Extracellular Vesicle Associated Non-Coding RNAs in Lung Infections and Injury
Abstract
1. Introduction
2. Extracellular Vesicles (EVs)
EV Cargo
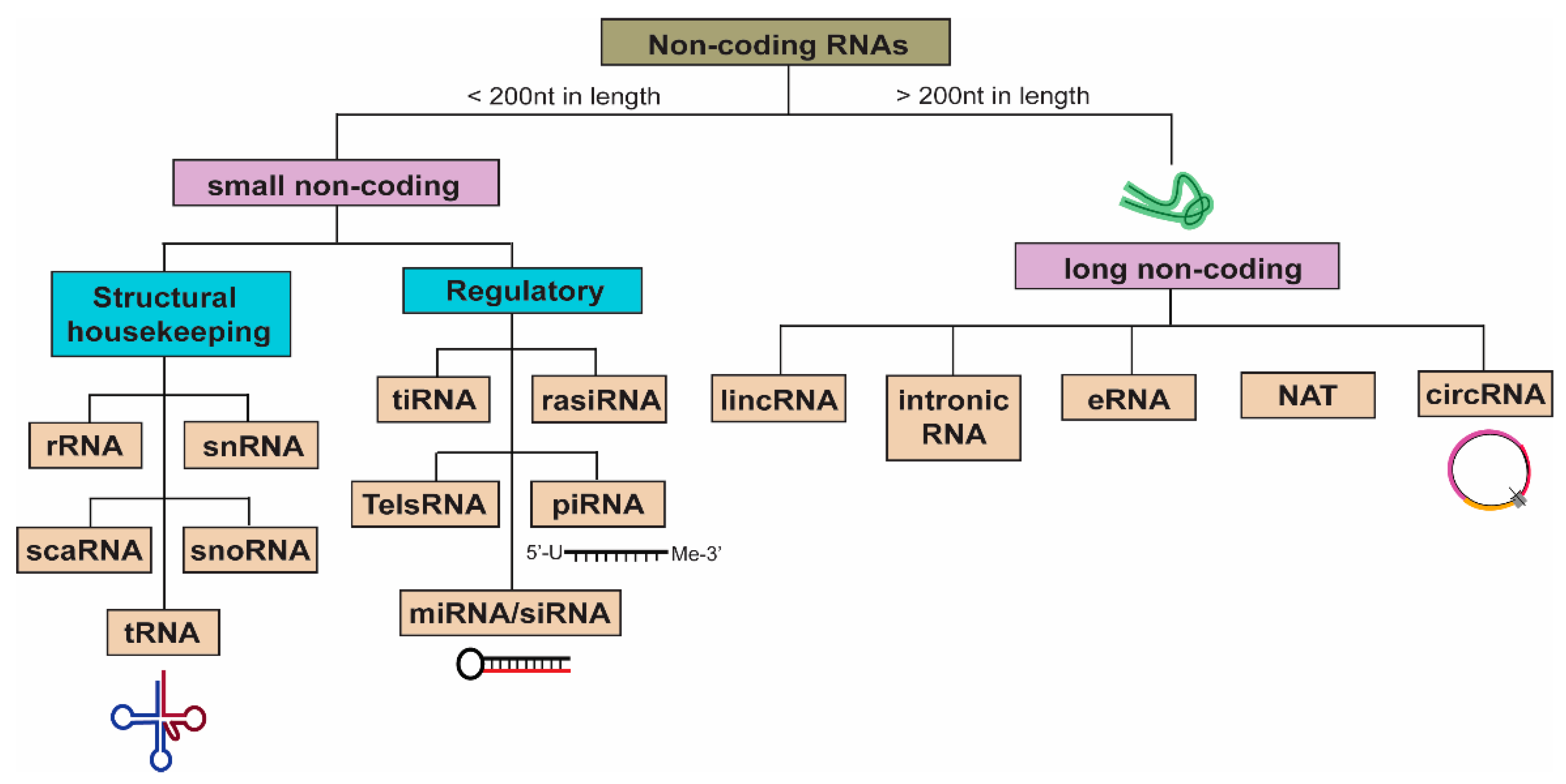
3. Non-Coding RNAs (ncRNAs)
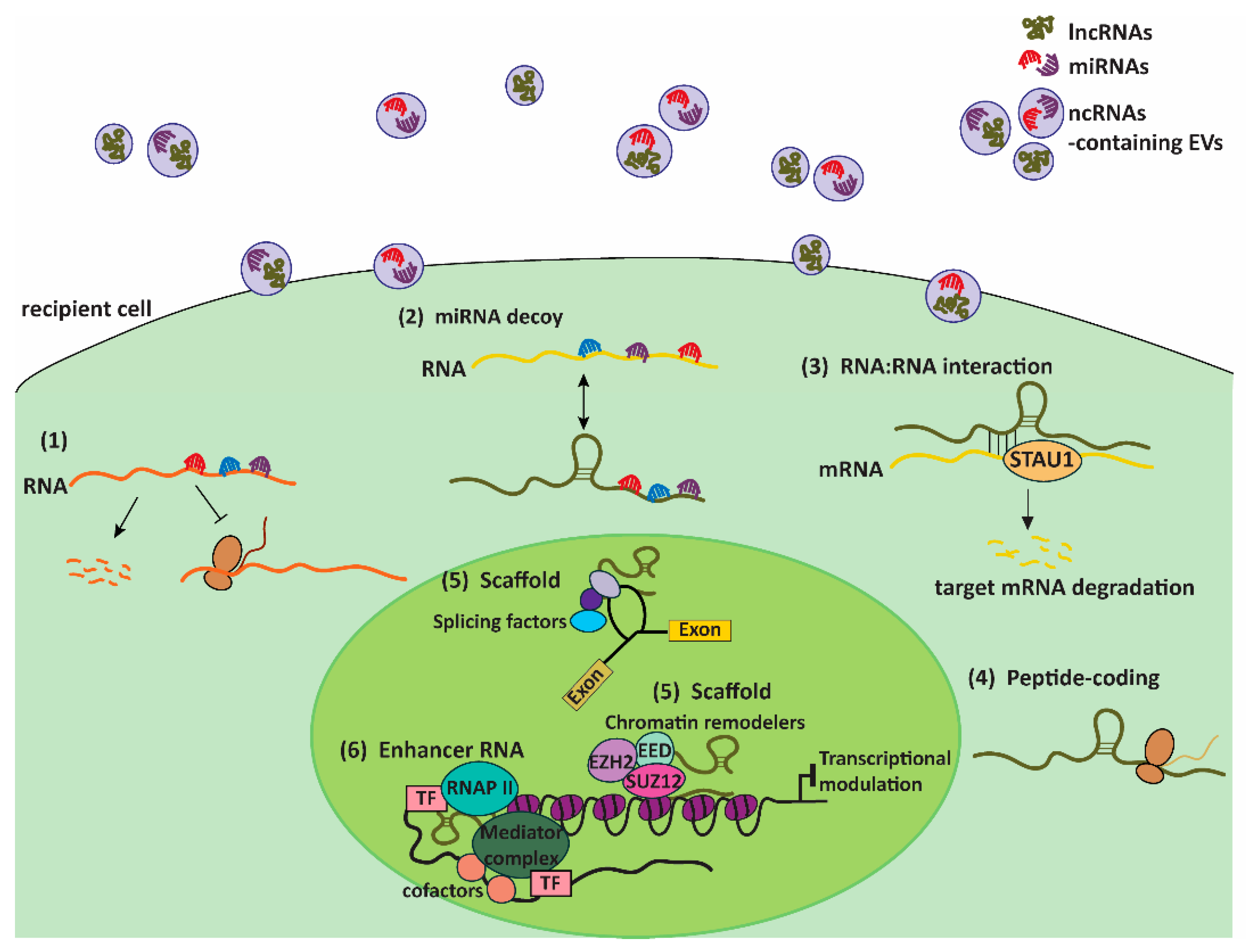
4. EVs: Couriers of ncRNAs
5. EV-ncRNAs in Lung Infections and Injury
5.1. Bacterial Infections
5.2. Viral Infections
5.3. Acute Lung Injury (ALI) and Acute Respiratory Distress Syndrome (ARDS)
5.4. Chronic Lung Injury
5.5. EV-miRNAs as Diagnostic and Therapeutic Tools in Lung Infections and Injury: Promises and Challenges
6. Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- GBD Chronic Respiratory Disease Collaborators. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med. 2020, 8, 585–596. [Google Scholar] [CrossRef]
- Lo Cicero, A.; Stahl, P.D.; Raposo, G. Extracellular vesicles shuffling intercellular messages: For good or for bad. Curr. Opin. Cell Biol. 2015, 35, 69–77. [Google Scholar] [CrossRef]
- Latifkar, A.; Hur, Y.H.; Sanchez, J.C.; Cerione, R.A.; Antonyak, M.A. New insights into extracellular vesicle biogenesis and function. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Kakarla, R.; Hur, J.; Kim, Y.J.; Kim, J.; Chwae, Y.J. Apoptotic cell-derived exosomes: Messages from dying cells. Exp. Mol. Med. 2020, 52, 1–6. [Google Scholar] [CrossRef]
- Tricarico, C.; Clancy, J.; D’Souza-Schorey, C. Biology and biogenesis of shed microvesicles. Small GTPases 2017, 8, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Soekmadji, C.; Hill, A.F.; Wauben, M.H.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Lotvall, J.; Sahoo, S.; Witwer, K.W. Towards mechanisms and standardization in extracellular vesicle and extracellular RNA studies: Results of a worldwide survey. J. Extracell. Vesicles 2018, 7, 1535745. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Sedgwick, A.E.; D’Souza-Schorey, C. The biology of extracellular microvesicles. Traffic 2018, 19, 319–327. [Google Scholar] [CrossRef]
- Juan, T.; Furthauer, M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev. Biol. 2018, 74, 66–77. [Google Scholar] [CrossRef]
- Fares, J.; Kashyap, R.; Zimmermann, P. Syntenin: Key player in cancer exosome biogenesis and uptake? Cell Adh. Migr. 2017, 11, 124–126. [Google Scholar] [CrossRef]
- Verderio, C.; Gabrielli, M.; Giussani, P. Role of sphingolipids in the biogenesis and biological activity of extracellular vesicles. J. Lipid Res. 2018, 59, 1325–1340. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed]
- Kwok, Z.H.; Wang, C.; Jin, Y. Extracellular Vesicle Transportation and Uptake by Recipient Cells: A Critical Process to Regulate Human Diseases. Processes 2021, 9, 273. [Google Scholar] [CrossRef]
- Carnino, J.M.; Ni, K.; Jin, Y. Post-translational Modification Regulates Formation and Cargo-Loading of Extracellular Vesicles. Front. Immunol. 2020, 11, 948. [Google Scholar] [CrossRef]
- Ni, K.; Wang, C.; Carnino, J.M.; Jin, Y. The Evolving Role of Caveolin-1: A Critical Regulator of Extracellular Vesicles. Med. Sci. 2020, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Kooijmans, S.A.; Aleza, C.G.; Roffler, S.R.; van Solinge, W.W.; Vader, P.; Schiffelers, R.M. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles 2016, 5, 31053. [Google Scholar] [CrossRef]
- Laulagnier, K.; Javalet, C.; Hemming, F.J.; Chivet, M.; Lachenal, G.; Blot, B.; Chatellard, C.; Sadoul, R. Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell. Mol. Life Sci. 2018, 75, 757–773. [Google Scholar] [CrossRef]
- Sancho-Albero, M.; Navascues, N.; Mendoza, G.; Sebastian, V.; Arruebo, M.; Martin-Duque, P.; Santamaria, J. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J. Nanobiotechnol. 2019, 17, 16. [Google Scholar] [CrossRef]
- Jurgielewicz, B.J.; Yao, Y.; Stice, S.L. Kinetics and Specificity of HEK293T Extracellular Vesicle Uptake using Imaging Flow Cytometry. Nanoscale Res. Lett. 2020, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Pazos, R.; Royo, F.; Gonzalez, E.; Roura-Ferrer, M.; Martinez, A.; Gamiz, J.; Reichardt, N.C.; Falcon-Perez, J.M. Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci. Rep. 2019, 9, 11920. [Google Scholar] [CrossRef] [PubMed]
- Mir, B.; Goettsch, C. Extracellular Vesicles as Delivery Vehicles of Specific Cellular Cargo. Cells 2020, 9, 1601. [Google Scholar] [CrossRef] [PubMed]
- Baglio, S.R.; Rooijers, K.; Koppers-Lalic, D.; Verweij, F.J.; Perez Lanzon, M.; Zini, N.; Naaijkens, B.; Perut, F.; Niessen, H.W.; Baldini, N.; et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015, 6, 127. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Malkin, E.Z.; Bratman, S.V. Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 2020, 11, 584. [Google Scholar] [CrossRef]
- Turchinovich, A.; Drapkina, O.; Tonevitsky, A. Transcriptome of Extracellular Vesicles: State-of-the-Art. Front. Immunol. 2019, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Veziroglu, E.M.; Mias, G.I. Characterizing Extracellular Vesicles and Their Diverse RNA Contents. Front. Genet. 2020, 11, 700. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- De Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Astrom, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Gao, Y.; Mu, D.G.; Fu, E.Q. MiR-23a-5p modulates mycobacterial survival and autophagy during mycobacterium tuberculosis infection through TLR2/MyD88/NF-kappaB pathway by targeting TLR2. Exp. Cell Res. 2017, 354, 71–77. [Google Scholar] [CrossRef]
- Podsiad, A.; Standiford, T.J.; Ballinger, M.N.; Eakin, R.; Park, P.; Kunkel, S.L.; Moore, B.B.; Bhan, U. MicroRNA-155 regulates host immune response to postviral bacterial pneumonia via IL-23/IL-17 pathway. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 310, L465–L475. [Google Scholar] [CrossRef]
- Gu, W.; Yuan, Y.; Yang, H.; Wu, H.; Wang, L.; Tang, Z.; Li, Q. Role of miR-195 in cigarette smoke-induced chronic obstructive pulmonary disease. Int. Immunopharmacol. 2018, 55, 49–54. [Google Scholar] [CrossRef]
- Xie, W.; Lu, Q.; Wang, K.; Lu, J.; Gu, X.; Zhu, D.; Liu, F.; Guo, Z. miR-34b-5p inhibition attenuates lung inflammation and apoptosis in an LPS-induced acute lung injury mouse model by targeting progranulin. J. Cell. Physiol. 2018, 233, 6615–6631. [Google Scholar] [CrossRef]
- Kwok, Z.H.; Tay, Y. Long noncoding RNAs: Lincs between human health and disease. Biochem. Soc. Trans. 2017, 45, 805–812. [Google Scholar] [CrossRef]
- Loda, A.; Heard, E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019, 15, e1008333. [Google Scholar] [CrossRef]
- Choi, S.W.; Kim, H.W.; Nam, J.W. The small peptide world in long noncoding RNAs. Brief. Bioinform. 2019, 20, 1853–1864. [Google Scholar] [CrossRef]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef]
- Zhang, D.; Lee, H.; Haspel, J.A.; Jin, Y. Long noncoding RNA FOXD3-AS1 regulates oxidative stress-induced apoptosis via sponging microRNA-150. FASEB J. 2017, 31, 4472–4481. [Google Scholar] [CrossRef]
- Lu, Q.; Guo, Z.; Xie, W.; Jin, W.; Zhu, D.; Chen, S.; Ren, T. The lncRNA H19 Mediates Pulmonary Fibrosis by Regulating the miR-196a/COL1A1 Axis. Inflammation 2018, 41, 896–903. [Google Scholar] [CrossRef]
- Sork, H.; Corso, G.; Krjutskov, K.; Johansson, H.J.; Nordin, J.Z.; Wiklander, O.P.B.; Lee, Y.X.F.; Westholm, J.O.; Lehtio, J.; Wood, M.J.A.; et al. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci. Rep. 2018, 8, 10813. [Google Scholar] [CrossRef]
- Pigati, L.; Yaddanapudi, S.C.; Iyengar, R.; Kim, D.J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 2010, 5, e13515. [Google Scholar] [CrossRef]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; t Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [PubMed]
- Chiou, N.T.; Kageyama, R.; Ansel, K.M. Selective Export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation. Cell Rep. 2018, 25, 3356–3370.e3354. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, Q.; Zhang, J.; Li, C.; Miao, Y.R.; Lei, Q.; Li, Q.; Guo, A.Y. EVmiRNA: A database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019, 47, D89–D93. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Li, C.; Zhang, Y.; Zhang, D.; Otterbein, L.E.; Jin, Y. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 2019, 216, 2202–2220. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.; Kaul, D. Cancer cells govern miR-2909 exosomal recruitment through its 3’-end post-transcriptional modification. Cell Biochem. Funct. 2018, 36, 106–111. [Google Scholar] [CrossRef]
- Liang, J.; Zhang, X.; He, S.; Miao, Y.; Wu, N.; Li, J.; Gan, Y. Sphk2 RNAi nanoparticles suppress tumor growth via downregulating cancer cell derived exosomal microRNA. J. Control. Release 2018, 286, 348–357. [Google Scholar] [CrossRef]
- Gezer, U.; Ozgur, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Cha, D.J.; Franklin, J.L.; Higginbotham, J.N.; Jeppesen, D.K.; Weaver, A.M.; Prasad, N.; Levy, S.; Coffey, R.J.; Patton, J.G.; et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 2016, 6, 37982. [Google Scholar] [CrossRef] [PubMed]
- Kossinova, O.A.; Gopanenko, A.V.; Tamkovich, S.N.; Krasheninina, O.A.; Tupikin, A.E.; Kiseleva, E.; Yanshina, D.D.; Malygin, A.A.; Ven’yaminova, A.G.; Kabilov, M.R.; et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim. Biophys. Acta Proteins Proteom 2017, 1865, 664–673. [Google Scholar] [CrossRef]
- Yanshina, D.D.; Kossinova, O.A.; Gopanenko, A.V.; Krasheninina, O.A.; Malygin, A.A.; Venyaminova, A.G.; Karpova, G.G. Structural features of the interaction of the 3’-untranslated region of mRNA containing exosomal RNA-specific motifs with YB-1, a potential mediator of mRNA sorting. Biochimie 2018, 144, 134–143. [Google Scholar] [CrossRef]
- Zhang, D.; Lee, H.; Wang, X.; Groot, M.; Sharma, L.; Dela Cruz, C.S.; Jin, Y. A potential role of microvesicle-containing miR-223/142 in lung inflammation. Thorax 2019, 74, 865–874. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef]
- Lucero, R.; Zappulli, V.; Sammarco, A.; Murillo, O.D.; Cheah, P.S.; Srinivasan, S.; Tai, E.; Ting, D.T.; Wei, Z.; Roth, M.E.; et al. Glioma-Derived miRNA-Containing Extracellular Vesicles Induce Angiogenesis by Reprogramming Brain Endothelial Cells. Cell Rep. 2020, 30, 2065–2074.e2064. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Maas, S.L.N.; Nieland, L.; Wei, Z.; Cheah, P.S.; Tai, E.; Kolsteeg, C.J.; Dusoswa, S.A.; Ting, D.T.; Hickman, S.; et al. Glioblastoma-Associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular miR-21. Cell Rep. 2019, 28, 3105–3119.e3107. [Google Scholar] [CrossRef]
- Kenneweg, F.; Bang, C.; Xiao, K.; Boulanger, C.M.; Loyer, X.; Mazlan, S.; Schroen, B.; Hermans-Beijnsberger, S.; Foinquinos, A.; Hirt, M.N.; et al. Long Noncoding RNA-Enriched Vesicles Secreted by Hypoxic Cardiomyocytes Drive Cardiac Fibrosis. Mol. Ther. Nucleic Acids 2019, 18, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, J.; Yang, L.; Liu, J.; Zhang, X.; Zhang, Y.; Tu, Q.; Yin, D.; Lin, D.; Wong, P.P.; et al. Extracellular vesicle-packaged HIF-1alpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 2019, 21, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.M.; Tseng, C.H.; Chen, Y.C.; Yu, W.Y.; Ho, M.Y.; Ho, C.Y.; Lai, M.M.C.; Su, W.C. Exosome-delivered and Y RNA-derived small RNA suppresses influenza virus replication. J. Biomed. Sci. 2019, 26, 58. [Google Scholar] [CrossRef] [PubMed]
- Maemura, T.; Fukuyama, S.; Sugita, Y.; Lopes, T.J.S.; Nakao, T.; Noda, T.; Kawaoka, Y. Lung-Derived Exosomal miR-483-3p Regulates the Innate Immune Response to Influenza Virus Infection. J. Infect. Dis. 2018, 217, 1372–1382. [Google Scholar] [CrossRef]
- Huang, F.; Bai, J.; Zhang, J.; Yang, D.; Fan, H.; Huang, L.; Shi, T.; Lu, G. Identification of potential diagnostic biomarkers for pneumonia caused by adenovirus infection in children by screening serum exosomal microRNAs. Mol. Med. Rep. 2019, 19, 4306–4314. [Google Scholar] [CrossRef]
- Alipoor, S.D.; Mortaz, E.; Tabarsi, P.; Farnia, P.; Mirsaeidi, M.; Garssen, J.; Movassaghi, M.; Adcock, I.M. Bovis Bacillus Calmette-Guerin (BCG) infection induces exosomal miRNA release by human macrophages. J. Transl. Med. 2017, 15, 105. [Google Scholar] [CrossRef]
- Alipoor, S.D.; Tabarsi, P.; Varahram, M.; Movassaghi, M.; Dizaji, M.K.; Folkerts, G.; Garssen, J.; Adcock, I.M.; Mortaz, E. Serum Exosomal miRNAs Are Associated with Active Pulmonary Tuberculosis. Dis. Markers 2019, 2019, 1907426. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Wei, L.; Han, Z.; Chen, Z. Mesenchymal stromal cells-derived exosomes alleviate ischemia/reperfusion injury in mouse lung by transporting anti-apoptotic miR-21-5p. Eur. J. Pharmacol. 2019, 852, 68–76. [Google Scholar] [CrossRef]
- Yi, X.; Wei, X.; Lv, H.; An, Y.; Li, L.; Lu, P.; Yang, Y.; Zhang, Q.; Yi, H.; Chen, G. Exosomes derived from microRNA-30b-3p-overexpressing mesenchymal stem cells protect against lipopolysaccharide-induced acute lung injury by inhibiting SAA3. Exp. Cell Res. 2019, 383, 111454. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Araya, J.; Ito, S.; Kobayashi, K.; Kosaka, N.; Yoshioka, Y.; Kadota, T.; Hara, H.; Kuwano, K.; Ochiya, T. Suppression of autophagy by extracellular vesicles promotes myofibroblast differentiation in COPD pathogenesis. J. Extracell. Vesicles 2015, 4, 28388. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ling, M.; Xue, J.; Dai, X.; Sun, Q.; Chen, C.; Liu, Y.; Zhou, L.; Liu, J.; Luo, F.; et al. Exosomal microRNA-21 derived from bronchial epithelial cells is involved in aberrant epithelium-fibroblast cross-talk in COPD induced by cigarette smoking. Theranostics 2018, 8, 5419–5433. [Google Scholar] [CrossRef]
- Levanen, B.; Bhakta, N.R.; Torregrosa Paredes, P.; Barbeau, R.; Hiltbrunner, S.; Pollack, J.L.; Skold, C.M.; Svartengren, M.; Grunewald, J.; Gabrielsson, S.; et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J. Allergy Clin. Immunol. 2013, 131, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Gon, Y.; Maruoka, S.; Inoue, T.; Kuroda, K.; Yamagishi, K.; Kozu, Y.; Shikano, S.; Soda, K.; Lotvall, J.; Hashimoto, S. Selective release of miRNAs via extracellular vesicles is associated with house-dust mite allergen-induced airway inflammation. Clin. Exp. Allergy 2017, 47, 1586–1598. [Google Scholar] [CrossRef]
- Makiguchi, T.; Yamada, M.; Yoshioka, Y.; Sugiura, H.; Koarai, A.; Chiba, S.; Fujino, N.; Tojo, Y.; Ota, C.; Kubo, H.; et al. Serum extracellular vesicular miR-21-5p is a predictor of the prognosis in idiopathic pulmonary fibrosis. Respir. Res. 2016, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Lal, C.V.; Olave, N.; Travers, C.; Rezonzew, G.; Dolma, K.; Simpson, A.; Halloran, B.; Aghai, Z.; Das, P.; Sharma, N.; et al. Exosomal microRNA predicts and protects against severe bronchopulmonary dysplasia in extremely premature infants. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Grief, S.N.; Loza, J.K. Guidelines for the Evaluation and Treatment of Pneumonia. Prim. Care 2018, 45, 485–503. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Groot, M.; Pinilla-Vera, M.; Fredenburgh, L.E.; Jin, Y. Identification of miRNA-rich vesicles in bronchoalveolar lavage fluid: Insights into the function and heterogeneity of extracellular vesicles. J. Control. Release 2019, 294, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Ruuskanen, O.; Lahti, E.; Jennings, L.C.; Murdoch, D.R. Viral pneumonia. Lancet 2011, 377, 1264–1275. [Google Scholar] [CrossRef]
- Yoon, B.W.; Song, Y.G.; Lee, S.H. Severe community-acquired adenovirus pneumonia treated with oral ribavirin: A case report. BMC Res. Notes 2017, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, S.; Bihl, J. Exosome-Mediated Transfer of ACE2 (Angiotensin-Converting Enzyme 2) from Endothelial Progenitor Cells Promotes Survival and Function of Endothelial Cell. Oxid. Med. Cell Longev. 2020, 2020, 4213541. [Google Scholar] [CrossRef]
- Nersisyan, S.; Engibaryan, N.; Gorbonos, A.; Kirdey, K.; Makhonin, A.; Tonevitsky, A. Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ 2020, 8, e9994. [Google Scholar] [CrossRef]
- Singh, P.P.; Li, L.; Schorey, J.S. Exosomal RNA from Mycobacterium tuberculosis-Infected Cells Is Functional in Recipient Macrophages. Traffic 2015, 16, 555–571. [Google Scholar] [CrossRef]
- Fan, E.; Brodie, D.; Slutsky, A.S. Acute Respiratory Distress Syndrome: Advances in Diagnosis and Treatment. JAMA 2018, 319, 698–710. [Google Scholar] [CrossRef]
- Lee, H.Y.; Hong, I.S. Double-edged sword of mesenchymal stem cells: Cancer-promoting versus therapeutic potential. Cancer Sci. 2017, 108, 1939–1946. [Google Scholar] [CrossRef] [PubMed]
- Matthay, M.A.; Calfee, C.S.; Zhuo, H.; Thompson, B.T.; Wilson, J.G.; Levitt, J.E.; Rogers, A.J.; Gotts, J.E.; Wiener-Kronish, J.P.; Bajwa, E.K.; et al. Treatment with allogeneic mesenchymal stromal cells for moderate to severe acute respiratory distress syndrome (START study): A randomised phase 2a safety trial. Lancet Respir. Med. 2019, 7, 154–162. [Google Scholar] [CrossRef]
- Russell, R.J.; Brightling, C. Pathogenesis of asthma: Implications for precision medicine. Clin. Sci. 2017, 131, 1723–1735. [Google Scholar] [CrossRef]
- Qiao, Y.; Liang, X.; Yan, Y.; Lu, Y.; Zhang, D.; Yao, W.; Wu, W.; Yan, Z. Identification of Exosomal miRNAs in Rats with Pulmonary Neutrophilic Inflammation Induced by Zinc Oxide Nanoparticles. Front. Physiol. 2018, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Di Pietro, G.M.; Esposito, S. Bronchopulmonary dysplasia: Clinical aspects and preventive and therapeutic strategies. J. Transl. Med. 2018, 16, 36. [Google Scholar] [CrossRef]
- Guo, H.; Su, Y.; Deng, F. Effects of Mesenchymal Stromal Cell-Derived Extracellular Vesicles in Lung Diseases: Current Status and Future Perspectives. Stem Cell Rev. Rep. 2021, 17, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Abreu, S.C.; Lopes-Pacheco, M.; Weiss, D.J.; Rocco, P.R.M. Mesenchymal Stromal Cell-Derived Extracellular Vesicles in Lung Diseases: Current Status and Perspectives. Front. Cell Dev. Biol. 2021, 9, 600711. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, R.; Xu, Q.; Zheng, G.; Qiu, G.; Ge, M.; Shu, Q.; Xu, J. Mesenchymal Stem Cell-Derived Extracellular Vesicles Alleviate Acute Lung Injury Via Transfer of miR-27a-3p. Crit. Care Med. 2020, 48, e599–e610. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Gudapati, V.; Monsel, A.; Park, J.H.; Hu, S.; Kato, H.; Lee, J.H.; Zhou, L.; He, H.; Lee, J.W. Mesenchymal Stem Cell-Derived Extracellular Vesicles Decrease Lung Injury in Mice. J. Immunol. 2019, 203, 1961–1972. [Google Scholar] [CrossRef]

| Lung Diseases | EV-ncRNA | Possible Function(s)/Significance | References |
|---|---|---|---|
| Bacterial infection | BALF-MV miRNA-223/142 | Potential biomarker for lung inflammation and macrophage activation. | [59] |
| Influenza viral infection | Exosomal hsa-miR-1975 | Suppresses influenza virus replication in recipient cell. | [65] |
| BALF-exosomal miR-483-3p | Promotes innate immune response against influenza. | [66] | |
| Adenovirus infection | Serum–exosomal miR‑450a‑5p/miR‑103a‑3p and miR‑103b‑5p/miR‑98‑5p | Potential biomarker for adenovirus pneumonia. | [67] |
| TB | Human monocyte-derived macrophages-exosomal miR-1224, -1293, -425, -4467, -4732, -484, -5094, -6848, -6849, -96, and -4488 | Plays a role in host–pathogen interaction. | [68] |
| Serum-exosomal miR-484, -425, and -96 | Potential biomarkers for TB diagnosis. | [69] | |
| ALI/ARDS | MSC-exosomal miR-21-5p | Targets PTEN and PDCD4 to inhibit apoptosis. | [70] |
| MSC-exosomal miR-30b-3p | Targets SAA3 to inhibit apoptosis. | [71] | |
| COPD | HBEC-derived EV miR-210 | Promotes differentiation of lung myofibroblasts. | [72] |
| HBEC-derived exosomal and serum exosomal miR-21 | Potential biomarker for COPD. | [73] | |
| Asthma | BALF-exosomal let-7a, miRNA- 21, miRNA-658, miRNA-24, miRNA-26a, miRNA-99a, miRNA-200c, miRNA-1268, miR-1827, miR-346, and miR-574-5p | Potential biomarkers for asthma. | [74,75] |
| IDF | Serum-EV miR-21-5p | Potential biomarker for treatment responsiveness. | [76] |
| BPD | BALF-exosomal miR-876-3p | Potential biomarker for BPD. | [77] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwok, Z.H.; Ni, K.; Jin, Y. Extracellular Vesicle Associated Non-Coding RNAs in Lung Infections and Injury. Cells 2021, 10, 965. https://doi.org/10.3390/cells10050965
Kwok ZH, Ni K, Jin Y. Extracellular Vesicle Associated Non-Coding RNAs in Lung Infections and Injury. Cells. 2021; 10(5):965. https://doi.org/10.3390/cells10050965
Chicago/Turabian StyleKwok, Zhi Hao, Kareemah Ni, and Yang Jin. 2021. "Extracellular Vesicle Associated Non-Coding RNAs in Lung Infections and Injury" Cells 10, no. 5: 965. https://doi.org/10.3390/cells10050965
APA StyleKwok, Z. H., Ni, K., & Jin, Y. (2021). Extracellular Vesicle Associated Non-Coding RNAs in Lung Infections and Injury. Cells, 10(5), 965. https://doi.org/10.3390/cells10050965





