Viroids as a Tool to Study RNA-Directed DNA Methylation in Plants
Abstract
:1. Viroids and RNAi
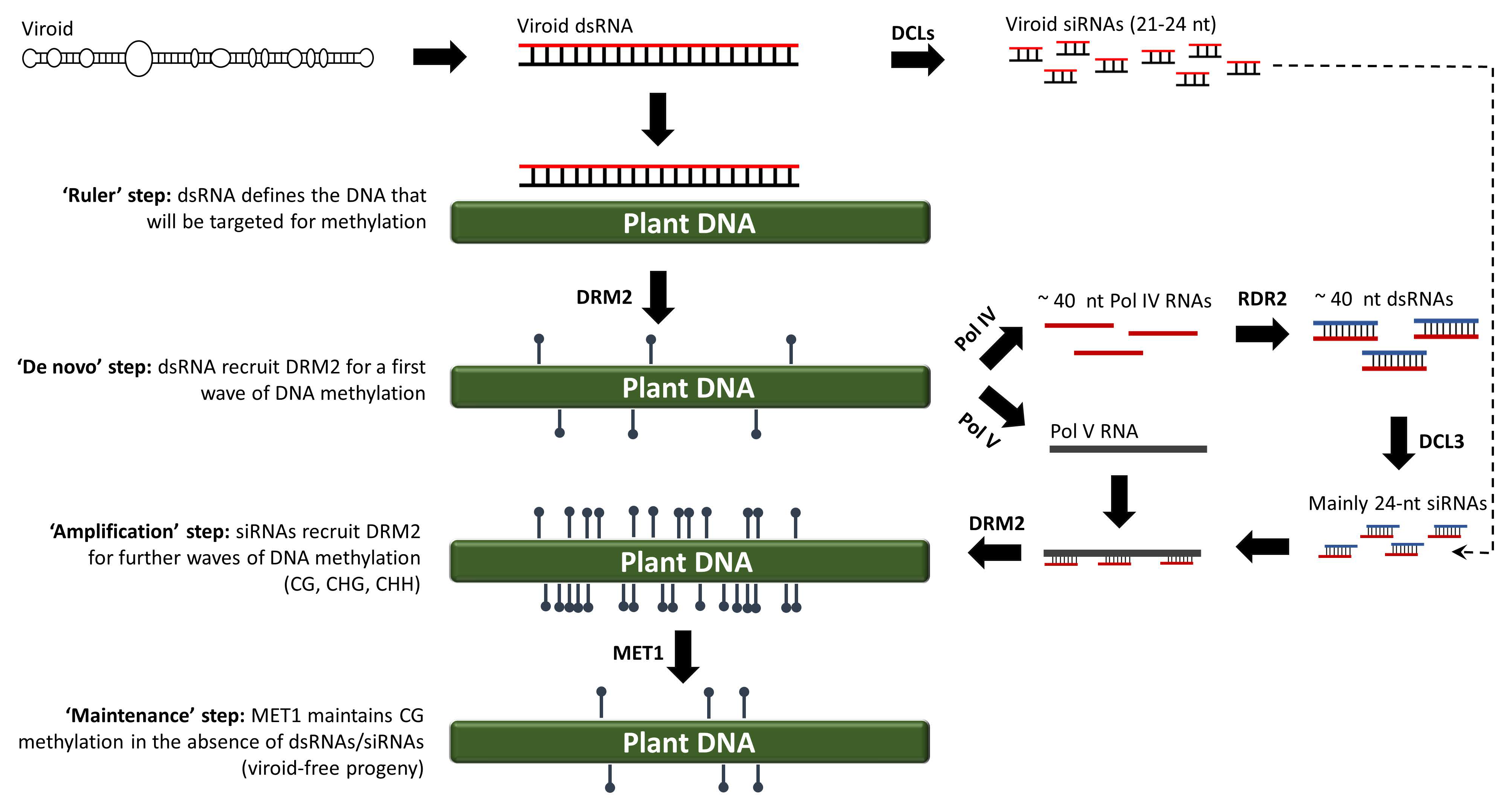
2. RNA-Directed DNA Methylation
3. Discovery of RdDM in Viroid-Infected Plants
4. CHH Is the Hallmark of the RNA-Directed de novo DNA Methylation
5. CG Methylation Can Be Maintained in the Absence of dsRNAs/siRNAs
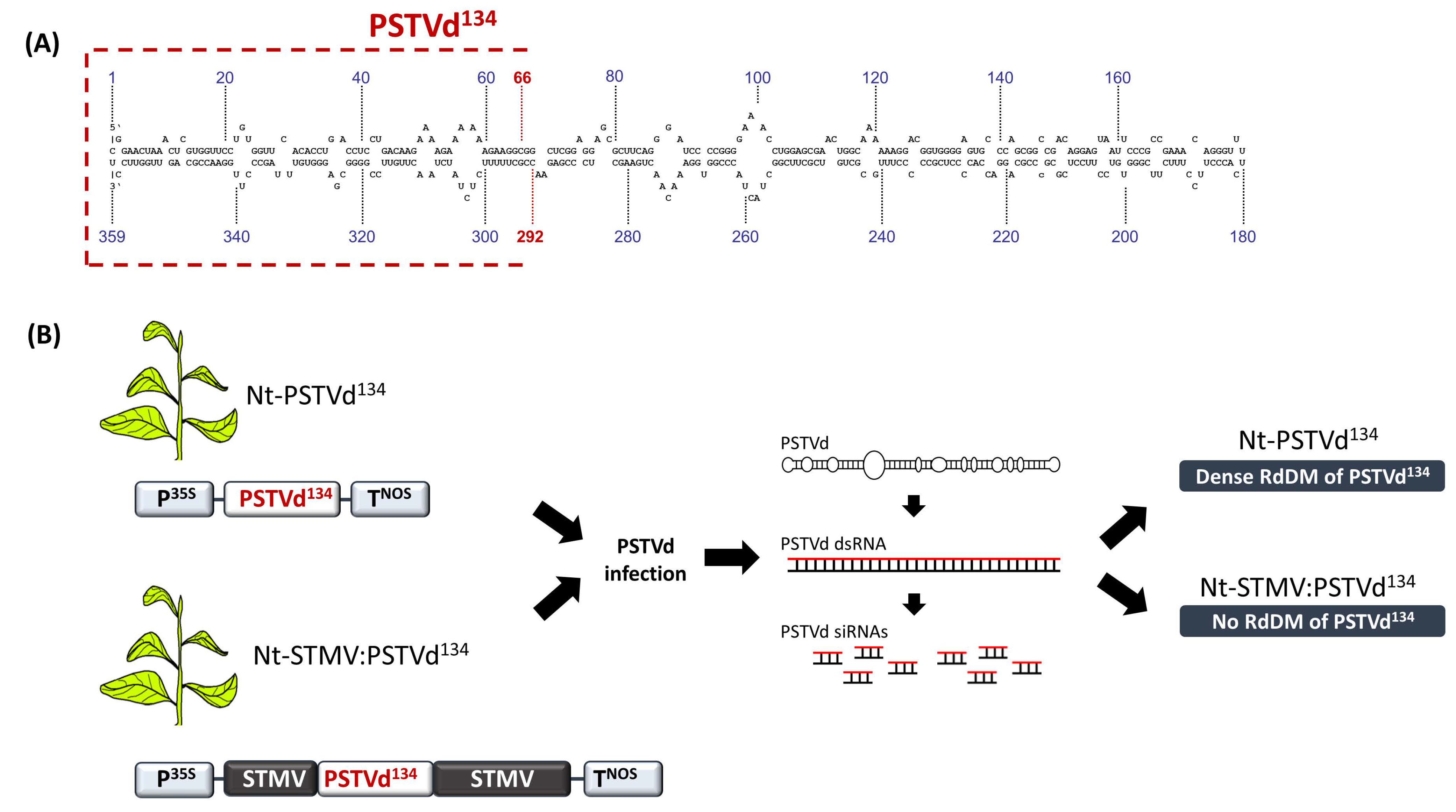
6. 30 bp Is the Minimum Target Size for RdDM
7. Local DNA Features Inhibit RdDM
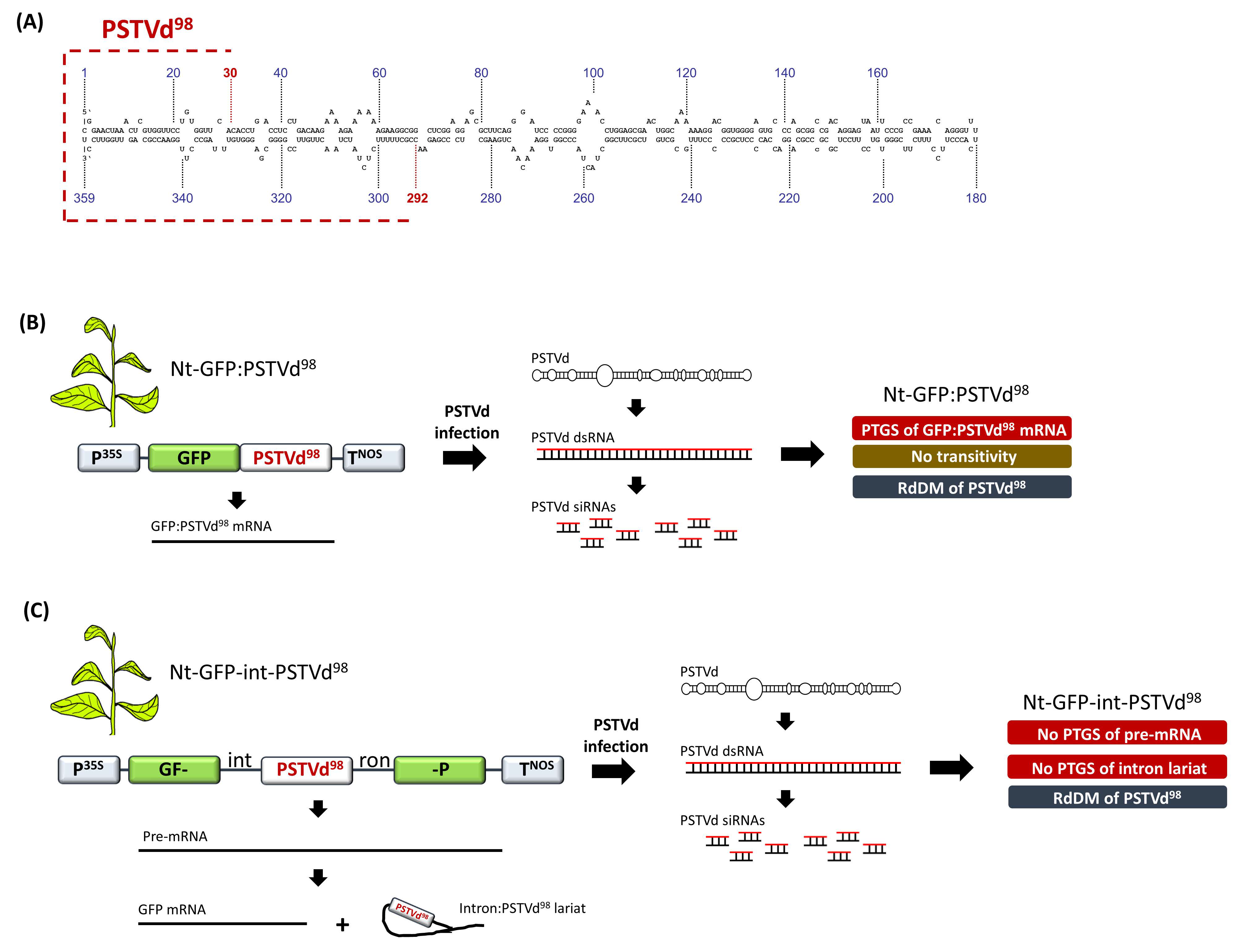
8. RdDM Is Not Necessarily Coupled to PTGS and Transitivity
9. RdDM Efficiency Depends on Sequence Identity between Trigger RNA and Target DNA
10. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Diener, T.O. Potato spindle tuber “virus”: IV. A replicating, low molecular weight RNA. Virology 1971, 45, 411–428. [Google Scholar] [CrossRef]
- Flores, R.; Gago-Zachert, S.; Serra, P.; Sanjuan, R.; Elena, S.F. Viroids: Survivors from the RNA world? Annu. Rev. Microbiol. 2014, 68, 395–414. [Google Scholar] [CrossRef] [Green Version]
- Tabler, M.; Tsagris, M. Viroids: Petite RNA pathogens with distinguished talents. Trends Plant Sci. 2004, 9, 339–348. [Google Scholar] [CrossRef]
- Wang, Y. Current view and perspectives in viroid replication. Curr. Opin. Virol. 2021, 47, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Hernandez, C.; de Alba, A.E.M.; Daros, J.A.; di Serio, F. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 2005, 43, 117–139. [Google Scholar] [CrossRef]
- Owens, R.A.; Hammond, R.W. Viroid pathogenicity: One process, many faces. Viruses 2009, 1, 298–316. [Google Scholar] [CrossRef] [Green Version]
- Tsagris, E.M.; de Alba, A.E.M.; Gozmanova, M.; Kalantidis, K. Viroids. Cell. Microbiol. 2008, 10, 2168–2179. [Google Scholar] [CrossRef]
- Gozmanova, M.; Denti, M.A.; Minkov, I.N.; Tsagris, M.; Tabler, M. Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res. 2003, 31, 5534–5543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalantidis, K.; Denti, M.A.; Tzortzakaki, S.; Marinou, E.; Tabler, M.; Tsagris, M. Virp1 is a host protein with a major role in potato spindle tuber viroid infection in Nicotiana plants. J. Virol. 2007, 81, 12872–12880. [Google Scholar] [CrossRef] [Green Version]
- Branch, A.D.; Robertson, H.D. A replication cycle for viroids and other small infectious RNA’s. Science 1984, 223, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Mudiyanselage, S.D.D.; Wang, Y. Evidence supporting that RNA polymerase II catalyzes de novo transcription using potato spindle tuber viroid circular RNA templates. Viruses 2020, 12, 371. [Google Scholar] [CrossRef] [Green Version]
- Nohales, M.A.; Flores, R.; Daros, J.A. Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. USA 2012, 109, 13805–13810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, B.; Kwon, M.O.; Hammond, R.; Owens, R. Cell-to-cell movement of potato spindle tuber viroid. Plant J. 1997, 12, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Qi, Y.; Xun, Y.; Owens, R.; Ding, B. Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 2002, 130, 138–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, R.A.; Diener, T.O. RNA intermediates in potato spindle tuber viroid replication. Proc. Natl. Acad. Sci. USA 1982, 79, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Grill, L.K.; Semancik, J.S. RNA sequences complementary to citrus exocortis viroid in nucleic acid preparations from infected Gynura aurantiaca. Proc. Natl. Acad. Sci. USA 1978, 75, 896–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadidi, A.; Hashimoto, J.; Diener, T.O. Potato spindle tuber viroid-specific double-stranded RNA in extracts from infected tomato leaves. Annales de l’Institut Pasteur Virologie 1982, 133, 15–31. [Google Scholar] [CrossRef]
- Dalakouras, A.; Dadami, E.; Wassenegger, M. Engineering viroid resistance. Viruses 2015, 7, 634–646. [Google Scholar] [CrossRef] [Green Version]
- Itaya, A.; Folimonov, A.; Matsuda, Y.; Nelson, R.S.; Ding, B. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant Microbe Interact. 2001, 14, 1332–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.B.; Bian, X.Y.; Wu, L.M.; Liu, L.X.; Smith, N.A.; Isenegger, D.; Wu, R.M.; Masuta, C.; Vance, V.B.; Watson, J.M.; et al. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 2004, 101, 3275–3280. [Google Scholar] [CrossRef] [Green Version]
- Dadami, E.; Boutla, A.; Vrettos, N.; Tzortzakaki, S.; Karakasilioti, I.; Kalantidis, K. Dicer-like 4 but not dicer-like 2 may have a positive effect on potato spindle tuber viroid accumulation in Nicotiana benthamiana. Mol. Plant 2013, 6, 232–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsarou, K.; Mavrothalassiti, E.; Dermauw, W.; van Leeuwen, T.; Kalantidis, K. Combined activity of DCL2 and DCL3 is crucial in the defense against potato spindle tuber viroid. PLoS Pathog. 2016, 12, e1005936. [Google Scholar] [CrossRef] [PubMed]
- Minoia, S.; Carbonell, A.; di Serio, F.; Gisel, A.; Carrington, J.C.; Navarro, B.; Flores, R. Specific argonautes selectively bind small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo. J. Virol. 2014, 88, 11933–11945. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.M.; Chen, L.T.; Patel, K.; Li, Y.H.; Baulcombe, D.C.; Wu, S.H. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 2010, 107, 15269–15274. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Li, B.; Iwakawa, H.O.; Pan, Y.; Tang, X.; Ling-Hu, Q.; Liu, Y.; Sheng, S.; Feng, L.; Zhang, H.; et al. Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature 2020, 581, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.; Zilberman, D.; Xie, Z.; Johansen, L.K.; Carrington, J.C.; Jacobsen, S.E. RNA silencing genes control de novo DNA methylation. Science 2004, 303, 1336. [Google Scholar] [CrossRef]
- Martin, R.; Arenas, C.; Daros, J.A.; Covarrubias, A.; Reyes, J.L.; Chua, N.H. Characterization of small RNAs derived from citrus exocortis viroid (CEVd) in infected tomato plants. Virology 2007, 367, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Itaya, A.; Zhong, X.; Bundschuh, R.; Qi, Y.; Wang, Y.; Takeda, R.; Harris, A.R.; Molina, C.; Nelson, R.S.; Ding, B. A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 2007, 81, 2980–2994. [Google Scholar] [CrossRef] [Green Version]
- Adkar-Purushothama, C.R.; Brosseau, C.; Giguere, T.; Sano, T.; Moffett, P.; Perreault, J.P. Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell 2015, 27, 2178–2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogt, U.; Pelissier, T.; Putz, A.; Razvi, F.; Fischer, R.; Wassenegger, M. Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. 2004, 38, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Schwind, N.; Zwiebel, M.; Itaya, A.; Ding, B.; Wang, M.B.; Krczal, G.; Wassenegger, M. RNAi-mediated resistance to potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. plant Pathol. 2009, 10, 459–469. [Google Scholar] [CrossRef]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef]
- Wassenegger, M.; Heimes, S.; Riedel, L.; Sanger, H.L. RNA-directed de novo methylation of genomic sequences in plants. Cell 1994, 76, 567–576. [Google Scholar] [CrossRef]
- Gallego-Bartolome, J. DNA methylation in plants: Mechanisms and tools for targeted manipulation. New Phytol. 2020, 227, 38–44. [Google Scholar] [CrossRef] [Green Version]
- Matzke, M.A.; Kanno, T.; Matzke, A.J. RNA-directed DNA Methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Liu, Z.W.; Shao, C.R.; Zhang, C.J.; Zhou, J.X.; Zhang, S.W.; Li, L.; Chen, S.; Huang, H.W.; Cai, T.; He, X.J. The SET domain proteins SUVH2 and SUVH9 are required for pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 2014, 10, e1003948. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.L.; Zhang, G.; Tang, K.; Li, J.; Yang, L.; Huang, H.; Zhang, H.; Zhu, J.K. Dicer-independent RNA-directed DNA methylation in arabidopsis. Cell Res. 2016, 26, 1264. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; He, X.; Wang, X.J.; Kohany, O.; Jurka, J.; Hannon, G.J. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 2006, 443, 1008–1012. [Google Scholar] [CrossRef]
- Zilberman, D.; Cao, X.; Johansen, L.K.; Xie, Z.; Carrington, J.C.; Jacobsen, S.E. Role of arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 2004, 14, 1214–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalakouras, A.; Dadami, E.; Wassenegger, M. Viroid-induced DNA methylation in plants. Biomol. Concepts 2013, 4, 557–565. [Google Scholar] [CrossRef]
- Dalakouras, A.; Papadopoulou, K.K. Epigenetic modifications: An unexplored facet of exogenous RNA application in plants. Plants 2020, 9, 673. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Wassenegger, M. Revisiting RNA-directed DNA methylation. RNA Biol. 2013, 10, 453–455. [Google Scholar] [CrossRef] [Green Version]
- Dalakouras, A.; Ganopoulos, I. Induction of promoter DNA methylation upon high-pressure spraying of double-stranded RNA in plants. Agronomy 2021, 11, 789. [Google Scholar] [CrossRef]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA triplex formation by long noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef] [Green Version]
- Zhai, J.; Bischof, S.; Wang, H.; Feng, S.; Lee, T.F.; Teng, C.; Chen, X.; Park, S.Y.; Liu, L.; Gallego-Bartolome, J.; et al. A one precursor one siRNA model for Pol IV-pependent siRNA biogenesis. Cell 2015, 163, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Wierzbicki, A.T.; Haag, J.R.; Pikaard, C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008, 135, 635–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wierzbicki, A.T.; Ream, T.S.; Haag, J.R.; Pikaard, C.S. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009, 41, 630–634. [Google Scholar] [CrossRef]
- Pelissier, T.; Thalmeir, S.; Kempe, D.; Sanger, H.L.; Wassenegger, M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999, 27, 1625–1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aufsatz, W.; Mette, M.F.; Matzke, A.J.; Matzke, M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol. Biol. 2004, 54, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Lindroth, A.M.; Cao, X.; Jackson, J.P.; Zilberman, D.; McCallum, C.M.; Henikoff, S.; Jacobsen, S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001, 292, 2077–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Matzke, M.; Aufsatz, W.; Kanno, T.; Daxinger, L.; Papp, I.; Mette, M.F.; Matzke, A.J. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim. Biophys. Acta 2004, 1677, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Dadami, E.; Zwiebel, M.; Krczal, G.; Wassenegger, M. Transgenerational maintenance of transgene body CG but not CHG and CHH methylation. Epigenetics 2012, 7, 1071–1078. [Google Scholar] [CrossRef] [Green Version]
- Jones, L.; Ratcliff, F.; Baulcombe, D.C. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 2001, 11, 747–757. [Google Scholar] [CrossRef] [Green Version]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef] [Green Version]
- Bewick, A.J.; Schmitz, R.J. Gene body DNA methylation in plants. Curr. Opin. Plant Biol. 2017, 36, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Pelissier, T.; Wassenegger, M. A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. RNA 2000, 6, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Dalakouras, A.; Moser, M.; Krczal, G.; Wassenegger, M. A chimeric satellite transgene sequence is inefficiently targeted by viroid-induced DNA methylation in tobacco. Plant Mol. Biol. 2010, 73, 439–447. [Google Scholar] [CrossRef]
- Fischer, U.; Kuhlmann, M.; Pecinka, A.; Schmidt, R.; Mette, M.F. Local DNA features affect RNA-directed transcriptional gene silencing and DNA methylation. Plant J. 2008, 53, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.; Hamilton, A.J.; Voinnet, O.; Thomas, C.L.; Maule, A.J.; Baulcombe, D.C. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 1999, 11, 2291–2301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dadami, E.; Dalakouras, A.; Zwiebel, M.; Krczal, G.; Wassenegger, M. An endogene-resembling transgene is resistant to DNA methylation and systemic silencing. RNA Biol. 2014, 11, 934–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dadami, E.; Moser, M.; Zwiebel, M.; Krczal, G.; Wassenegger, M.; Dalakouras, A. An endogene-resembling transgene delays the onset of silencing and limits siRNA accumulation. FEBS Lett. 2013, 587, 706–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaistij, F.E.; Jones, L. Compromised virus-induced gene silencing in RDR6-deficient plants. Plant Physiol. 2009, 149, 1399–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwach, F.; Vaistij, F.E.; Jones, L.; Baulcombe, D.C. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005, 138, 1842–1852. [Google Scholar] [CrossRef] [Green Version]
- Dalakouras, A.; Dadami, E.; Bassler, A.; Zwiebel, M.; Krczal, G.; Wassenegger, M. Replicating potato spindle tuber viroid mediates de novo methylation of an intronic viroid sequence but no cleavage of the corresponding pre-mRNA. RNA Biol. 2015, 12, 268–275. [Google Scholar] [CrossRef] [Green Version]
- Hoffer, P.; Ivashuta, S.; Pontes, O.; Vitins, A.; Pikaard, C.; Mroczka, A.; Wagner, N.; Voelker, T. Posttranscriptional gene silencing in nuclei. Proc. Natl. Acad. Sci. USA 2011, 108, 409–414. [Google Scholar] [CrossRef] [Green Version]
- Dalakouras, A.; Dadami, E.; Wassenegger, M.; Krczal, G.; Wassenegger, M. RNA-directed DNA methylation efficiency depends on trigger and target sequence identity. Plant J. 2016, 87, 202–214. [Google Scholar] [CrossRef] [Green Version]
- Diermann, N.; Matousek, J.; Junge, M.; Riesner, D.; Steger, G. Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato. Biol. Chem. 2010, 391, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Eamens, A.L.; Smith, N.A.; Dennis, E.S.; Wassenegger, M.; Wang, M.B. In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology 2014, 450, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; di Serio, F. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012, 70, 991–1003. [Google Scholar] [CrossRef]
- Martinez, G.; Castellano, M.; Tortosa, M.; Pallas, V.; Gomez, G. A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic Acids Res. 2014, 42, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Leister, D.; Kleine, T. Chloroplast development and genomes uncoupled signaling are independent of the RNA-directed DNA methylation pathway. Sci. Rep. 2020, 10, 15412. [Google Scholar] [CrossRef]
- Gomez, G.; Pallas, V. Studies on subcellular compartmentalization of plant pathogenic noncoding RNAs give new insights into the intracellular RNA-traffic mechanisms. Plant Physiol. 2012, 159, 558–564. [Google Scholar] [CrossRef] [Green Version]
- De Alba, A.E.M.; Flores, R.; Hernandez, C. Two chloroplastic viroids induce the accumulation of small RNAs associated with posttranscriptional gene silencing. J. Virol. 2002, 76, 13094–13096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daros, J.A. Eggplant latent viroid: A friendly experimental system in the family Avsunviroidae. Mol. Plant Pathol. 2016, 17, 1170–1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wassenegger, M.; Dalakouras, A. Viroids as a Tool to Study RNA-Directed DNA Methylation in Plants. Cells 2021, 10, 1187. https://doi.org/10.3390/cells10051187
Wassenegger M, Dalakouras A. Viroids as a Tool to Study RNA-Directed DNA Methylation in Plants. Cells. 2021; 10(5):1187. https://doi.org/10.3390/cells10051187
Chicago/Turabian StyleWassenegger, Michael, and Athanasios Dalakouras. 2021. "Viroids as a Tool to Study RNA-Directed DNA Methylation in Plants" Cells 10, no. 5: 1187. https://doi.org/10.3390/cells10051187
APA StyleWassenegger, M., & Dalakouras, A. (2021). Viroids as a Tool to Study RNA-Directed DNA Methylation in Plants. Cells, 10(5), 1187. https://doi.org/10.3390/cells10051187





