Modulation of Endocannabinoid Tone in Osteoblastic Differentiation of MC3T3-E1 Cells and in Mouse Bone Tissue over Time
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cells Culture and Differentiation
2.3. LC-MS Quantification of Endocannabinoids and Related N-Acylethanolamines
2.4. RNA Isolation and Quantitative Polymerase Chain Reaction (qPCR)
2.5. Statistical Analysis
3. Results
3.1. mRNA Expression of Early and Late Markers during MC3T3-E1 Osteoblast Differentiation and Alizarin Red Matrix Mineralization Quantification
3.2. Quantification of Endocannabinoids and EC-Related Molecules in MC3T3-E1 Cells during Differentiation
3.3. Expression of 2-AG, AEA, OEA, and PEA Metabolic Enzymes during Osteoblast Differentiation in MC3T3-E1 Cells
3.4. Quantification of Endocannabinoids and EC-Related Molecules in Femurs of C57BL6 Male Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Kim, J.-M.; Lin, C.; Stavre, Z.; Greenblatt, M.B.; Shim, J.-H. Osteoblast-Osteoclast Communication and Bone Homeostasis. Cells 2020, 9, 2073. [Google Scholar] [CrossRef] [PubMed]
- Kassem, M.; Marie, P.J. Senescence-associated intrinsic mechanisms of osteoblast dysfunctions. Aging Cell 2011, 10, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.-L.; Xiao, S.-M.; Kung, A.W.C. Genetic epidemiology of age-related osteoporosis and its clinical applications. Nat. Rev. Rheumatol. 2010, 6, 507–517. [Google Scholar] [CrossRef]
- Ensrud, K.E. Epidemiology of Fracture Risk with Advancing Age. J. Gerontol. Ser. A Boil. Sci. Med. Sci. 2013, 68, 1236–1242. [Google Scholar] [CrossRef] [PubMed]
- Jilka, R.L.; Weinstein, R.S.; Takahashi, K.; Parfitt, A.M.; Manolagas, S.C. Linkage of decreased bone mass with impaired osteoblastogenesis in a murine model of accelerated senescence. J. Clin. Investig. 1996, 97, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Glatt, V.; Canalis, E.; Stadmeyer, L.; Bouxsein, M.L. Age-Related Changes in Trabecular Architecture Differ in Female and Male C57BL/6J Mice. J. Bone Miner. Res. 2007, 22, 1197–1207. [Google Scholar] [CrossRef]
- Manolagas, S.C. From Estrogen-Centric to Aging and Oxidative Stress: A Revised Perspective of the Pathogenesis of Osteoporosis. Endocr. Rev. 2010, 31, 266–300. [Google Scholar] [CrossRef]
- Kuipers, E.N.; Kantae, V.; Maarse, B.C.E.; Berg, S.M.V.D.; Van Eenige, R.; Nahon, K.J.; Reifel-Miller, A.; Coskun, T.; De Winther, M.P.J.; Lutgens, E.; et al. High Fat Diet Increases Circulating Endocannabinoids Accompanied by Increased Synthesis Enzymes in Adipose Tissue. Front. Physiol. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Jilka, R.L. The Relevance of Mouse Models for Investigating Age-Related Bone Loss in Humans. J. Gerontol. Ser. A Boil. Sci. Med. Sci. 2013, 68, 1209–1217. [Google Scholar] [CrossRef]
- Halloran, B.P.; Ferguson, V.L.; Simske, S.J.; Burghardt, A.; Venton, L.L.; Majumdar, S. Changes in Bone Structure and Mass With Advancing Age in the Male C57BL/6J Mouse. J. Bone Miner. Res. 2002, 17, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Ligresti, A.; De Petrocellis, L.; Di Marzo, V. From Phytocannabinoids to Cannabinoid Receptors and Endocannabinoids: Pleiotropic Physiological and Pathological Roles through Complex Pharmacology. Physiol. Rev. 2016, 96, 1593–1659. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V. Targeting the endocannabinoid system: To enhance or reduce? Nat. Rev. Drug Discov. 2008, 7, 438–455. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V.; Melck, D.; Bisogno, T.; De Petrocellis, L. Endocannabinoids: Endogenous cannabinoid receptor ligands with neuromodulatory action. Trends Neurosci. 1998, 21, 521–528. [Google Scholar] [CrossRef]
- Iannotti, F.A.; Di Marzo, V.; Petrosino, S. Endocannabinoids and endocannabinoid-related mediators: Targets, metabolism and role in neurological disorders. Prog. Lipid Res. 2016, 62, 107–128. [Google Scholar] [CrossRef] [PubMed]
- Veilleux, A.; Di Marzo, V.; Silvestri, C. The Expanded Endocannabinoid System/Endocannabinoidome as a Potential Target for Treating Diabetes Mellitus. Curr. Diabetes Rep. 2019, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Muccioli, G.G. Endocannabinoid biosynthesis and inactivation, from simple to complex. Drug Discov. Today 2010, 15, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Fezza, F.; Bari, M.; Florio, R.; Talamonti, E.; Feole, M.; Maccarrone, M. Endocannabinoids, Related Compounds and Their Metabolic Routes. Molecules 2014, 19, 17078–17106. [Google Scholar] [CrossRef]
- Leung, D.; Saghatelian, A.; Simon, G.M.; Cravatt, B.F. Inactivation of N-Acyl Phosphatidylethanolamine Phospholipase D Reveals Multiple Mechanisms for the Biosynthesis of Endocannabinoids. Biochemistry 2006, 45, 4720–4726. [Google Scholar] [CrossRef]
- Simon, G.M.; Cravatt, B.F. Endocannabinoid Biosynthesis Proceeding through Glycerophospho-N-acyl Ethanolamine and a Role for α/β-Hydrolase 4 in This Pathway. J. Biol. Chem. 2006, 281, 26465–26472. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Harvey-White, J.; Osei-Hyiaman, D.; Razdan, R.; Gong, Q.; Chan, A.C.; Zhou, Z.; Huang, B.X.; Kim, H.-Y.; et al. A biosynthetic pathway for anandamide. Proc. Natl. Acad. Sci. USA 2006, 103, 13345–13350. [Google Scholar] [CrossRef]
- Higgs, H.N.; Glomset, J.A. Identification of a phosphatidic acid-preferring phospholipase A1 from bovine brain and testis. Proc. Natl. Acad. Sci. USA 1994, 91, 9574–9578. [Google Scholar] [CrossRef]
- Nakane, S.; Oka, S.; Arai, S.; Waku, K.; Ishima, Y.; Tokumura, A.; Sugiura, T. 2-Arachidonoyl-sn-glycero-3-phosphate, an arachidonic acid-containing lysophosphatidic acid: Occurrence and rapid enzymatic conversion to 2-arachidonoyl-sn-glycerol, a cannabinoid receptor ligand, in rat brain. Arch. Biochem. Biophys. 2002, 402, 51–58. [Google Scholar] [CrossRef]
- Marrs, W.R.; Blankman, J.L.; Horne, E.A.; Thomazeau, A.; Lin, Y.H.; Coy, J.; Bodor, A.L.; Muccioli, G.G.; Hu, S.S.-J.; Woodruff, G.; et al. The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat. Neurosci. 2010, 13, 951–957. [Google Scholar] [CrossRef]
- Savinainen, J.R.; Saario, S.M.; Laitinen, J.T. The serine hydrolases MAGL, ABHD6 and ABHD12 as guardians of 2-arachidonoylglycerol signalling through cannabinoid receptors. Acta Physiol. 2011, 204, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Moody, J.S.; Kozak, K.R.; Ji, C.; Marnett, L.J. Selective Oxygenation of the Endocannabinoid 2-Arachidonylglycerol by Leukocyte-Type 12-Lipoxygenase. Biochemistry 2001, 40, 861–866. [Google Scholar] [CrossRef] [PubMed]
- Kozak, K.R.; Gupta, R.A.; Moody, J.S.; Ji, C.; Boeglin, W.E.; DuBois, R.N.; Brash, A.R.; Marnett, L.J. 15-Lipoxygenase Metabolism of 2-Arachidonylglycerol. J. Biol. Chem. 2002, 277, 23278–23286. [Google Scholar] [CrossRef] [PubMed]
- Van der Stelt, M.; van Kuik, J.A.; Bari, M.; van Zadelhoff, G.; Leeflang, B.R.; Veldink, G.A.; Finazzi-Agrò, A.; Vliegenthart, J.F.G.; Maccarrone, M. Oxygenated Metabolites of Anandamide and 2-Arachidonoylglycerol: Conformational Analysis and Interaction with Cannabinoid Receptors, Membrane Transporter, and Fatty Acid Amide Hydrolase. J. Med. Chem. 2002, 45, 3709–3720. [Google Scholar] [CrossRef]
- Bab, I.; Zimmer, A.; Melamed, E. Cannabinoids and the skeleton: From marijuana to reversal of bone loss. Ann. Med. 2009, 41, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.I.; Ralston, S.H. Cannabinoids and Bone: Friend or Foe? Calcif. Tissue Int. 2010, 87, 285–297. [Google Scholar] [CrossRef]
- Tam, J.; Trembovler, V.; Di Marzo, V.; Petrosino, S.; Leo, G.; Alexandrovich, A.; Regev, E.; Casap, N.; Shteyer, A.; Ledent, C.; et al. The cannabinoid CB1 receptor regulates bone formation by modulating adrenergic signaling. FASEB J. 2007, 22, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Deis, S.; Srivastava, R.K.; de Azua, I.R.; Bindila, L.; Baraghithy, S.; Lutz, B.; Bab, I.; Tam, J. Age-related regulation of bone formation by the sympathetic cannabinoid CB1 receptor. Bone 2018, 108, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.I.; Sophocleous, A.; Landao-Bassonga, E.; Hof, R.J.V.; Ralston, S.H. Regulation of Bone Mass, Osteoclast Function, and Ovariectomy-Induced Bone Loss by the Type 2 Cannabinoid Receptor. Endocrinology 2008, 149, 5619–5626. [Google Scholar] [CrossRef] [PubMed]
- Ofek, O.; Karsak, M.; Leclerc, N.; Fogel, M.; Frenkel, B.; Wright, K.; Tam, J.; Attar-Namdar, M.; Kram, V.; Shohami, E.; et al. Peripheral cannabinoid receptor, CB2, regulates bone mass. Proc. Natl. Acad. Sci. USA 2006, 103, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Sophocleous, A.; Marino, S.; Kabir, D.; Ralston, S.H.; Idris, A.I. Combined deficiency of the Cnr1 and Cnr2 receptors protects against age-related bone loss by osteoclast inhibition. Aging Cell 2017, 16, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.I.; Sophocleous, A.; Landao-Bassonga, E.; Canals, M.; Milligan, G.; Baker, D.; Hof, R.J.V.; Ralston, S.H. Cannabinoid Receptor Type 1 Protects against Age-Related Osteoporosis by Regulating Osteoblast and Adipocyte Differentiation in Marrow Stromal Cells. Cell Metab. 2009, 10, 139–147. [Google Scholar] [CrossRef]
- Lozano-Ondoua, A.N.; Wright, C.; Vardanyan, A.; King, T.; Largent-Milnes, T.M.; Nelson, M.; Jimenez-Andrade, J.M.; Mantyh, P.W.; Vanderah, T.W. A cannabinoid 2 receptor agonist attenuates bone cancer-induced pain and bone loss. Life Sci. 2010, 86, 646–653. [Google Scholar] [CrossRef]
- Ofek, O.; Attar-Namdar, M.; Kram, V.; Dvir-Ginzberg, M.; Mechoulam, R.; Zimmer, A.; Frenkel, B.; Shohami, E.; Bab, I. CB2 cannabinoid receptor targets mitogenic Gi protein-cyclin D1 axis in osteoblasts. J. Bone Miner. Res. 2011, 26, 308–316. [Google Scholar] [CrossRef]
- Bab, I.; Ofek, O.; Tam, J.; Rehnelt, J.; Zimmer, A. Endocannabinoids and the Regulation of Bone Metabolism. J. Neuroendocr. 2008, 20 (Suppl. 1), 69–74. [Google Scholar] [CrossRef]
- Rossi, F.; Siniscalco, D.; Luongo, L.; De Petrocellis, L.; Bellini, G.; Petrosino, S.; Torella, M.; Santoro, C.; Nobili, B.; Perrotta, S.; et al. The endovanilloid/endocannabinoid system in human osteoclasts: Possible involvement in bone formation and resorption. Bone 2009, 44, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Scutt, A.; Williamson, E.M. Cannabinoids Stimulate Fibroblastic Colony Formation by Bone Marrow Cells Indirectly via CB2 Receptors. Calcif. Tissue Int. 2007, 80, 50–59. [Google Scholar] [CrossRef]
- Smith, M.; Wilson, R.; O’Brien, S.; Tufarelli, C.; Anderson, S.I.; O’Sullivan, S.E. The Effects of the Endocannabinoids Anandamide and 2-Arachidonoylglycerol on Human Osteoblast Proliferation and Differentiation. PLoS ONE 2015, 10, e0136546. [Google Scholar] [CrossRef] [PubMed]
- Piscitelli, F.; Carta, G.; Bisogno, T.; Murru, E.; Cordeddu, L.; Berge, K.; Tandy, S.; Cohn, J.S.; Griinari, M.; Banni, S.; et al. Effect of dietary krill oil supplementation on the endocannabinoidome of metabolically relevant tissues from high-fat-fed mice. Nutr. Metab. 2011, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Rutkovskiy, A.; Stensløkken, K.-O.; Vaage, I.J. Osteoblast Differentiation at a Glance. Med. Sci. Monit. Basic Res. 2016, 22, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Hwang, P.W.; Horton, J.A. Variable osteogenic performance of MC3T3-E1 subclones impacts their utility as models of osteoblast biology. Sci. Rep. 2019, 9, 8299. [Google Scholar] [CrossRef] [PubMed]
- Czekanska, E.M.; Stoddart, M.J.; Richards, R.G.; Hayes, J.S. In search of an osteoblast cell model for in vitro research. Eur. Cells Mater. 2012, 24, 1–17. [Google Scholar] [CrossRef]
- Kodama, H.-A.; Amagai, Y.; Sudo, H.; Kasai, S.; Yamamoto, S. Establishment of a clonal osteogenic cell line from newborn mouse calvaria. Jpn. J. Oral Biol. 1981, 23, 899–901. [Google Scholar] [CrossRef]
- Sudo, H.; Kodama, H.A.; Amagai, Y.; Yamamoto, S.; Kasai, S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J. Cell Biol. 1983, 96, 191–198. [Google Scholar] [CrossRef]
- Wang, D.; Christensen, K.; Chawla, K.; Xiao, G.; Krebsbach, P.H.; Franceschi, R.T. Isolation and Characterization of MC3T3-E1 Preosteoblast Subclones with Distinct In Vitro and In Vivo Differentiation/Mineralization Potential. J. Bone Miner. Res. 1999, 14, 893–903. [Google Scholar] [CrossRef]
- Luttrell, L.M.; Dar, M.S.; Gesty-Palmer, D.; El-Shewy, H.M.; Robinson, K.M.; Haycraft, C.J.; Barth, J.L. Transcriptomic characterization of signaling pathways associated with osteoblastic differentiation of MC-3T3E1 cells. PLoS ONE 2019, 14, e0204197. [Google Scholar] [CrossRef]
- Sugawara, Y.; Suzuki, K.; Koshikawa, M.; Ando, M.; Iida, J. Necessity of Enzymatic Activity of Alkaline Phosphatase for Mineralization of Osteoblastic Cells. Jpn. J. Pharmacol. 2002, 88, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Alekos, N.S.; Moorer, M.C.; Riddle, R.C. Dual Effects of Lipid Metabolism on Osteoblast Function. Front. Endocrinol. (Lausanne) 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rendina-Ruedy, E.; Guntur, A.R.; Rosen, C.J. Intracellular lipid droplets support osteoblast function. Adipocyte 2017, 6, 250–258. [Google Scholar] [CrossRef]
- Guntur, A.R.; Gerencser, A.A.; Le, P.T.; DeMambro, V.E.; Bornstein, S.A.; Mookerjee, S.A.; Maridas, D.E.; Clemmons, D.E.; Brand, M.D.; Rosen, C.J. Osteoblast-like MC3T3-E1 Cells Prefer Glycolysis for ATP Production but Adipocyte-like 3T3-L1 Cells Prefer Oxidative Phosphorylation. J. Bone Miner. Res. 2018, 33, 1052–1065. [Google Scholar] [CrossRef]
- Symmank, J.; Chorus, M.; Appel, S.; Marciniak, J.; Knaup, I.; Bastian, A.; Hennig, C.-L.; Döding, A.; Schulze-Späte, U.; Jacobs, C.; et al. Distinguish fatty acids impact survival, differentiation and cellular function of periodontal ligament fibroblasts. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Smoum, R.; Bar, A.; Tan, B.; Milman, G.; Attar-Namdar, M.; Ofek, O.; Stuart, J.M.; Bajayo, A.; Tam, J.; Kram, V.; et al. Oleoyl serine, an endogenous N-acyl amide, modulates bone remodeling and mass. Proc. Natl. Acad. Sci. USA 2010, 107, 17710–17715. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Bellini, G.; Tortora, C.; Bernardo, M.E.; Luongo, L.; Conforti, A.; Starc, N.; Manzo, I.; Nobili, B.; Locatelli, F.; et al. CB2 and TRPV1 receptors oppositely modulate in vitro human osteoblast activity. Pharmacol. Res. 2015, 99, 194–201. [Google Scholar] [CrossRef]
- Hutchins, H.L.; Li, Y.; Hannon, K.; Watkins, B.A. Eicosapentaenoic acid decreases expression of anandamide synthesis enzyme and cannabinoid receptor 2 in osteoblast-like cells. J. Nutr. Biochem. 2011, 22, 195–200. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Harvey-White, J.; Huang, B.X.; Kim, H.-Y.; Luquet, S.; Palmiter, R.D.; Krystal, G.; Rai, R.; Mahadevan, A.; et al. Multiple pathways involved in the biosynthesis of anandamide. Neuropharmacology 2008, 54, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-X.; Tsuboi, K.; Okamoto, Y.; Tonai, T.; Murakami, M.; Kudo, I.; Ueda, N. Biosynthesis of anandamide and N-palmitoylethanolamine by sequential actions of phospholipase A2 and lysophospholipase D. Biochem. J. 2004, 380, 749–756. [Google Scholar] [CrossRef]
- Higashi, S.; Ohishi, H.; Kudo, I. Augmented prostaglandin E 2 generation resulting from increased activities of cytosolic and secretory phospholipase A 2 and induction of cyclooxygenase-2 in interleukin-1 β-stimulated rat calvarial cells during the mineralizing phase. Inflamm. Res. 2000, 49, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Ueda, N.; Tsuboi, K.; Uyama, T. Metabolism of endocannabinoids and related N-acylethanolamines: Canonical and alternative pathways. FEBS J. 2013, 280, 1874–1894. [Google Scholar] [CrossRef]
- Kozak, K.R.; Rowlinson, S.W.; Marnett, L.J. Oxygenation of the Endocannabinoid, 2-Arachidonylglycerol, to Glyceryl Prostaglandins by Cyclooxygenase-2. J. Biol. Chem. 2000, 275, 33744–33749. [Google Scholar] [CrossRef]
- Kozak, K.R.; Crews, B.C.; Morrow, J.D.; Wang, L.-H.; Ma, Y.H.; Weinander, R.; Jakobsson, P.-J.; Marnett, L.J. Metabolism of the Endocannabinoids, 2-Arachidonylglycerol and Anandamide, into Prostaglandin, Thromboxane, and Prostacyclin Glycerol Esters and Ethanolamides. J. Biol. Chem. 2002, 277, 44877–44885. [Google Scholar] [CrossRef] [PubMed]
- Wasnik, S.; Lakhan, R.; Baylink, D.J.; Rundle, C.H.; Xu, Y.; Zhang, J.; Qin, X.; Lau, K.-H.W.; Carreon, E.E.; Tang, X. Cyclooxygenase 2 augments osteoblastic but suppresses chondrocytic differentiation of CD90+ skeletal stem cells in fracture sites. Sci. Adv. 2019, 5, eaaw2108. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Hu, B.; Lv, X.; Zhu, S.; Zhen, G.; Wan, M.; Jain, A.; Gao, B.; Chai, Y.; Yang, M.; et al. Prostaglandin E2 mediates sensory nerve regulation of bone homeostasis. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kellinsalmi, M.; Parikka, V.; Risteli, J.; Hentunen, T.; Leskelä, H.-V.; Lehtonen, S.; Selander, K.; Väänänen, K.; Lehenkari, P. Inhibition of cyclooxygenase-2 down-regulates osteoclast and osteoblast differentiation and favours adipocyte formation in vitro. Eur. J. Pharmacol. 2007, 572, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Dunn, S.L.; Wilkinson, J.M.; Crawford, A.; Bunning, R.A.; Le Maitre, C.L. Expression of Cannabinoid Receptors in Human Osteoarthritic Cartilage: Implications for Future Therapies. Cannabis Cannabinoid Res. 2016, 1, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Whyte, L.S.; Ryberg, E.; Sims, N.A.; Ridge, S.A.; Mackie, K.; Greasley, P.J.; Ross, R.A.; Rogers, M.J. The putative cannabinoid receptor GPR55 affects osteoclast function in vitro and bone mass in vivo. Proc. Natl. Acad. Sci. USA 2009, 106, 16511–16516. [Google Scholar] [CrossRef]
- Gregory, L.S.; Kelly, W.L.; Reid, R.C.; Fairlie, D.P.; Forwood, M.R. Inhibitors of cyclo-oxygenase-2 and secretory phospholipase A2 preserve bone architecture following ovariectomy in adult rats. Bone 2006, 39, 134–142. [Google Scholar] [CrossRef]
- Zygmunt, P.M.; Petersson, J.; Andersson, D.A.; Chuang, H.-H.; Sørgård, M.; Di Marzo, V.; Julius, D.; Högestätt, E.D. Vanilloid receptors on sensory nerves mediate the vasodilator action of anandamide. Nat. Cell Biol. 1999, 400, 452–457. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Bisogno, T.; Maccarrone, M.; Davis, J.B.; Finazzi-Agrò, A.; Di Marzo, V. The Activity of Anandamide at Vanilloid VR1 Receptors Requires Facilitated Transport across the Cell Membrane and Is Limited by Intracellular Metabolism. J. Biol. Chem. 2001, 276, 12856–12863. [Google Scholar] [CrossRef] [PubMed]
- He, L.-H.; Liu, M.; He, Y.; Xiao, E.; Zhao, L.; Zhang, T.; Yang, H.-Q.; Zhang, Y. TRPV1 deletion impaired fracture healing and inhibited osteoclast and osteoblast differentiation. Sci. Rep. 2017, 7, srep42385. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Tortora, C.; Punzo, F.; Bellini, G.; Argenziano, M.; Di Paola, A.; Torella, M.; Perrotta, S. The Endocannabinoid/Endovanilloid System in Bone: From Osteoporosis to Osteosarcoma. Int. J. Mol. Sci. 2019, 20, 1919. [Google Scholar] [CrossRef] [PubMed]
- Whyte, L.S.; Ford, L.; Ridge, S.A.; Cameron, G.A.; Rogers, M.J.; Ross, R.A. Cannabinoids and bone: Endocannabinoids modulate human osteoclast function in vitro. Br. J. Pharmacol. 2011, 165, 2584–2597. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Lee, D.-K.; Jin, X.; Che, X.; Choi, J.-Y. Oleoylethanolamide Exhibits GPR119-Dependent Inhibition of Osteoclast Function and GPR119-Independent Promotion of Osteoclast Apoptosis. Mol. Cells 2020, 43, 340–349. [Google Scholar] [PubMed]

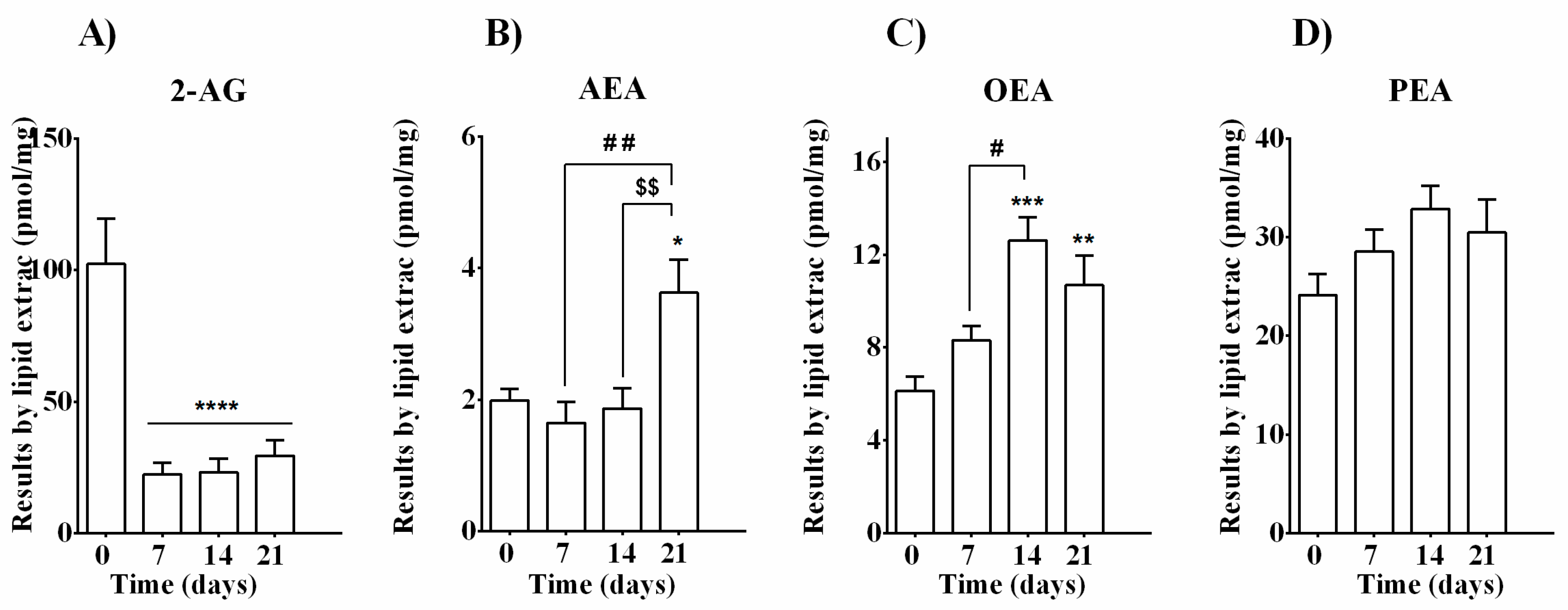
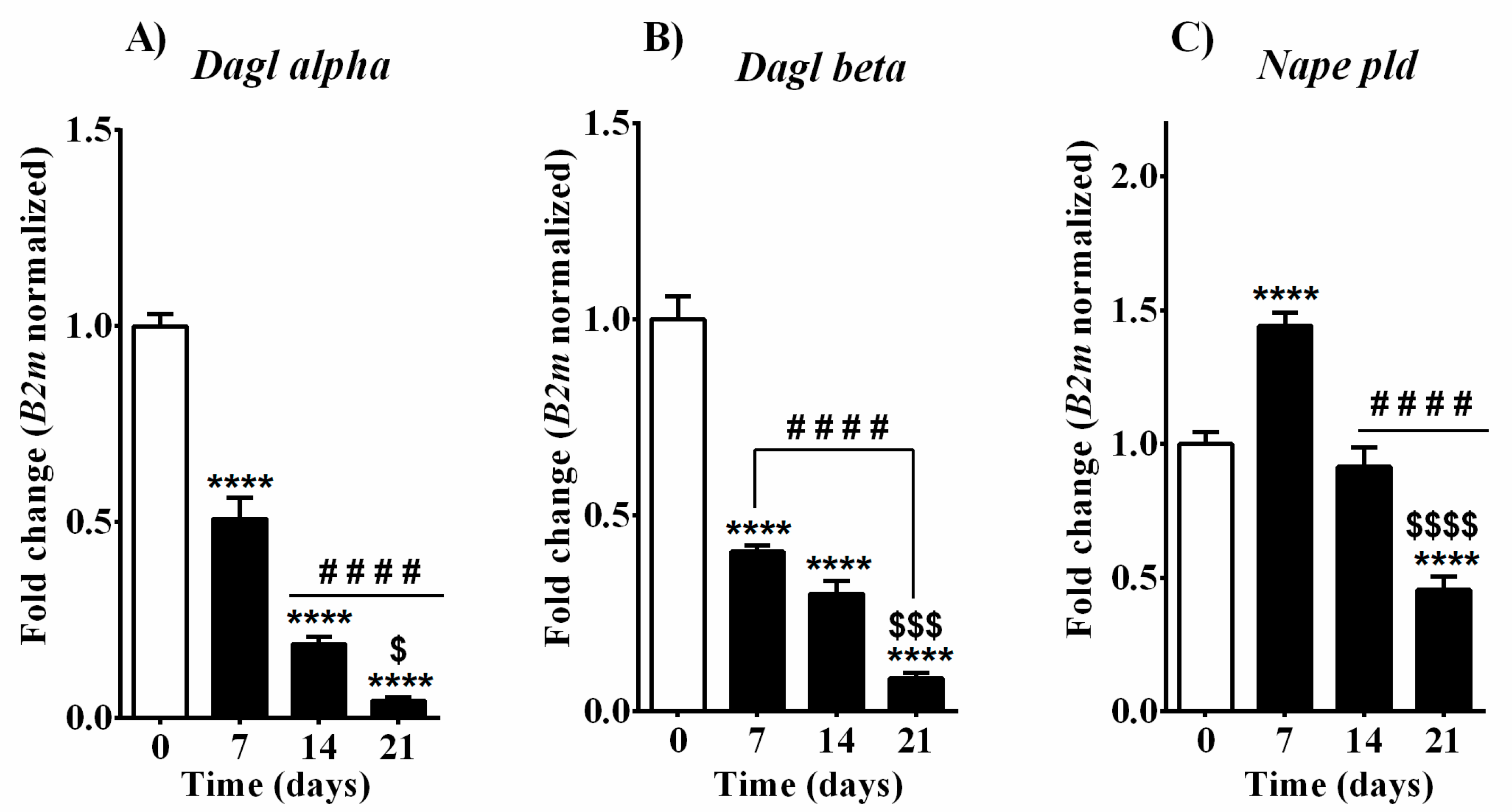
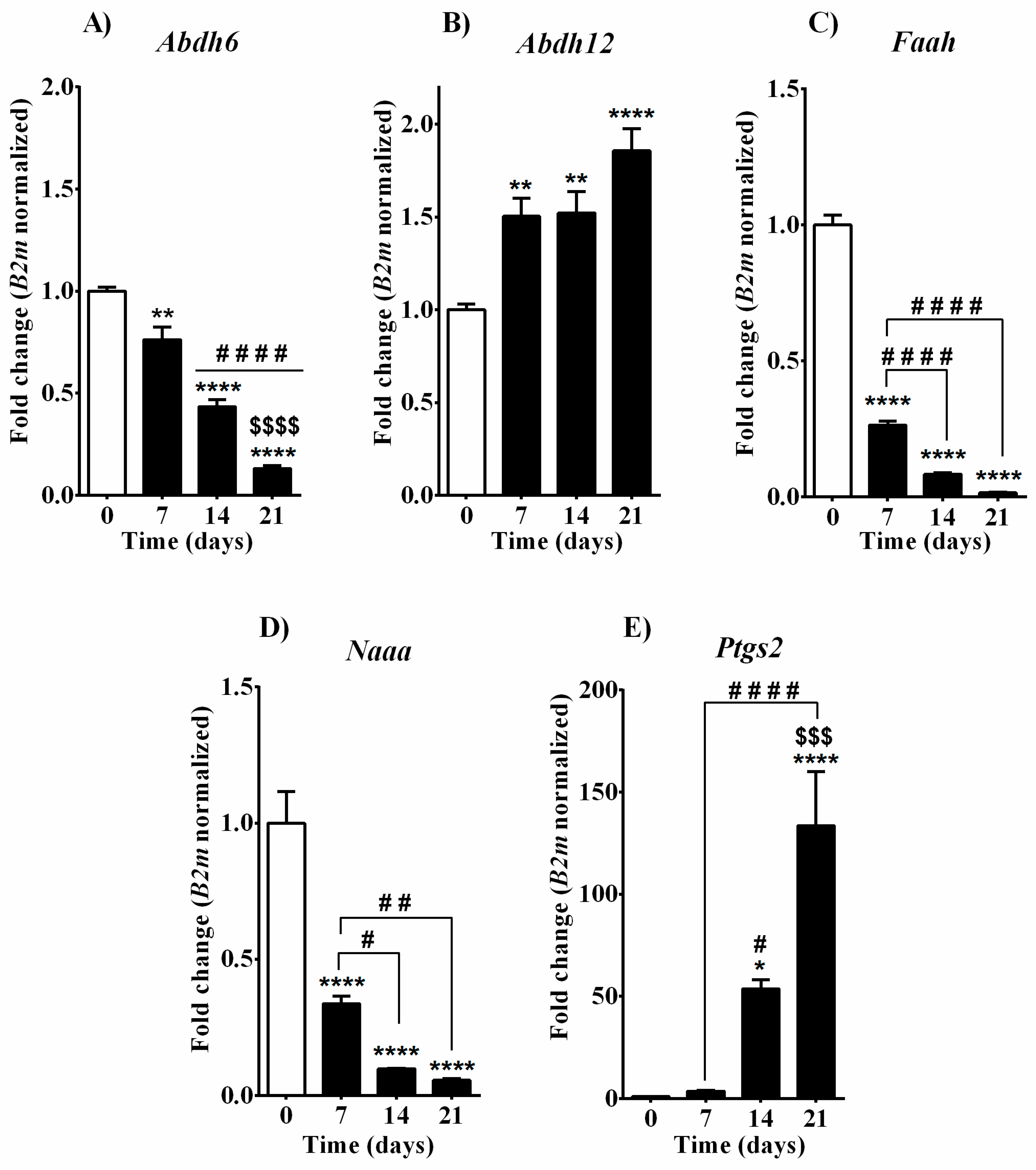
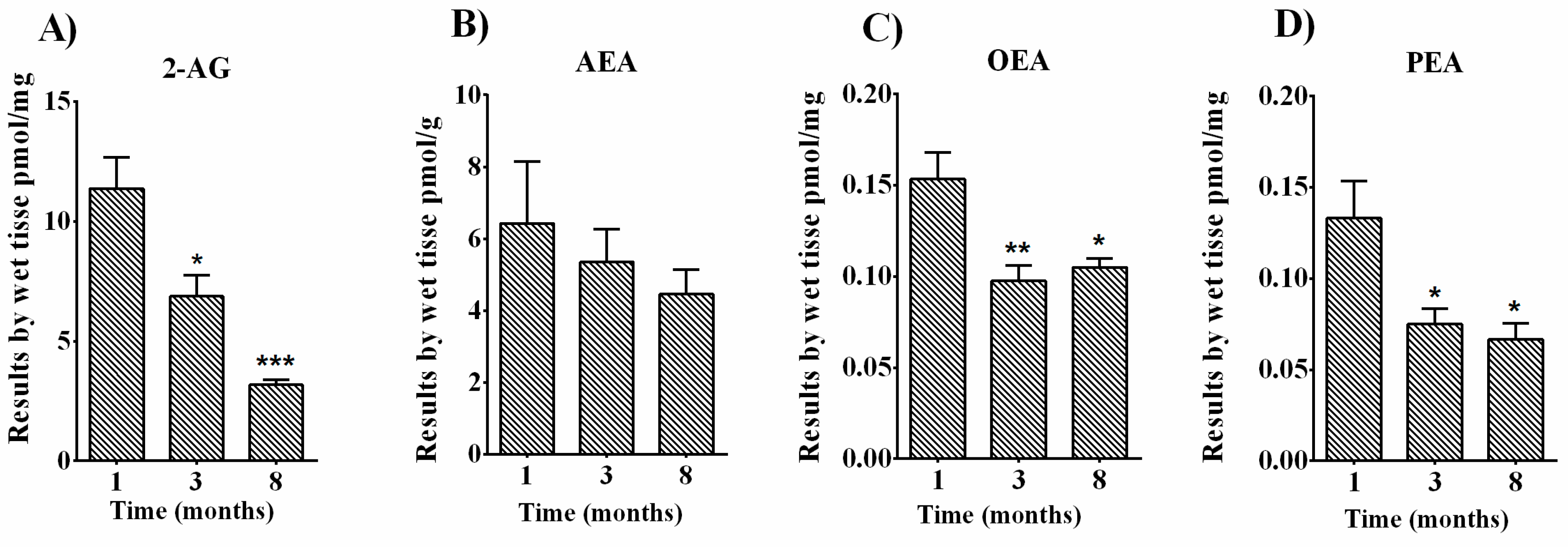
| Days of Osteoblast Differentiation | ||||
|---|---|---|---|---|
| Investigated Group/Target | Day 7 | Day 14 | Day 21 | |
| EC and NAE levels | 2-AG | ↓↓↓↓ | ↓↓↓↓ | ↓↓↓↓ |
| AEA | - | - | ↑ | |
| OEA | - | ↑↑↑ | ↑↑ | |
| PEA | - | - | - | |
| EC synthesis | Dagl alpha | ↓↓↓↓ | ↓↓↓↓ | ↓↓↓↓ |
| Dagl beta | ↓↓↓↓ | ↓↓↓↓ | ↓↓↓↓ | |
| Naple pld | ↑↑↑↑ | - | ↓↓↓↓ | |
| EC degradation | Abdh 6 | ↓↓ | ↓↓↓↓ | ↓↓↓↓ |
| Abdh 12 | ↑↑ | ↑↑ | ↑↑↑↑ | |
| Faah | ↓↓↓↓ | ↓↓↓↓ | ↓↓↓↓ | |
| Naaa | ↓↓↓↓ | ↓↓↓↓ | ↓↓↓↓ | |
| Ptgs2 | - | ↑ | ↑↑↑↑ | |
| Age of B6D2 Male Mice (Months) | |||
|---|---|---|---|
| Investigated Group/Target | 3 | 8 | |
| EC and NAE levels | 2-AG | ↓ | ↓↓↓ |
| AEA | - | - | |
| OEA | ↓↓ | ↓ | |
| PEA | ↓ | ↓ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostrzewa, M.; Mahmoud, A.M.; Verde, R.; Scotto di Carlo, F.; Gianfrancesco, F.; Piscitelli, F.; Ligresti, A. Modulation of Endocannabinoid Tone in Osteoblastic Differentiation of MC3T3-E1 Cells and in Mouse Bone Tissue over Time. Cells 2021, 10, 1199. https://doi.org/10.3390/cells10051199
Kostrzewa M, Mahmoud AM, Verde R, Scotto di Carlo F, Gianfrancesco F, Piscitelli F, Ligresti A. Modulation of Endocannabinoid Tone in Osteoblastic Differentiation of MC3T3-E1 Cells and in Mouse Bone Tissue over Time. Cells. 2021; 10(5):1199. https://doi.org/10.3390/cells10051199
Chicago/Turabian StyleKostrzewa, Magdalena, Ali Mokhtar Mahmoud, Roberta Verde, Federica Scotto di Carlo, Fernando Gianfrancesco, Fabiana Piscitelli, and Alessia Ligresti. 2021. "Modulation of Endocannabinoid Tone in Osteoblastic Differentiation of MC3T3-E1 Cells and in Mouse Bone Tissue over Time" Cells 10, no. 5: 1199. https://doi.org/10.3390/cells10051199
APA StyleKostrzewa, M., Mahmoud, A. M., Verde, R., Scotto di Carlo, F., Gianfrancesco, F., Piscitelli, F., & Ligresti, A. (2021). Modulation of Endocannabinoid Tone in Osteoblastic Differentiation of MC3T3-E1 Cells and in Mouse Bone Tissue over Time. Cells, 10(5), 1199. https://doi.org/10.3390/cells10051199








