Extracellular Vesicles and Pancreatic Cancer: Insights on the Roles of miRNA, lncRNA, and Protein Cargos in Cancer Progression
Abstract
1. Introduction
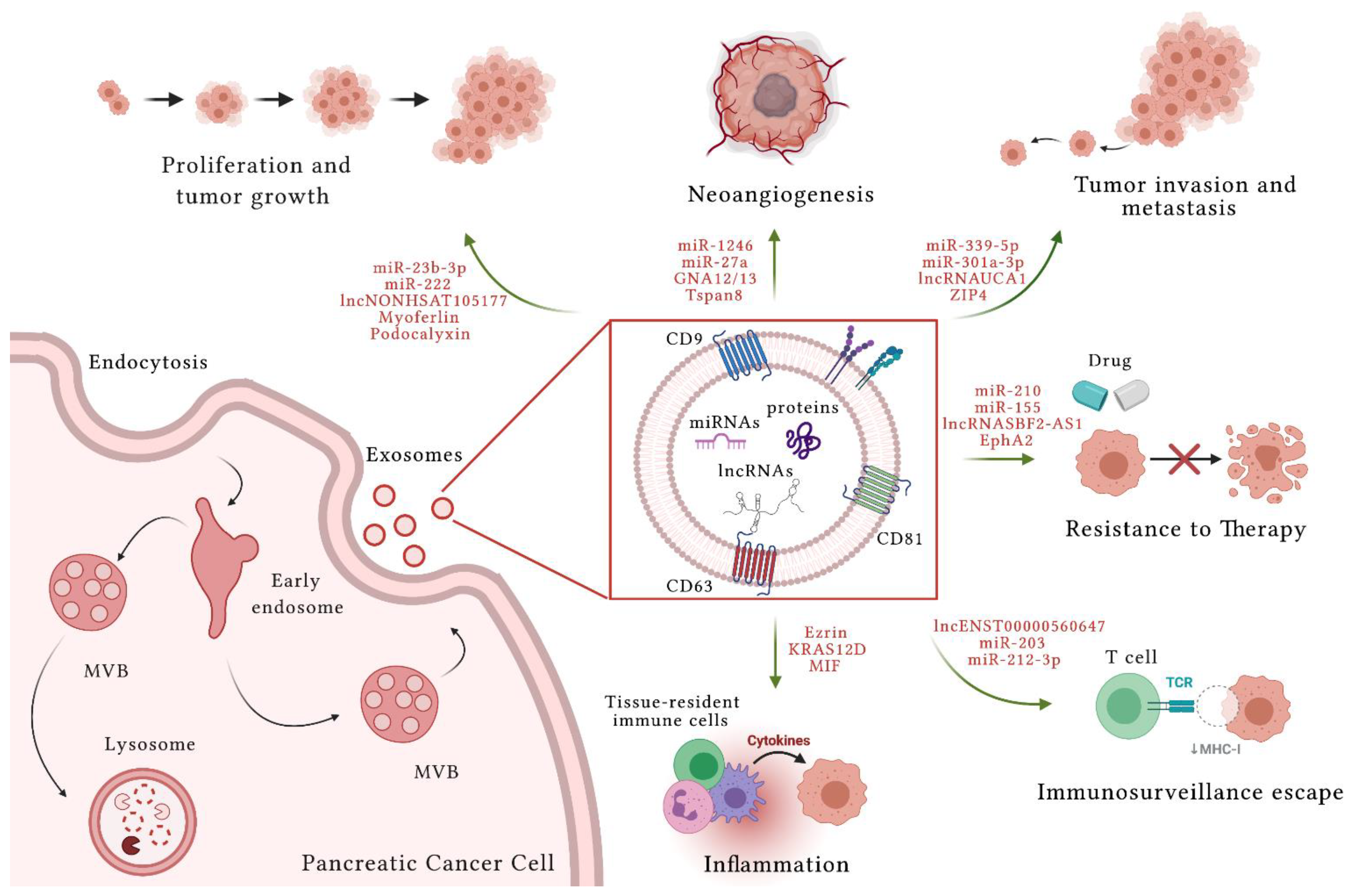
2. Exosomes
3. Extracellular Vesicles in Pancreatic Cancer
3.1. Pancreatic Cancer and Extracellular Vesicle miRNAs
3.2. Pancreatic Cancer and Extracellular Vesicle lncRNAs
3.3. Pancreatic Cancer and Extracellular Vesicle Proteins
4. Extracellular Vesicles and Inflammation in Pancreatic Cancer
5. Extracellular Vesicles and Tumor Microenvironment: From CAFs to PC
6. Extracellular Vesicles in Pancreatic Cancer Diagnosis and Prognosis: Methodologies of Purification and Clinical Implications
6.1. Methodologies for High-Quality Purification of Extracellular Vesicles
6.2. Extracellular Vesicles as Diagnostic and Prognostic Biomarkers of Pancreatic Cancer
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cameron, J.L.; He, J. Two thousand consecutive pancreaticoduodenectomies. J. Am. Coll. Surg. 2015, 220, 530–536. [Google Scholar] [CrossRef]
- Bisht, S.; Feldmann, G. Novel targets in pancreatic cancer therapy—Current status and ongoing translational efforts. Oncol. Res. Treat. 2018, 41, 596–602. [Google Scholar] [CrossRef]
- Deplanque, G.; Demartines, N. Pancreatic cancer: Are more chemotherapy and surgery needed? Lancet 2017, 389, 985–986. [Google Scholar] [CrossRef]
- Ducreux, M.; Sa Cuhna, A.; Caramella, C.; Hollebecque, A.; Burtin, P.; Goéré, D.; Seufferlein, T.; Haustermans, K.; Van Laethem, J.L.; Conroy, T.; et al. Cancer of the pancreas: Esmo clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, v56–v68. [Google Scholar] [CrossRef]
- Cho, I.R.; Kang, H.; Jo, J.H.; Lee, H.S.; Chung, M.J.; Park, J.Y.; Park, S.W.; Song, S.Y.; An, C.; Park, M.-S.; et al. Folfirinox vs gemcitabine/nab-paclitaxel for treatment of metastatic pancreatic cancer: Single-center cohort study. World J. Gastrointest. Oncol. 2020, 12, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Sohal, D.P.S.; Mangu, P.B.; Khorana, A.A.; Shah, M.A.; Philip, P.A.; O’Reilly, E.M.; Uronis, H.E.; Ramanathan, R.K.; Crane, C.H.; Engebretson, A.; et al. Metastatic pancreatic cancer: American society of clinical oncology clinical practice guideline. J. Clin. Oncol. 2016, 34, 2784–2796. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardière, C.; et al. Folfirinox versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Yachida, S.; Iacobuzio-Donahue, C.A. Evolution and dynamics of pancreatic cancer progression. Oncogene 2013, 32, 5253–5260. [Google Scholar] [CrossRef] [PubMed]
- Maitra, A.; Fukushima, N.; Takaori, K.; Hruban, R.H. Precursors to invasive pancreatic cancer. Adv. Anat. Pathol. 2005, 12, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef]
- Hu, H.-F.; Ye, Z.; Qin, Y.; Xu, X.-W.; Yu, X.-J.; Zhuo, Q.-F.; Ji, S.-R. Mutations in key driver genes of pancreatic cancer: Molecularly targeted therapies and other clinical implications. Acta Pharmacol. Sin. 2021, in press. [Google Scholar] [CrossRef]
- Hruban, R.H.; Goggins, M.; Parsons, J.; Kern, S.E. Progression model for pancreatic cancer. Clin. Cancer Res. 2000, 6, 2969–2977. [Google Scholar] [PubMed]
- Yako, Y.Y.; Kruger, D.; Smith, M.; Brand, M. Cytokines as biomarkers of pancreatic ductal adenocarcinoma: A systematic review. PLoS ONE 2016, 11, e0154016. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Brehm, C.U.; Gress, T.M.; Buchholz, M.; Alashkar Alhamwe, B.; von Strandmann, E.P.; Slater, E.P.; Bartsch, J.W.; Bauer, C.; Lauth, M. The immune microenvironment in pancreatic cancer. Int. J. Mol. Sci. 2020, 21, 7307. [Google Scholar] [CrossRef]
- Padoan, A.; Plebani, M.; Basso, D. Inflammation and pancreatic cancer: Focus on metabolism, cytokines, and immunity. Int. J. Mol. Sci. 2019, 20, 676. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (misev2018): A position statement of the international society for extracellular vesicles and update of the misev2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xia, W.; Lv, Z.; Ni, C.; Xin, Y.; Yang, L. Liquid biopsy for cancer: Circulating tumor cells, circulating free DNA or exosomes? Cell. Physiol. Biochem. 2017, 41, 755–768. [Google Scholar] [CrossRef]
- Yan, Y.; Fu, G.; Ming, L. Role of exosomes in pancreatic cancer. Oncol. Lett. 2018, 15, 7479–7488. [Google Scholar] [PubMed]
- Wang, C.C.; Zhao, Y.M.; Wang, H.Y.; Zhao, Y.P. New insight into the role of exosomes in pancreatic cancer. Ann. Clin. Lab. Sci. 2019, 49, 385–392. [Google Scholar] [PubMed]
- Sun, W.; Ren, Y.; Lu, Z.; Zaho, X. The potential role of exosomes in pancreatic cancer initiation and metastasis. Mol. Cancer 2020, 19, 135. [Google Scholar] [CrossRef] [PubMed]
- Battke, C.; Ruiss, R.; Welsch, U.; Wimberger, P.; Lang, S.; Jochum, S.; Zeidler, R. Tumour exosomes inhibit binding of tu-mour-reactive antibodies to tumour cells and reduce ADCC. Cancer Immunol. Immunother. 2011, 60, 639–648. [Google Scholar] [CrossRef]
- Lan, B.; Zeng, S.; Grützmann, R.; Pilarsky, C. The role of exosomes in pancreatic cancer. Int. J. Mol. Med. 2019, 20, 4332. [Google Scholar]
- Moeng, S.; Son, S.W.; Lee, J.S.; Lee, H.Y.; Kim, T.H.; Choi, S.Y.; Kuh, H.J.; Park, J.K. Extracellular vesicles (evs) and pancreatic cancer: From the role of evs to the interference with ev-mediated reciprocal communication. Biomedicines 2020, 8, 267. [Google Scholar] [CrossRef] [PubMed]
- De Lellis, L.; Florio, R.; Di Bella, M.C.; Brocco, D.; Guidotti, F.; Tinari, N.; Grassadonia, A.; Lattanzio, R.; Cama, A.; Veschi, S. Exosomes as pleiotropic players in pancreatic cancer. Biomedicines 2021, 9, 275. [Google Scholar] [CrossRef]
- Picca, A.; Guerra, F.; Calvani, R.; Coelho-Júnior, H.J.; Landi, F.; Bernabei, R.; Romano, R.; Bucci, C.; Marzetti, E. Extracellular vesicles and damage-associated molecular patterns: A pandora’s box in health and disease. Front. Immunol. 2020, 11, 601740. [Google Scholar] [CrossRef] [PubMed]
- Meldolesi, J. Exosomes and ectosomes in intercellular communication. Curr. Biol. 2018, 28, R435–R444. [Google Scholar] [CrossRef] [PubMed]
- Gruenberg, J.; Stenmark, H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004, 5, 317–323. [Google Scholar] [CrossRef]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The escrt pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Thery, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Charrin, S.; Jouannet, S.; Boucheix, C.; Rubinstein, E. Tetraspanins at a glance. J. Cell Sci. 2014, 127, 3641–3648. [Google Scholar] [CrossRef]
- Perez-Hernandez, D.; Gutierrez-Vazquez, C.; Jorge, I.; Lopez-Martin, S.; Ursa, A.; Sanchez-Madrid, F.; Vazquez, J.; Yanez-Mo, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 11649–11661. [Google Scholar] [CrossRef]
- Hullin-Matsuda, F.; Taguchi, T.; Greimel, P.; Kobayashi, T. Lipid compartmentalization in the endosome system. Semin. Cell Dev. Biol. 2014, 31, 48–56. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Jalalian, S.H.; Ramezani, M.; Jalalian, S.A.; Abnous, K.; Taghdisi, S.M. Exosomes, new biomarkers in early cancer detection. Anal. Biochem. 2019, 571, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Fader, C.M.; Sánchez, D.G.; Mestre, M.B.; Colombo, M.I. Ti-vamp/vamp7 and vamp3/cellubrevin: Two v-snare proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta 2009, 1793, 1901–1916. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.; Morohashi, Y.; Yoshimura, S.I.; Manrique-Hoyos, N.; Jung, S.Y.; Lauterbach, M.A.; Bakhti, M.; Grønborg, M.; Möbius, W.; Rhee, J.; et al. Regulation of exosome secretion by rab35 and its gtpase-activating proteins tbc1d10a-c. J. Cell Biol. 2010, 189, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Guerra, F.; Paiano, A.; Migoni, D.; Girolimetti, G.; Perrone, A.M.; De Iaco, P.; Fanizzi, F.P.; Gasparre, G.; Bucci, C. Modulation of rab7a protein expression determines resistance to cisplatin through late endocytic pathway impairment and extracellular vesicular secretion. Cancers 2019, 11, 52. [Google Scholar] [CrossRef]
- Guerra, F.; Bucci, C. Role of the rab7 protein in tumor progression and cisplatin chemoresistance. Cancers 2019, 11, 1096. [Google Scholar] [CrossRef]
- Jadli, A.S.; Ballasy, N.; Edalat, P.; Patel, V.B. Inside (sight) of tiny communicator: Exosome biogenesis, secretion, and uptake. Mol. Cell. Biochem. 2020, 467, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Schey, K.L.; Luther, J.M.; Rose, K.L. Proteomics characterization of exosome cargo. Methods 2015, 87, 75–82. [Google Scholar] [CrossRef]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Simeone, P.; Bologna, G.; Lanuti, P.; Pierdomenico, L.; Guagnano, M.T.; Pieragostino, D.; Del Boccio, P.; Vergara, D.; Marchisio, M.; Miscia, S.; et al. Extracellular vesicles as signaling mediators and disease biomarkers across biological barriers. Int. J. Mol. Sci. 2020, 21, 2514. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. Rna maps reveal new rna classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the seed supports microrna targeting specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. Elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microrna expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Ding, G.; Zhou, L.; Qian, Y.; Fu, M.; Chen, J.; Chen, J.; Xiang, J.; Wu, Z.; Jiang, G.; Cao, L. Pancreatic cancer-derived exosomes transfer mirnas to dendritic cells and inhibit rfxap expression via mir-212-3p. Oncotarget 2015, 6, 29877–29888. [Google Scholar] [CrossRef]
- Madhavan, B.; Yue, S.; Galli, U.; Rana, S.; Gross, W.; Müller, M.; Giese, N.A.; Kalthoff, H.; Becker, T.; Büchler, M.W.; et al. Combined evaluation of a panel of protein and mirna serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer 2015, 136, 2616–2627. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of micrornas detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous rnas modulate microrna sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Micrornas as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. Microrna dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- Ma, Y.; Yu, S.; Zhao, W.; Lu, Z.; Chen, J. Mir-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting sprouty2. Cancer Lett. 2010, 298, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Shang, D.; Xie, C.; Hu, J.; Tan, J.; Yuan, Y.; Liu, Z.; Yang, Z. Pancreatic cancer cell-derived exosomal microrna-27a promotes angiogenesis of human microvascular endothelial cells in pancreatic cancer via btg2. J. Cell. Mol. Med. 2020, 24, 588–604. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhao, S.; Wang, L.; Wang, J.; Zhou, J. Mirna-339-5p plays an important role in invasion and migration of pancreatic cancer cells. Med. Sci Monit. 2019, 25, 7509–7517. [Google Scholar] [CrossRef] [PubMed]
- Patton, M.C.; Zubair, H.; Khan, M.A.; Singh, S.; Singh, A.P. Hypoxia alters the release and size distribution of extracellular vesicles in pancreatic cancer cells to support their adaptive survival. J. Cell Biochem. 2019, 121, 828–839. [Google Scholar] [CrossRef]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic tumor-derived exosomal mir-301a mediates m2 macrophage polarization via pten/pi3kgamma to promote pancreatic cancer metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef]
- Zhai, L.Y.; Li, M.X.; Pan, W.L.; Chen, Y.; Li, M.M.; Pang, J.X.; Zheng, L.; Chen, J.X.; Duan, W.J. In situ detection of plasma exosomal microrna-1246 for breast cancer diagnostics by a au nanoflare probe. ACS Appl. Mater. Interfaces 2018, 10, 39478–39486. [Google Scholar] [CrossRef]
- Bhagirath, D.; Yang, T.L.; Bucay, N.; Sekhon, K.; Majid, S.; Shahryari, V.; Dahiya, R.; Tanaka, Y.; Saini, S. Microrna-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res. 2018, 78, 1833–1844. [Google Scholar] [CrossRef]
- Hasegawa, S.; Eguchi, H.; Nagano, H.; Konno, M.; Tomimaru, Y.; Wada, H.; Hama, N.; Kawamoto, K.; Kobayashi, S.; Nishida, N.; et al. Microrna-1246 expression associated with ccng2-mediated chemoresistance and stemness in pancreatic cancer. Br. J. Cancer 2014, 111, 1572–1580. [Google Scholar] [CrossRef]
- Patel, G.K.; Khan, M.A.; Bhardwaj, A.; Srivastava, S.K.; Zubair, H.; Patton, M.C.; Singh, S.; Khushman, M.; Singh, A.P. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ros detoxification and mir-155-mediated suppression of key gemcitabine-metabolising enzyme, dck. Br. J. Cancer 2017, 116, 609–619. [Google Scholar] [CrossRef]
- Mikamori, M.; Yamada, D.; Eguchi, H.; Hasegawa, S.; Kishimoto, T.; Tomimaru, Y.; Asaoka, T.; Noda, T.; Wada, H.; Kawamoto, K.; et al. Microrna-155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci. Rep. 2017, 7, 4233. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhao, N.; Cui, J.; Wu, H.; Xiong, J.; Peng, T. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering mir-210. Cell. Oncol. 2020, 43, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The mir-200 family determines the epithelial phenotype of cancer cells by targeting the e-cadherin repressors zeb1 and zeb2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, X.; Xia, M.; Wu, F.; Ding, J.; Jiao, Y.; Zhan, Q.; An, F. Upregulated exosomic mir-23b-3p plays regulatory roles in the progression of pancreatic cancer. Oncol. Rep. 2017, 38, 2182–2188. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.-J.; Chen, Y.-Y.; Dai, J.-J.; Gu, D.-N.; Mei, Z.; Liu, F.-R.; Huang, Q.; Tian, L. Dying tumor cell-derived exosomal mir-194-5p potentiates survival and repopulation of tumor repopulating cells upon radiotherapy in pancreatic cancer. Mol. Cancer 2020, 19, 68. [Google Scholar] [CrossRef]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncrnas regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef]
- Pandya, G.; Kirtonia, A.; Sethi, G.; Pandey, A.K.; Garg, M.T. He implication of long non-coding rnas in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188423. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, X.; Zhou, Y.; Liu, Y.; Zhou, Q.; Ye, H.; Wang, Y.; Zeng, J.; Song, Y.; Gao, W.; et al. The long non- coding rna hottip promotes progression and gemcitabine resistance by regulating hoxa13 in pancreatic cancer. J. Transl. Med. 2015, 13, 84. [Google Scholar] [CrossRef]
- Chien, W.; Sudo, M.; Ding, L.W.; Sun, Q.Y.; Wuensche, P.; Lee, K.L.; Hattori, N.; Garg, M.; Xu, L.; Zheng, Y.; et al. Functional genome-wide screening identifies targets and pathways sensitizing pancreatic cancer cells to dasatinib. J. Cancer 2018, 9, 4762–4773. [Google Scholar] [CrossRef]
- Huang, X.; Zhi, X.; Gao, Y.; Ta, N.; Jiang, H.; Zheng, J. Lncrnas in pancreatic cancer. Oncotarget 2016, 7, 57379–57390. [Google Scholar] [CrossRef]
- Yin, Z.; Zhou, Y.; Ma, T.; Chen, S.; Shi, N.; Zou, Y.; Hou, B.; Zhang, C. Down-regulated lncrna sbf2-as1 in m2 macrophage-derived exosomes elevates mir-122-5p to restrict xiap, thereby limiting pancreatic cancer development. J. Cell Mol. Med. 2020, 24, 5028–5038. [Google Scholar] [CrossRef]
- Wang, X.; Li, H.; Lu, X.; Wen, C.; Huo, Z.; Shi, M.; Tang, X.; Chen, H.; Peng, C.; Fang, Y.; et al. Melittin-induced long non-coding rna nonhsat105177 inhibits proliferation and migration of pancreatic ductal adenocarcinoma. Cell Death Dis. 2018, 9, 940. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, X.; Yang, Y.; Chen, W.; Zhang, K.; Teng, B.; Huang, C.; Zhao, Q.; Qiu, Z. Hypoxic tumor-derived exosomal long noncoding rna uca1 promotes angiogenesis via mir-96-5p/amotl2 in pancreatic cancer. Mol. Ther. Nucleic Acids 2020, 22, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, P.; Li, J.; Peng, M.; Zhao, X.; Zhang, X.; Chen, K.; Zhang, Y.; Liu, H.; Gan, L.; et al. Tumor-derived exosomal lnc-sox2ot promotes emt and stemness by acting as a cerna in pancreatic ductal adenocarcinoma. Oncogene 2018, 37, 3822–3838. [Google Scholar] [CrossRef]
- Sun, C.; Wang, P.; Dong, W.; Liu, H.; Sun, J.; Zhao, L. Lncrna pvt1 promotes exosome secretion through ykt6, rab7, and vamp3 in pancreatic cancer. Aging 2020, 12, 10427–10440. [Google Scholar] [CrossRef]
- Stefanius, K.; Servage, K.; de Souza Santos, M.; Gray, H.F.; Toombs, J.E.; Chimalapati, S.; Kim, M.S.; Malladi, V.S.; Brekken, R.; Orth, K. Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. eLife 2019, 8, e40226. [Google Scholar] [CrossRef]
- Servage, K.A.; Stefanius, K.; Gray, H.F.; Orth, K. Proteomic profiling of small extracellular vesicles secreted by human pancreatic cancer cells implicated in cellular transformation. Sci. Rep. 2020, 10, 7713. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.R.; Minden, A.; Karin, M.; Brown, J.H. Galpha12 stimulates c-jun nh2-terminal kinase through the small g proteins ras and rac. J. Biol. Chem. 1996, 271. [Google Scholar] [CrossRef]
- Mitsui, H.; Takuwa, N.; Kurokawa, K.; Exton, J.H.; Takuwa, Y. Dependence of activated galpha12-induced g1 to s phase cell cycle progression on both ras/mitogen-activated protein kinase and ras/rac1/jun n-terminal kinase cascades in nih3t3 fibroblasts. J. Biol. Chem. 1997, 272, 4903–4910. [Google Scholar] [CrossRef] [PubMed]
- Buhl, A.M.; Johnson, N.L.; Dhanasekaran, N.; Johnson, G.L. G alpha 12 and g alpha 13 stimulate rho-dependent stress fiber formation and focal adhesion assembly. J. Biol. Chem. 1995, 270, 24631–24634. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, M.D.M.; Dharmawardhane, S. Targeting rac and cdc42 gtpases in cancer. Cancer Res. 2018, 78, 3101–3111. [Google Scholar] [CrossRef]
- Chiu, Y.H.; Macmillan, J.B.; Chen, Z.J. Rna polymerase iii detects cytosolic DNA and induces type i interferons through the rig-i pathway. Cell 2009, 138, 576–591. [Google Scholar] [CrossRef] [PubMed]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta 2012, 1826, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Szczepanski, M.J.; Szajnik, M.; Welsh, A.; Whiteside, T.L.; Boyiadzis, M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica 2011, 96, 1302–1309. [Google Scholar] [CrossRef]
- Jin, H.; Liu, P.; Wu, Y.; Meng, X.; Wu, M.; Han, J.; Tan, X. Exosomal zinc transporter zip4 promotes cancer growth and is a novel diagnostic biomarker for pancreatic cancer. Cancer Sci. 2018, 109, 2946–2956. [Google Scholar] [CrossRef]
- Yan, Q.; Yuan, W.B.; Sun, X.; Zhang, M.J.; Cen, F.; Zhou, S.Y.; Wu, W.B.; Xu, Y.C.; Tong, L.H.; Ma, Z.H. Asparaginyl endopeptidase enhances pancreatic ductal adenocarcinoma cell invasion in an exosome-dependent manner and correlates with poor prognosis. Int. J. Oncol. 2018, 52, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; von Au, A.; Schnölzer, M.; Hackert, T.; Zöller, M. Cd44v6-competent tumor exosomes promote motility, invasion and cancer-initiating cell marker expression in pancreatic and colorectal cancer cells. Oncotarget 2016, 7, 55409–55436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.F.; Zhou, Y.Z.; Zhang, B.; Huang, S.F.; Li, P.P.; He, X.M.; Cao, G.D.; Kang, M.X.; Dong, X.; Wu, Y.L. Pancreatic cancer-derived exosomes promoted pancreatic stellate cells recruitment by pancreatic cancer. J. Cancer 2019, 10, 4397–4407. [Google Scholar] [CrossRef]
- Xu, Z.; Vonlaufen, A.; Phillips, P.A.; Fiala-Beer, E.; Zhang, X.; Yang, L.; Biankin, A.V.; Goldstein, D.; Pirola, R.C.; Wilson, J.S.; et al. Role of pancreatic stellate cells in pancreatic cancer metastasis. Am. J. Pathol. 2010, 177, 2585–2596. [Google Scholar] [CrossRef]
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zöller, M. Cell surface tetraspanin tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Mu, W.; Erb, U.; Zöller, M. The tetraspanins cd151 and tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget 2015, 6, 2366–2384. [Google Scholar] [CrossRef] [PubMed]
- Rademaker, G.; Hennequière, V.; Brohée, L.; Nokin, M.J.; Lovinfosse, P.; Durieux, F.; Gofflot, S.; Bellier, J.; Costanza, B.; Herfs, M.; et al. Myoferlin controls mitochondrial structure and activity in pancreatic ductal adenocarcinoma, and affects tumor aggressiveness. Oncogene 2018, 37, 4398–4412. [Google Scholar] [CrossRef]
- Leung, C.; Yu, C.; Lin, M.I.; Tognon, C.; Bernatchez, P. Expression of myoferlin in human and murine carcinoma tumors: Role in membrane repair, cell proliferation, and tumorigenesis. Am. J. Pathol. 2013, 182, 1900–1909. [Google Scholar] [CrossRef]
- Blomme, A.; Fahmy, K.; Peulen, O.; Costanza, B.; Fontaine, M.; Struman, I.; Baiwir, D.; de Pauw, E.; Thiry, M.; Bellahcène, A.; et al. Myoferlin is a novel exosomal protein and fuinctional regulator of cancer-derived exosomes. Oncotarget 2016, 7, 83669–83683. [Google Scholar] [CrossRef]
- Novo, D.; Heath, N.; Mitchell, L.; Caligiuri, G.; MacFarlane, A.; Reijmer, D.; Charlton, L.; Knight, J.; Calka, M.; McGhee, E.; et al. Mutant p53s generate pro-invasive niches by influencing exosome podocalyxin levels. Nat. Commun. 2018, 9, 5069. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Wei, Q.; Koay, E.J.; Liu, Y.; Ning, B.; Bernard, P.W.; Zhang, N.; Han, H.; Katz, M.H.; Zhao, Z.; et al. Chemoresistance transmission via exosome-mediated epha2 transfer in pancreatic cancer. Theranostics 2018, 8, 5986–5994. [Google Scholar] [CrossRef] [PubMed]
- Manohar, M.; Verma, A.K.; Venkateshaiah, S.U.; Sanders, N.L.; Mishra, A. Pathogenic mechanisms of pancreatitis. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, S.; Lowenfels, A.B.; Morselli-Labate, A.M.; Maisonneuve, P.; Pezzilli, R. Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection. Best Pract. Res. Clin. Gastroenterol. 2010, 24, 349–358. [Google Scholar] [CrossRef]
- Bonjoch, L.; Casas, V.; Carrascal, M.; Closa, D. Involvement of exosomes in lung inflammation associated with experimental acute pancreatitis. J. Pathol. 2016, 240, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.B.; Sun, H.Y.; Luo, Z.L.; Cheng, L.; Duan, X.M.; Ren, J.D. Plasma-derived exosomes contribute to pancreatitis-associated lung injury by triggering nlrp3-dependent pyroptosis in alveolar macrophages. Biochim. Biophys. Acta 2020, 1866, 165685. [Google Scholar] [CrossRef]
- Jiménez-Alesanco, A.; Marcuello, M.; Pastor-Jiménez, M.; López-Puerto, L.; Bonjoch, L.; Gironella, M.; Carrascal, M.; Abian, J.; de-Madaria, E.; Closa, D. Acute pancreatitis promotes the generation of two different exosome populations. Sci. Rep. 2019, 9, 19887. [Google Scholar] [CrossRef]
- Ansari, D.; Friess, H.; Bauden, M.; Samnegård, J.; Andersson, R. Pancreatic cancer: Disease dynamics, tumor biology and the role of the microenvironment. Oncotarget 2018, 9, 6644–6651. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Huang, B.; Hanash, S.M.; Onuchic, J.N.; Ben-Jacob, E. Modeling putative therapeutic implications of exosome exchange between tumor and immune cells. Proc. Natl. Acad. Sci. USA 2014, 111, E4165–E4174. [Google Scholar] [CrossRef]
- Tucci, M.; Passarelli, A.; Mannavola, F.; Felici, C.; Stucci, L.S.; Cives, M.; Silvestris, F. Immune system evasion as hallmark of melanoma progression: The role of dendritic cells. Front. Oncol. 2019, 9, 1148. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Chen, J.; Zhou, L.; Chen, W.; Ding, G.; Cao, L. Pancreatic cancer derived exosomes regulate the expression of tlr4 in dendritic cells via mir-203. Cell. Immunol. 2014, 292, 65–69. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal pd-l1 contributes to immunosuppression and is associated with anti-pd-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Ning, B.; Spiegel, S.; Lyon, C.J.; Hu, T.Y. Tumor-derived exosomes (tdes): How to avoid the sting in the tail. Med. Res. Rev. 2020, 40, 385–412. [Google Scholar] [CrossRef]
- Chen, J.; Wang, S.; Jia, S.; Ding, G.; Jiang, G.; Cao, L. Integrated analysis of long non- coding rna and mrna expression profile in pancreatic cancer derived exosomes treated dendritic cells by microarray analysis. J. Cancer 2018, 9, 21–31. [Google Scholar] [CrossRef]
- Capello, M.; Vykoukal, J.V.; Katayama, H.; Bantis, L.E.; Wang, H.; Kundnani, D.L.; Aguilar-Bonavides, C.; Aguilar, M.; Tripathi, S.C.; Dhillon, D.S.; et al. Exosomes harbor b cell targets in pancreatic adenocarcinoma and exert decoy function against complement-mediated cytotoxicity. Nat. Commun. 2019, 10, 254. [Google Scholar] [CrossRef]
- Zech, D.; Rana, S.; Büchler, M.W.; Zöller, M. Tumor-exosomes and leukocyte activation: An ambivalent crosstalk. Cell Commun. Signal. 2012, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Zeitouni, D.; Pylayeva-Gupta, Y.; Der, C.J.; Bryant, K.L. Kras mutant pancreatic cancer: No lone path to an effective treatment. Cancers 2016, 8, 45. [Google Scholar] [CrossRef]
- Dai, E.; Han, L.; Liu, J.; Xie, Y.; Kroemer, G.; Klionsky, D.J.; Zeh, H.J.; Kang, R.; Wang, J.; Tang, D. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic kras protein. Autophagy 2020, 16, 2069–2083. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.T.; Peng, H.Y.; Hu, C.M.; Huang, S.C.; Tien, S.C.; Jeng, Y.M. Pancreatic cancer-derived small extracellular vesical ezrin regulates macrophage polarization and promotes metastasis. Am. J. Cancer Res. 2020, 10, 12–37. [Google Scholar] [PubMed]
- Helm, O.; Held-Feindt, J.; Schäfer, H.; Sebens, S. M1 and m2: There is no “good” and “bad”-how macrophages promote malignancy-associated features in tumorigenesis. Oncoimmunology 2014, 3, e946818. [Google Scholar] [CrossRef][Green Version]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Morine, Y.; Ikemoto, T.; Imura, S.; Iwahashi, S.; Saito, Y.; Shimada, M. Nab-paclitaxel interrupts cancer-stromal interaction through c-x-c motif chemokine 10-mediated interleukin-6 downregulation in vitro. Cancer Sci. 2018, 109, 2509–2519. [Google Scholar] [CrossRef]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef]
- Ishii, N.; Araki, K.; Yokobori, T.; Hagiwara, K.; Gantumur, D.; Yamanaka, T.; Handa, T.; Tsukagoshi, M.; Igarashi, T.; Watanabe, A.; et al. Conophylline suppresses pancreatic cancer desmoplasia and cancer-promoting cytokines produced by cancer-associated fibroblasts. Cancer Sci. 2019, 110, 334–344. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. eLife 2016, 5, e10250. [Google Scholar] [CrossRef]
- Basso, D.; Gnatta, E.; Padoan, A.; Fogar, P.; Furlanello, S.; Aita, A.; Bozzato, D.; Zambon, C.F.; Arrigoni, G.; Frasson, C.; et al. Pdac-derived exosomes enrich the microenvironment in mdscs in a smad4-dependent manner through a new calcium related axis. Oncotarget 2017, 8, 84928–84944. [Google Scholar] [CrossRef] [PubMed]
- Landskron, G.; De la Fuente, M.; Thuwajit, P.; Thuwajit, C.; Hermoso, M.A. Chronic inflammation and cytokines in the tumor microenvironment. J. Immunol. Res. 2014, 2014, 149185. [Google Scholar] [CrossRef] [PubMed]
- Pop, V.V.; Seicean, A.; Lupan, I.; Samasca, G.; Burz, C.C. Il-6 roles—Molecular pathway and clinical implication in pancreatic cancer—A systemic review. Immunol. Lett. 2017, 181, 45–80. [Google Scholar] [CrossRef] [PubMed]
- Basso, D.; Bozzato, D.; Padoan, A.; Moz, S.; Zambon, C.F.; Fogar, P.; Greco, E.; Scorzeto, M.; Simonato, F.; Navaglia, F.; et al. Inflammation and pancreatic cancer: Molecular and functional interactions between s100a8, s100a9, nt-s100a8 and tgfβ1. Cell Commun. Signal. 2014, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Karakhanova, S.; Link, J.; Heinrich, M.; Shevchenko, I.; Yang, Y.; Hassenpflug, M.; Bunge, H.; von Ahn, K.; Brecht, R.; Mathes, A.; et al. Characterization of myeloid leukocytes and soluble mediators in pancreatic cancer: Importance of myeloid-derived suppressor cells. Oncoimmunology 2015, 4, e998519. [Google Scholar] [CrossRef]
- Helm, O.; Held-Feindt, J.; Grage-Griebenow, E.; Reiling, N.; Ungefroren, H.; Vogel, I.; Krüger, U.; Becker, T.; Ebsen, M.; Röcken, C.; et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer 2014, 135, 843–861. [Google Scholar] [CrossRef]
- Balkwill, F. Tumour necrosis factor and cancer. Nat. Rev. Cancer 2009, 9, 361–371. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, G.; Li, Q.; Wang, Z.; Hu, W.; Li, P.; Li, S.; Wu, H.; Kong, X.; Gao, J.; et al. Hedgehog signaling non-canonical activated by pro-inflammatory cytokines in pancreatic ductal adenocarcinoma. J. Cancer 2016, 7, 2067–2076. [Google Scholar] [CrossRef]
- Chopra, M.; Lang, I.; Salzmann, S.; Pachel, C.; Kraus, S.; Bäuerlein, C.A.; Brede, C.; Garrote, A.L.; Mattenheimer, K.; Ritz, M.; et al. Tumor necrosis factor induces tumor promoting and anti-tumoral effects on pancreatic cancer via tnfr1. PLoS ONE 2013, 8, e75737. [Google Scholar] [CrossRef]
- Zhao, X.; Fan, W.; Xu, Z.; Chen, H.; He, Y.; Yang, G.; Yang, G.; Hu, H.; Tang, S.; Wang, P.; et al. Inhibiting tumor necrosis factor-alpha diminishes desmoplasia and inflammation to overcome chemoresistance in pancreatic ductal adenocarcinoma. Oncotarget 2016, 7, 81110–81122. [Google Scholar] [CrossRef]
- Kali, A.; Ostapchuk, Y.O.; Belyaev, N.N. Tnfα and tgfβ-1 synergistically increase the cancer stem cell properties of miapaca-2 cells. Oncol. Lett. 2017, 14, 4647–4658. [Google Scholar] [CrossRef]
- Weniger, M.; Honselmann, K.C.; Liss, A.S. The extracellular matrix and pancreatic cancer: A complex relationship. Cancers 2018, 10, 316. [Google Scholar] [CrossRef]
- Kleeff, J.; Beckhove, P.; Esposito, I.; Herzig, S.; Huber, P.E.; Lohr, J.M.; Friess, H. Pancreatic cancer microenvironment. Int. J. Cancer 2007, 121, 699–705. [Google Scholar] [CrossRef]
- Neesse, A.; Krug, S.; Gress, T.M.; Tuveson, D.A.; Michl, P. Emerging concepts in pancreatic cancer medicine: Targeting the tumor stroma. OncoTargets Ther. 2013, 7, 33–43. [Google Scholar] [CrossRef]
- Apte, M.V.; Wilson, J.S.; Lugea, A.; Pandol, S.J. A starring role for stellate cells in the pancreatic cancer microenvironment. Gastroenterology 2013, 144, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Secq, V.; Leca, J.; Bressy, C.; Guillaumond, F.; Skrobuk, P.; Nigri, J.; Lac, S.; Lavaut, M.N.; Bui, T.T.; Thakur, A.K.; et al. Stromal slit2 impacts on pancreatic cancer-associated neural remodeling. Cell Death Dis. 2015, 6, e1592. [Google Scholar] [CrossRef]
- Duluc, C.; Moatassim-Billah, S.; Chalabi-Dchar, M.; Perraud, A.; Samain, R.; Breibach, F.; Gayral, M.; Cordelier, P.; Delisle, M.-B.; Bousquet-Dubouch, M.-P.; et al. Pharmacological targeting of the protein synthesis mtor/4e-bp1 pathway in cancer-associated fibroblasts abrogates pancreatic tumor chemoresistance. EMBO Mol. Med. 2015, 7, 735–753. [Google Scholar] [CrossRef]
- Richards, K.E.; Zeleniak, A.E.; Fishel, M.L.; Wu, J.; Littlepage, L.E.; Hill, R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene 2016, 36, 1770–1778. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Zhou, W.; Rong, Y.; Kuang, T.; Xu, X.; Wu, W.; Wang, D.; Lou, W. Exosomal mirna-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp. Cell Res. 2019, 383, 111543. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Li, L.; Wang, G.; Deng, X.; Li, Z.; Kong, X. Vdr signaling inhibits cancer-associated-fibroblasts’ release of exosomal mir-10a-5p and limits their supportive effects on pancreatic cancer cells. Gut 2018, 68, 950–951. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.H.; Yu, R.T.; Engle, D.D.; Ding, N.; Atkins, A.R.; Tiriac, H.; Collisson, E.A.; Connor, F.; Van Dyke, T.; Kozlov, S.; et al. Vitamin d receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 2014, 159, 80–93. [Google Scholar] [CrossRef]
- Leca, J.; Martinez, S.; Lac, S.; Nigri, J.; Secq, V.; Rubis, M.; Bressy, C.; Sergé, A.; Lavaut, M.-N.; Dusetti, N.; et al. Cancer-associated fibroblast-derived annexin a6+ extracellular vesicles support pancreatic cancer aggressiveness. J. Clin. Invest. 2016, 126, 4140–4156. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Gonzalo, D.H.; Feely, M.; Rinaldi, C.; Belsare, S.; Zhai, H.; Kalra, K.; Gerber, M.H.; Forsmark, C.E.; Hughes, S.J. Stroma-derived extracellular vesicles deliver tumor-suppressive mirnas to pancreatic cancer cells. Oncotarget 2017, 9, 5764–5777. [Google Scholar] [CrossRef] [PubMed]
- Kurian, T.K.; Banik, S.; Gopal, S.; Chakrabarti, S.; Mazumder, N. Elucidating methods for isolation and quantification of exosomes: A review. Mol. Biotecnol. 2021, 63, 249–266. [Google Scholar] [CrossRef]
- An, M.; Wu, J.; Zhu, J.; Lubman, D.M. Comparison of an optimized ultracentrifugation method versus size-exclusion chromatography for isolation of exosomes from human serum. J. Proteome Res. 2018, 17, 3599–3605. [Google Scholar] [CrossRef]
- Barreiro, K.; Huber, T.B.; Holthofer, H. Isolating urinary extracellular vesicles as biomarkers for diabetic disease. In Diabetic Nephropathy: Methods in Molecular Biology; Long, L.G.D., Ed.; Humana: New York, NY, USA, 2020. [Google Scholar]
- Feng, Y.; Huang, W.; Wani, M.; Yu, X.; Ashraf, M. Ischemic preconditioning potentiates the protective effect of stem cells through secretion of exosomes by targeting mecp2 via mir-22. PLoS ONE 2014, 9, 1–8. [Google Scholar] [CrossRef]
- Rider, M.A.; Hurwitz, S.N.; Meckes, D.G.J. ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 2016, 6, 23978. [Google Scholar] [CrossRef]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef]
- Qiu, J.; Yang, G.; Feng, M.; Zheng, S.; Cao, Z.; You, L.; Zheng, L.; Zhang, T.; Zhao, Y. Extracellular vesicles as mediators of the progression and chemoresistance of pancreatic cancer and their potential clinical applications. Mol. Cancer 2018, 17, 2. [Google Scholar] [CrossRef]
- Kulasingam, V.; Pavlou, M.P.; Diamandis, E.P. Integrating high-throughput technologies in the quest for effective biomarkers for ovarian cancer. Nat. Rev. Cancer 2010, 10, 371–378. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.; Li, H.; Wu, Y.; Zhang, H.; Chen, W. Tumor markers ca19-9, ca242 and cea in the diagnosis of pancreatic cancer: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 11683–11691. [Google Scholar]
- Ali, S.; Dubaybo, H.; Brand, R.E.; Sarkar, F.H. Differential expression of micrornas in tissues and plasma co-exists as a biomarker for pancreatic cancer. J. Cancer Sci. 2015, 7, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.K.; Deitz-McElyea, S.; Liyanage, T.; Lawrence, K.; Mali, S.; Sardar, R.; Korc, M. Label-free nanoplasmonic-based short noncoding rna sensing at attomolar concentrations allows for quantitative and highly specific assay of microrna-10b in biological fluids and circulating exosomes. ACS Nano 2015, 9, 11075–11089. [Google Scholar] [CrossRef]
- Xu, Y.-F.; Hannafon, B.N.; Zhao, Y.D.; Postier, R.G.; Ding, W.-Q. Plasma exosome mir-196a and mir-1246 are potential indicators of localized pancreatic cancer. Oncotarget 2017, 8, 77028–77040. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Wang, M.; McElyea, S.D.; Sherman, S.; House, M.; Korc, M. A microrna signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017, 393, 86–93. [Google Scholar] [CrossRef]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of serum exosomal micrornas and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef]
- Yoshizawa, N.; Sugimoto, K.; Tameda, M.; Inagaki, Y.; Ikejiri, M.; Inoue, H.; Usui, M.; Ito, M.; Takei, Y. Mir-3940-5p/mir-8069 ratio in urine exosomes is a novel diagnostic biomarker for pancreatic ductal adenocarcinoma. Oncol. Lett. 2020, 19, 2677–2684. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Wei, Q.; Li, Z.; Feng, H.; Ren, L. Serum exosomal epha2 is a prognostic biomarker in patients with pancreatic cancer. Cancer Manag. Res. 2021, 13, 3675–3683. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Flippot, R.; Malouf, G.G.; Su, X.; Mouawad, R.; Spano, J.P.; Khayat, D. Cancer subtypes classification using long non-coding rna. Oncotarget 2016, 7, 54082–54093. [Google Scholar] [CrossRef]
- Zhou, M.; Diao, Z.; Yue, X.; Chen, Y.; Zhao, H.; Cheng, H.; Sun, J. Construction and analysis of dysregulated lncrna-associated cerna network identified novel lncrna biomarkers for early diagnosis of human pancreatic cancer. Oncotarget 2016, 7, 56383–56394. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Li, Y.; Liao, Z.; Wang, Z.; Wang, Z.; Li, Y.; Qian, L.; Zhao, J.; Zong, H.; Kang, B.; et al. Plasma extracellular vesicle long rna profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut 2020, 69, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.R.; Kimchi, E.T.; Manjunath, Y.; Gajagowni, S.; Stuckel, A.J.; Kaifi, J.T. Rna cargos in extracellular vesicles derived from blood serum in pancreas associated conditions. Sci. Rep. 2020, 10, 2800. [Google Scholar] [CrossRef]
- Takahasi, K.; Iinuma, H.; Wada, K.; Minezaki, S.; Kawamura, S.; Kainuma, M.; Ikeda, Y.; Shibuya, M.; Miura, F.; Sano, K. Usefulness of exosome-encapsulated microrna-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J. Hepatobiliary Pancreat. Sci. 2018, 25, 155–161. [Google Scholar] [CrossRef] [PubMed]
| EV Content | Role in PC | Mechanism | Diagnostic/ Prognostic Marker | Ref. |
|---|---|---|---|---|
| miR-27a | Induce proliferation, invasion and angiogenesis | Suppression of BTG2 expression | - | [63] |
| miR-339-5p | Inhibit migration and invasion | Downregulation of ZNF689 | - | [63] |
| miR-222 | Induce proliferation and invasion | Downregulation, phosphorylation and nuclear exit of p27 through PPP2R2A/Akt pathway | - | [64] |
| miR-301a-3p | Promote invasion and metastasis | PTEN/PI3K signaling cascade | - | [66] |
| miR-1246 | Induce resistance to gemcitabine, metastasis, invasion, cancer stemness and angiogenesis. Detected in the patient’s serum | Inhibition of CCNG2 expression | Yes | [69] [57,164] |
| miR-155 | Induce resistance to gemcitabine | Downregulation of DCK | - | [70] |
| miR-210 | Induce resistance to gemcitabine, invasion and metastasis | Induction of ZEB1 and ZEB2 expression | - | [73] |
| miR-23b-3p | Promote proliferation, migration and invasion | Correlate with diagnosis of PC and with CA19-9 levels | - | [74] |
| miR-196b-5p/194-5p | Promote resistance to radiation, cell survival | Regulation of E2F3 and HMGA2 | - | [75] |
| miR-203 | Induce immunosurveillance escape | Inhibition of TLR4, TNF-α and IL12 expression, DC dysfunction | - | [115] |
| miR-212-3p | Induce immunosurveillance escape | Inhibit the DC role of APCs towards T lymphocytes by reducing the expression of the MHC II and associated regulatory factors | - | [56] |
| miR-17-p and miR-21 | High levels are associated to PC | Correlate with metastasis and advanced PC | Yes | [166] |
| miR-10b, miR-31, miR-30c, miR-181a, miR-221, miR-935, miR-508, miR-196a, | High levels in the plasma are associated to PC | Correlate with diagnosis of PC | Yes | [162,163,164,165] |
| miR-let7a | Low levels in the plasma are associated to PC | Correlate with diagnosis of PC | Yes | [165] |
| miR-451a | Associate to tumor size and stage of PC | Correlate with recurrence and survival | Yes | [175] |
| miR-3940-5p/miR-8069 ratio | High levels are specific in PC patient | Correlate with diagnosis of PC | Yes | [167] |
| miR-4306, miR-3976, miR-4644 | Detection in the patient’s serum | Allow to distinguish between patients with PC, CP and benign pancreatic lesions | Yes | [57] |
| lncRNA SBF2-AS1 | Inhibit of progression and increase of chemoresistance | Increase of miR-122-5p level | - | [81] |
| lncRNA NONHSAT105177 | Inhibit proliferation and migration | Modulation of cholesterol pathway | - | [82] |
| lncRNAUCA1 | Induce migration and tubulogenesis | Sponge of miR-96p | - | [83] |
| lncRNA SOX2OT | Promote EMT, invasion and metastasis | Associated with tumor stage | Yes | [84] |
| lncRNA HULC | Modulate cellular signaling, invasion and migration | Interact with TGF-β | Yes | [159] |
| lncRNA ENST00000560647 | Induce immunosurveillance escape | Induce DC dysfunction | - | [118] |
| lncRNA FGA, KRT19, HIST1H2BK, ITIH2, MARCH2, CLDN1, MAL2, TIM1 | Allow to distinguish between patients with stage I and stage III tumors | Diagnostic signature | Yes | [173] |
| lncRNA MALAT1 and CRNDE | High levels in serum of PC patient | Correlate with diagnosis of PC | Yes | [174] |
| GNA12/13 | Contribute to tumor growth, angiogenesis, EMT, migration and invasion | Activation of Ras, Rac, rho, CDC42 pathways | - | [[91] |
| ZIP4 | Enhance migration, invasion and chemoresistance | Not mentioned | - | [95] |
| AEP | Promote invasion | Activation of PI3K pathway | - | [96] |
| CD44v6 | Promote migration and invasion | Activate Wnt/β-Catenin pathway and increase PAI-1, MMP and TIM-1 | Yes | [97] [57] |
| Lin28B | Promote invasion and metastasis | Activation of Lin28B/let-7/HMGA2/PDGFB pathway | - | [108] |
| Integrin α6β4/ | Induce dissemination of metastasis to liver and lung tissues | Integrin-mediated organotropic incorporation of EVs | Yes | [170] |
| Tspan8, CD106, Cd49d | Promote neovascularization, tumor growth, metastasis | Increase expression of chemokine and receptor such as CXCR4 and EGFR3 | Yes | [100,101] [57] |
| Myoferlin | Induce migration, invasion, angiogenesis, proliferation | Regulation of mitochondrial organization and energy production, stimulate inclusion of VEGF into EVs | - | [102,103,104] |
| Podocalyxin | Induce migration and ECM organization | Correlation with TP53 mutations | - | [105] |
| EphA2 | Resistance to gemcitabine | Not mentioned | Yes | [106] |
| GPC1 | Associate to early stage of PC | Correlation with KRAS mutations | Yes | [168] |
| KRAS G12D | Correlate with poor survival | Stimulation of macrophage switch to an M2-like pro-tumor phenotype via STAT3-dependent fatty acid oxidation | - | [122] |
| Ezrin | ECM remodeling, angiogenesis, and metastasis | Stimulation of macrophage switch to an M2-like pro-tumor phenotype and release of TGF-β1 and IL-10 | - | [123,135] |
| MIF | Promote the liver pre-metastatic niche formation | Up-regulation of TGF-β expression and fibronectin secretion | Yes | [125] |
| ANXA6 | Improve PC cell survival and migration | Formation of ANAX6/LRP1/TSP1 complex | Yes | [151] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romano, R.; Picca, A.; Eusebi, L.H.U.; Marzetti, E.; Calvani, R.; Moro, L.; Bucci, C.; Guerra, F. Extracellular Vesicles and Pancreatic Cancer: Insights on the Roles of miRNA, lncRNA, and Protein Cargos in Cancer Progression. Cells 2021, 10, 1361. https://doi.org/10.3390/cells10061361
Romano R, Picca A, Eusebi LHU, Marzetti E, Calvani R, Moro L, Bucci C, Guerra F. Extracellular Vesicles and Pancreatic Cancer: Insights on the Roles of miRNA, lncRNA, and Protein Cargos in Cancer Progression. Cells. 2021; 10(6):1361. https://doi.org/10.3390/cells10061361
Chicago/Turabian StyleRomano, Roberta, Anna Picca, Leonardo Henry Umberto Eusebi, Emanuele Marzetti, Riccardo Calvani, Loredana Moro, Cecilia Bucci, and Flora Guerra. 2021. "Extracellular Vesicles and Pancreatic Cancer: Insights on the Roles of miRNA, lncRNA, and Protein Cargos in Cancer Progression" Cells 10, no. 6: 1361. https://doi.org/10.3390/cells10061361
APA StyleRomano, R., Picca, A., Eusebi, L. H. U., Marzetti, E., Calvani, R., Moro, L., Bucci, C., & Guerra, F. (2021). Extracellular Vesicles and Pancreatic Cancer: Insights on the Roles of miRNA, lncRNA, and Protein Cargos in Cancer Progression. Cells, 10(6), 1361. https://doi.org/10.3390/cells10061361











