang, R.; Sun, X. Enhancer LncRNAs influence chromatin interactions in different ways. Front. Genet. 2019, 10, 936. [Google Scholar] [CrossRef]
- Kim, K.; Noh, J.; Abdelmohsen, K.; Gorospe, M. Mitochondrial noncoding RNA transport. BMB Rep. 2017, 50, 164–174. [Google Scholar] [CrossRef]
- Rashid, F.; Shah, A.; Shan, G. Long Non-coding RNAs in the cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80. [Google Scholar] [CrossRef]
- Kim, J.; Noh, J.H.; Lee, S.K.; Munk, R.; Sharov, A.; Lehrmann, E.; Zhang, Y.; Wang, W.; Abdelmohsen, K.; Gorospe, M. LncRNA OIP5-AS1/cyrano suppresses GAK expression to control mitosis. Oncotarget 2017, 25, 49409–49420. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, L.; Liu, Q.; Gong, C.; Yao, Y.; Lv, X.; Lin, L.; Yao, H.; Su, F.; Li, D.; et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015, 27, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1471. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.L.; Beck, A.H.; Edris, B.; Sweeney, R.T.; Zhu, S.X.; Li, R.; Montgomery, K.; Varma, S.; Gilks, T.; Guo, X.; et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human Cancers. Genome Biol. 2012, 13, R75. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biology 2012, 9, 703–719. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Ma, J.; Xue, Y.; Wang, P.; Li, Z.; Liu, J.; Chen, L.; Xi, Z.; Teng, H.; Wang, Z.; et al. Knockdown of Long Non-Coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating MiR-152. Cancer Lett. 2015, 359, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long Non-Coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, L.; Wu, L.-M.; Lai, M.-C.; Xie, H.-Y.; Zhang, F.; Zheng, S.-S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011, 18, 1243–1250. [Google Scholar] [CrossRef]
- Yildirim, E.; Kirby, J.E.; Brown, D.E.; Mercier, F.E.; Sadreyev, R.I.; Scadden, D.T.; Lee, J.T. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 2013, 152, 727–742. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An estimate of the total number of true human MiRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Bill, R.; Christofori, G. The relevance of EMT in breast cancer metastasis: Correlation or causality? FEBS Lett. 2015, 589, 1577–1587. [Google Scholar] [CrossRef]
- Huang, Q.; Yan, J.; Agami, R. Long Non-Coding RNAs in Metastasis. Cancer Metastasis Rev. 2018, 37, 75–81. [Google Scholar] [CrossRef]
- Qu, X.; Alsager, S.; Zhuo, Y.; Shan, B. HOX Transcript Antisense RNA (HOTAIR) in Cancer. Cancer Lett. 2019, 454, 90–97. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta (BBA) Rev. Cancer 2015, 1856, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, T.; Talluri, S.; Akshaya, R.L.; Dunna, N.R. HOTAIR LncRNA: A novel oncogenic propellant in human cancer. Clin. Chim. Acta 2020, 503, 1–18. [Google Scholar] [CrossRef]
- Dong, L.; Hui, L. HOTAIR promotes proliferation, migration, and invasion of ovarian cancer SKOV3 cells through regulating PIK3R3. Med. Sci. Monit. 2016, 22, 325–331. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, J.; Wu, Y.; Qiu, J.; Sun, Y.; Tong, X. LncRNA HOTAIR Controls the Expression of Rab22a by Sponging MiR-373 in Ovarian Cancer. Mol. Med. Rep. 2016, 14, 2465–2472. [Google Scholar] [CrossRef]
- Chang, L.; Guo, R.; Yuan, Z.; Shi, H.; Zhang, D. LncRNA HOTAIR Regulates CCND1 and CCND2 expression by sponging MiR-206 in ovarian cancer. Cell Physiol. Biochem. 2018, 49, 1289–1303. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers 2019, 11, 216. [Google Scholar] [CrossRef]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A Druggable Long Non-Coding RNA for Targeted Anti-Cancer Approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Macias, S.; Plass, M.; Stajuda, A.; Michlewski, G.; Eyras, E.; Cáceres, J.F. DGCR8 HITS-CLIP Reveals Novel Functions for the Microprocessor. Nat. Struct. Mol. Biol. 2012, 19, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhu, C.; Jin, Y. The oncogenic and tumor suppressive functions of the long noncoding RNA MALAT1: An emerging controversy. Front. Genet. 2020, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.; Xue, M.; Zhang, L.; Lin, Z. Long Noncoding RNA MALAT1-Regulated MicroRNA 506 modulates ovarian cancer growth by targeting IASPP. OncoTargets Ther. 2016, 10, 35–46. [Google Scholar] [CrossRef]
- Wu, L.; Wang, X.; Guo, Y. Long Non-Coding RNA MALAT1 Is upregulated and involved in cell proliferation, migration and apoptosis in ovarian cancer. Exp. Ther. Med. 2017, 13, 3055–3060. [Google Scholar] [CrossRef]
- Lin, Q.; Guan, W.; Ren, W.; Zhang, L.; Zhang, J.; Xu, G. MALAT1 affects ovarian cancer cell behavior and patient survival. Oncol. Rep. 2018. [Google Scholar] [CrossRef]
- Cory, S.; Graham, M.; Webb, E.; Corcoran, L.; Adams, J.M. Variant (6;15) translocations in murine plasmacytomas involve a Chromosome 15 Locus at Least 72 Kb from the c-Myc Oncogene. EMBO J. 1985, 4, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Luo, P.; Wang, Q.; Ye, Y.; Wang, B. LncRNA PVT1 in cancer: A review and meta-analysis. Clin. Chim. Acta 2017, 474, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Li, Y.; Sang, Y.; Yu, B.; Lv, D.; Zhang, W.; Feng, H. LncRNA PVT1 Regulates triple-negative breast cancer through KLF5/Beta-catenin signaling. Oncogene 2018, 37, 4723–4734. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Zhang, E.; Yin, D.; You, L.; Xu, T.; Chen, W.; Xia, R.; Wan, L.; Sun, M.; Wang, Z.; et al. Long Noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating P15 and P16. Mol. Cancer 2015, 14, 82. [Google Scholar] [CrossRef]
- He, Y.; Jing, Y.; Wei, F.; Tang, Y.; Yang, L.; Luo, J.; Yang, P.; Ni, Q.; Pang, J.; Liao, Q.; et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018, 9, 235. [Google Scholar] [CrossRef]
- Yang, Q.; Yu, Y.; Sun, Z.; Pan, Y. Long Non-Coding RNA PVT1 Promotes cell proliferation and invasion through regulating MiR-133a in ovarian cancer. Biomed. Pharmacother. 2018, 106, 61–67. [Google Scholar] [CrossRef]
- Ding, Y.; Fang, Q.; Li, Y.; Wang, Y. Amplification of LncRNA PVT1 Promotes Ovarian Cancer Proliferation by Binding to MiR-140. Mamm. Genome 2019, 30, 217–225. [Google Scholar] [CrossRef]
- Kanduri, C. Kcnq1ot1: A chromatin regulatory RNA. Semin. Cell Dev. Biol. 2011, 22, 343–350. [Google Scholar] [CrossRef]
- Ren, Y.; Gao, X.-P.; Liang, H.; Zhang, H.; Hu, C.-Y. LncRNA KCNQ1OT1 contributes to oxygen-glucose-deprivation/reoxygenation-induced injury via sponging MiR-9 in cultured neurons to regulate MMP8. Exp. Mol. Pathol. 2020, 112, 104356. [Google Scholar] [CrossRef]
- Chen, Q.-H.; Li, B.; Liu, D.-G.; Zhang, B.; Yang, X.; Tu, Y.-L. LncRNA KCNQ1OT1 Sponges MiR-15a to promote immune evasion and malignant progression of prostate cancer via up-regulating PD-L1. Cancer Cell Int. 2020, 20, 394. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wei, M.; Wang, C.; Sun, D.; Liu, P.; Zhong, X.; Yu, W. Long noncoding RNA KCNQ1OT1 promotes colorectal carcinogenesis by enhancing aerobic glycolysis via hexokinase-2. Aging 2020, 12, 11685–11697. [Google Scholar] [CrossRef]
- Liu, H.; Chen, R.; Kang, F.; Lai, H.; Wang, Y. KCNQ1OT1 promotes ovarian cancer progression via modulating MIR-142-5p/CAPN10 Axis. Mol. Genet. Genom. Med. 2020, 8. [Google Scholar] [CrossRef]
- Lu, X.; Wang, F.; Fu, M.; Li, Y.; Wang, L. Long noncoding RNA KCNQ1OT1 accelerates the progression of ovarian cancer via MicroRNA-212-3/LCN2 Axis. Oncol. Res. 2020, 28, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Ye, H.; Huang, G.; Luo, F.; Liu, Y.; Liu, Y.; Yang, X.; Shen, J.; Liu, Q.; Zhang, J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via MiR-21. Tumor Biol. 2016, 37, 2691–2702. [Google Scholar] [CrossRef]
- Li, W.; Zhai, L.; Wang, H.; Liu, C.; Zhang, J.; Chen, W.; Wei, Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 2016, 7, 27778–27786. [Google Scholar] [CrossRef]
- Ma, N.; Li, S.; Zhang, Q.; Wang, H.; Qin, H.; Wang, S. Long Non-coding RNA GAS5 Inhibits Ovarian Cancer Cell Proliferation via the Control of MicroRNA-21 and SPRY2 Expression. Exp. Ther. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yu, H.; Zheng, J.; Ning, N.; Tang, F.; Yang, Y.; Wang, Y. Lowly-Expressed LncRNA GAS5 facilitates progression of ovarian cancer through targeting MiR-196-5p and thereby regulating HOXA5. Gynecol. Oncol. 2018, 151, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Yarmishyn, A.A.; Batagov, A.O.; Tan, J.Z.; Sundaram, G.M.; Sampath, P.; Kuznetsov, V.A.; Kurochkin, I.V. HOXD-AS1 Is a Novel LncRNA Encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genom. 2014, 15, S7. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.; Mao, M.; Shen, Z.; Chen, Y.; Chen, J.; Hou, W. HOXD-AS1 exerts oncogenic functions and promotes chemoresistance in cisplatin-resistant cervical cancer cells. Human Gene Ther. 2018, 29, 1438–1448. [Google Scholar] [CrossRef]
- Zhang, Y.; Dun, Y.; Zhou, S.; Huang, X.-H. LncRNA HOXD-AS1 Promotes Epithelial Ovarian Cancer Cells Proliferation and Invasion by Targeting MiR-133a-3p and Activating Wnt/β-Catenin Signaling Pathway. Biomed. Pharmacother. 2017, 96, 1216–1221. [Google Scholar] [CrossRef]
- Li, J.; Zhuang, C.; Liu, Y.; Chen, M.; Chen, Y.; Chen, Z.; He, A.; Lin, J.; Zhan, Y.; Liu, L.; et al. Synthetic tetracycline-controllable ShRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer. J. Exp. Clin. Cancer Res. 2016, 35, 99. [Google Scholar] [CrossRef]
- Dong, S.; Wang, R.; Wang, H.; Ding, Q.; Zhou, X.; Wang, J.; Zhang, K.; Long, Y.; Lu, S.; Hong, T.; et al. HOXD-AS1 Promotes the epithelial to mesenchymal transition of ovarian cancer cells by regulating MiR-186-5p and PIK3R3. J. Exp. Clin. Cancer Res. 2019, 38, 110. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jin, S.-D.; Zhu, Q.; Han, L.; Feng, J.; Lu, X.-Y.; Wang, W.; Wang, F.; Guo, R.-H. Long Non-Coding RNA LUCAT1 is associated with poor prognosis in human non-small cell lung cancer and regulates cell proliferation via epigenetically repressing P21 and P57 Expression. Oncotarget 2017, 8, 28297–28311. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; You, B.-H.; Park, C.H.; Kim, Y.J.; Nam, J.-W.; Lee, S.K. The Long Noncoding RNA LUCAT1 Promotes Tumorigenesis by Controlling Ubiquitination and Stability of DNA Methyltransferase 1 in Esophageal Squamous Cell Carcinoma. Cancer Lett. 2018, 417, 47–57. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhao, F.; Zhu, D.; Han, J.; Chen, H.; Cai, Y.; Chen, Z.; Xie, W. Long non-coding RNA LUCAT1 promotes proliferation and invasion in clear cell renal cell carcinoma through AKT/GSK-3β Signaling Pathway. Cell Physiol. Biochem. 2018, 48, 891–904. [Google Scholar] [CrossRef]
- Yu, H.; Xu, Y.; Zhang, D.; Liu, G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of MiR-612/HOXA13 Pathway. Biochem. Biophys. Res. Commun. 2018, 503, 2095–2100. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-Z.; Liu, G.-Y.; Pang, W.-W.; Zhang, H.; Zeng, Z.-J.; Wang, H.-J. LncRNA LUCAT1 Promotes Proliferation of Ovarian Cancer Cells by Regulating MiR-199a-5p Expression. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1682–1687. [Google Scholar] [CrossRef]
- Ozawa, T.; Matsuyama, T.; Toiyama, Y.; Takahashi, N.; Ishikawa, T.; Uetake, H.; Yamada, Y.; Kusunoki, M.; Calin, G.; Goel, A. CCAT1 and CCAT2 Long Noncoding RNAs, Located within the 8q.24.21 ‘Gene Desert’, Serve as Important Prognostic Biomarkers in Colorectal Cancer. Ann. Oncol. 2017, 28, 1882–1888. [Google Scholar] [CrossRef]
- Ma, M.-Z.; Chu, B.-F.; Zhang, Y.; Weng, M.-Z.; Qin, Y.-Y.; Gong, W.; Quan, Z.-W. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of MiRNA-218-5p. Cell Death Dis. 2015, 6, e1583. [Google Scholar] [CrossRef]
- Deng, L.; Yang, S.-B.; Xu, F.-F.; Zhang, J.-H. Long non-coding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as Let-7 Sponge. J. Exp. Clin. Cancer Res. 2015, 34, 18. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Liu, T.; Li, Y.; Li, S. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 9440–9445. [Google Scholar]
- Yu, Q.; Zhou, X.; Xia, Q.; Shen, J.; Yan, J.; Zhu, J.; Li, X.; Shu, M. Long non-coding RNA CCAT1 that can be activated by c-myc promotes pancreatic cancer cell proliferation and migration. Am. J. Transl. Res. 2016, 8, 5444–5454. [Google Scholar]
- Lai, X.-J.; Cheng, H.-F. LncRNA Colon Cancer-Associated Transcript 1 (CCAT1) promotes proliferation and metastasis of ovarian Cancer via MiR-1290. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Hua, F.; Li, C.-H.; Chen, X.-G.; Liu, X.-P. Long noncoding RNA CCAT2 knockdown suppresses tumorous progression by sponging mir-424 in epithelial ovarian cancer. Oncol. Res. 2018, 26, 241–247. [Google Scholar] [CrossRef]
- Lin, X.; Yang, F.; Qi, X.; Li, Q.; Wang, D.; Yi, T.; Yin, R.; Zhao, X.; Zhong, X.; Bian, C. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of MiR-145. Mol. Carcinog. 2019, 58, 2286–2296. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, Y.; Deng, X.; Luo, M.; Wang, X.; Hu, H.; Liu, C.; Zhong, M. Long noncoding RNA H19 promotes transforming growth factor-β-induced epithelial—Mesenchymal transition by acting as a competing endogenous RNA of mir-370-3p in ovarian cancer cells. OncoTargets Ther. 2018, 11, 427–440. [Google Scholar] [CrossRef]
- The UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; The UK Prostate testing for cancer and Treatment study (ProtecT Study) Collaborators; Al Olama, A.A.; Kote-Jarai, Z.; Giles, G.G.; Guy, M.; Morrison, J.; Severi, G.; Leongamornlert, D.A.; Tymrakiewicz, M.; et al. Multiple Loci on 8q24 Associated with Prostate Cancer Susceptibility. Nat. Genet. 2009, 41, 1058–1060. [Google Scholar] [CrossRef] [PubMed]
- Min, F.; Chu, G. Long Noncoding RNA PCAT-1 knockdown prevents the development of ovarian cancer cells via MicroRNA-124-3p. J. Cell Biochem. 2020, 121, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Wan, X.; Yu, H.; Chen, X.; Shan, X.; Miao, Y.; Fan, R.; Cha, W. LncRNA-AB209371 promotes the epithelial-mesenchymal transition of hepatocellular carcinoma cells. Oncol. Rep. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.-H.; Wu, D.-M.; Fan, S.-H.; Wen, X.; Han, X.-R.; Wang, S.; Wang, Y.-J.; Zhang, Z.-F.; Shan, Q.; Li, M.-Q.; et al. LncRNA AB209371 Up-Regulated Survivin Gene by down-Regulating MiR-203 in Ovarian Carcinoma. J. Ovarian Res. 2019, 12, 92. [Google Scholar] [CrossRef]
- Liu, X.; Wen, J.; Wang, H.; Wang, Y. Long Non-Coding RNA LINC00460 Promotes Epithelial Ovarian Cancer Progression by Regulating MicroRNA-338-3p. Biomed. Pharmacother. 2018, 108, 1022–1028. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, W. LncRNA SNHG12 regulates gastric cancer progression by acting as a molecular sponge of MiR-320. Mol. Med. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Fan, X.-H. LncRNA SNHG12 accelerates the progression of ovarian cancer via absorbing MiRNA-129 to Upregulate SOX4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2345–2352. [Google Scholar] [CrossRef] [PubMed]
- Yao, N.; Yu, L.; Zhu, B.; Gan, H.-Y.; Guo, B.-Q. LncRNA GIHCG promotes development of ovarian cancer by regulating microrna-429. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8127–8134. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Fu, X.; Lu, Z. Long Non-Coding RNA NEAT1 promoted ovarian cancer cells’ metastasis through regulation of MiR-382-3p/ROCK1 Axial. Cancer Sci. 2018, 109, 2188–2198. [Google Scholar] [CrossRef]
- Wang, A.; Jin, C.; Li, H.; Qin, Q.; Li, L. LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting mir-182-5p/foxf2 signaling pathway. Int. J. Biol. Macromol. 2018, 120, 1705–1713. [Google Scholar] [CrossRef] [PubMed]
- Pang, R.W.; Poon, R.T. Clinical implications of angiogenesis in cancers. Vasc. Health Risk Manag. 2006, 2, 97–108. [Google Scholar] [CrossRef]
- Song, R.; Liu, Z.; Lu, L.; Liu, F.; Zhang, B. Long Noncoding RNA SCAMP1 Targets MiR-137/CXCL12 axis to boost cell invasion and angiogenesis in ovarian cancer. DNA Cell Biol. 2020, 39, 1041–1050. [Google Scholar] [CrossRef]
- Ye, W.; Ni, Z.; Yicheng, S.; Pan, H.; Huang, Y.; Xiong, Y.; Liu, T. Anisomycin Inhibits Angiogenesis in Ovarian Cancer by Attenuating the Molecular Sponge Effect of the LncRNA-Meg3/MiR-421/PDGFRA Axis. Int. J. Oncol. 2019. [Google Scholar] [CrossRef]
- Li, Z.; Niu, H.; Qin, Q.; Yang, S.; Wang, Q.; Yu, C.; Wei, Z.; Jin, Z.; Wang, X.; Yang, A.; et al. LncRNA UCA1 mediates resistance to cisplatin by regulating the mir-143/fosl2-signaling pathway in ovarian cancer. Mol. Ther. Nucleic Acids 2019, 17, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhou, K.; Wu, Y.; Wang, L.; Lu, S. Linc00161 regulated the drug resistance of ovarian cancer by sponging MicroRNA-128 and modulating MAPK1. Mol. Carcinog. 2019, 58, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ai, H.; Fan, X.; Chen, S.; Wang, Y.; Liu, L. Knockdown of long non-coding RNA HOTAIR reverses cisplatin resistance of ovarian cancer cells through inhibiting MiR-138-5p-Regulated EZH2 and SIRT1. Biol. Res. 2020, 53, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Huang, J.; Geng, X.; Chu, W.; Li, S.; Chen, Z.-J.; Du, Y. Polycystic ovary syndrome: Novel and Hub LncRNAs in the insulin resistance-associated LncRNA–MRNA Network. Front. Genet. 2019, 10, 772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, J.; Xu, X.; Li, L. Curcumin suppresses cisplatin resistance development partly via modulating extracellular vesicle-mediated transfer of MEG3 and MiR-214 in ovarian cancer. Cancer Chemother. Pharm. 2017, 79, 479–487. [Google Scholar] [CrossRef]
- Shi, C.; Wang, M. LINC01118 modulates paclitaxel resistance of epithelial ovarian cancer by regulating MiR-134/ABCC1. Med. Sci. Monit. 2018, 24, 8831–8839. [Google Scholar] [CrossRef]
- Wang, J.; Ye, C.; Liu, J.; Hu, Y. UCA1 confers paclitaxel resistance to ovarian cancer through MiR-129/ABCB1 Axis. Biochem. Biophys. Res. Commun. 2018, 501, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-Y.; Wang, X.-L.; Dang, Y.; Zhu, X.-Z.; Zhang, Y.-H.; Cai, B.-X.; Zheng, L. Long Non-Coding RNA UCA1 Promotes the Progression of Paclitaxel Resistance in Ovarian Cancer by Regulating the MiR-654-5p/SIK2 Axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Li, Q.; Liu, H.; Lu, X.; Zhu, M. Long noncoding RNA TUG1 promotes autophagy-associated paclitaxel resistance by sponging MiR-29b-3p in ovarian cancer cells. OncoTargets Ther. 2020, 13, 2007–2019. [Google Scholar] [CrossRef]
- Xia, B.; Hou, Y.; Chen, H.; Yang, S.; Liu, T.; Lin, M.; Lou, G. Long Non-Coding RNA ZFAS1 Interacts with MiR-150-5p to regulate sp1 expression and ovarian cancer cell malignancy. Oncotarget 2017, 8, 19534–19546. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, Y.; Cheng, H.; Tian, J.; Yang, S. Roles of a TMPO-AS1/MicroRNA-200c/TMEFF2 CeRNA network in the malignant behaviors and 5-FU resistance of ovarian cancer cells. Exp. Mol. Pathol. 2020, 115, 104481. [Google Scholar] [CrossRef]
- Lu, J. The warburg metabolism fuels tumor metastasis. Cancer Metastasis Rev. 2019, 38, 157–164. [Google Scholar] [CrossRef]
- Zheng, X.; Zhou, Y.; Chen, W.; Chen, L.; Lu, J.; He, F.; Li, X.; Zhao, L. Ginsenoside 20(S)-Rg3 Prevents PKM2-Targeting MiR-324-5p from H19 sponging to antagonize the warburg effect in ovarian cancer cells. Cell Physiol. Biochem. 2018, 51, 1340–1353. [Google Scholar] [CrossRef]
- Liu, Y.; He, X.; Chen, Y.; Cao, D. Long non-coding RNA LINC00504 regulates the warburg effect in ovarian cancer through inhibition of MiR-1244. Mol. Cell Biochem. 2020, 464, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. IJMS 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed]
- Panoutsopoulou, K.; Avgeris, M.; Scorilas, A. MiRNA and long non-coding RNA: Molecular function and clinical value in breast and ovarian cancers. Expert Rev. Mol. Diagn. 2018, 18, 963–979. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Peng, Y.; Meng, Y.; Liu, Y.; Yang, S.; Jin, H.; Li, Q. Expression profiles analysis reveals an integrated MiRNA-LncRNA signature to predict survival in ovarian cancer patients with wild-type BRCA1/2. Oncotarget 2017, 8, 68483–68492. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meng, X.; Jin-Cheng, G.; Jue, Z.; Quan-Fu, M.; Bin, Y.; Xu-Feng, W. Protein-coding genes, long non-coding RNAs combined with MicroRNAs as a novel clinical multi-dimension transcriptome signature to predict prognosis in ovarian cancer. Oncotarget 2017, 8, 72847–72859. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vang, R.; Shih, I.-M.; Kurman, R.J. Ovarian low-grade and high-grade serous carcinoma: Pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv. Anat. Pathol. 2009, 16, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Jiang, H.; Cui, Z.; Wang, X.; Wang, L.; Han, Y. Gene microarray analysis of LncRNA and MRNA expression profiles in patients with high-grade ovarian serous cancer. Int. J. Mol. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bachmayr-Heyda, A.; Auer, K.; Sukhbaatar, N.; Aust, S.; Deycmar, S.; Reiner, A.T.; Polterauer, S.; Dekan, S.; Pils, D. Small RNAs and the competing endogenous rna network in high grade serous ovarian cancer tumor spread. Oncotarget 2016, 7, 39640–39653. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating MicroRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef]
- Li, W.; Ma, S.; Bai, X.; Pan, W.; Ai, L.; Tan, W. Long Noncoding RNA WDFY3-AS2 suppresses tumor progression by acting as a competing endogenous RNA of MicroRNA-18a in Ovarian Cancer. J. Cell Physiol. 2020, 235, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Han, L.; Ren, F.; Zhang, R.; Qin, G. Prognostic value of the tumor-specific CeRNA network in epithelial ovarian cancer. J. Cell Physiol. 2019, 234, 22071–22081. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zheng, X.; Xu, B.; Hu, W.; Huang, T.; Jiang, J. The identification and analysis of MRNA–LncRNA–MiRNA cliques from the integrative network of ovarian cancer. Front. Genet. 2019, 10, 751. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Camarillo, C.; Ruíz-García, E.; Salinas-Vera, Y.M.; Silva-Cázares, M.B.; Hernández-de la Cruz, O.N.; Marchat, L.A.; Gallardo-Rincón, D. Deciphering the Long Non-Coding RNAs and MicroRNAs Coregulation Networks in Ovarian Cancer Development: An Overview. Cells 2021, 10, 1407. https://doi.org/10.3390/cells10061407
López-Camarillo C, Ruíz-García E, Salinas-Vera YM, Silva-Cázares MB, Hernández-de la Cruz ON, Marchat LA, Gallardo-Rincón D. Deciphering the Long Non-Coding RNAs and MicroRNAs Coregulation Networks in Ovarian Cancer Development: An Overview. Cells. 2021; 10(6):1407. https://doi.org/10.3390/cells10061407
Chicago/Turabian StyleLópez-Camarillo, César, Erika Ruíz-García, Yarely M. Salinas-Vera, Macrina B. Silva-Cázares, Olga N. Hernández-de la Cruz, Laurence A. Marchat, and Dolores Gallardo-Rincón. 2021. "Deciphering the Long Non-Coding RNAs and MicroRNAs Coregulation Networks in Ovarian Cancer Development: An Overview" Cells 10, no. 6: 1407. https://doi.org/10.3390/cells10061407
APA StyleLópez-Camarillo, C., Ruíz-García, E., Salinas-Vera, Y. M., Silva-Cázares, M. B., Hernández-de la Cruz, O. N., Marchat, L. A., & Gallardo-Rincón, D. (2021). Deciphering the Long Non-Coding RNAs and MicroRNAs Coregulation Networks in Ovarian Cancer Development: An Overview. Cells, 10(6), 1407. https://doi.org/10.3390/cells10061407








