The Role of Protein Arginine Methylation as Post-Translational Modification on Actin Cytoskeletal Components in Neuronal Structure and Function
Abstract
1. Introduction
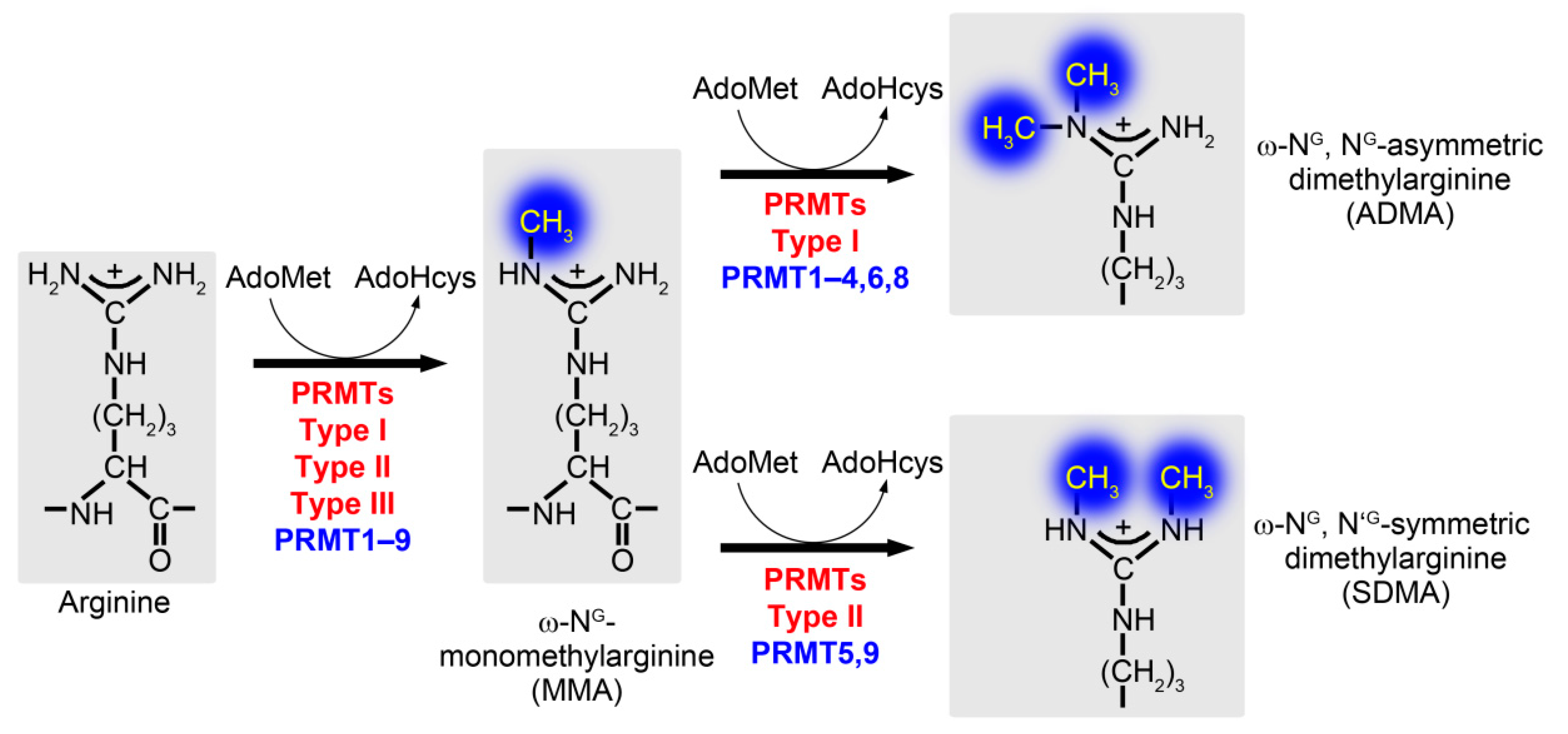
1.1. Protein Arginine Methylation
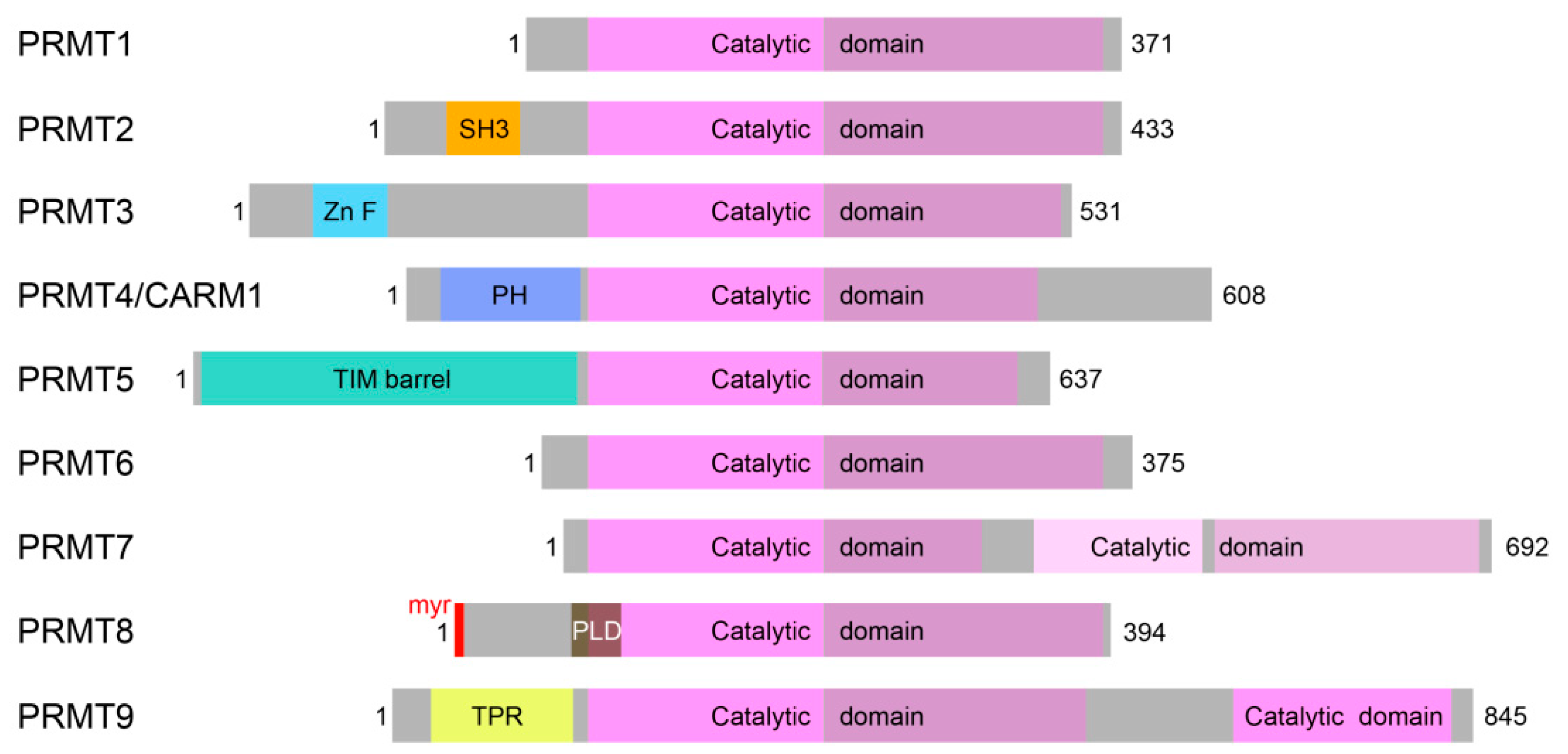
1.2. PRMTs in the Central Nervous System
1.3. Neuromorphogenesis Powered by Actin Cytoskeletal Forces
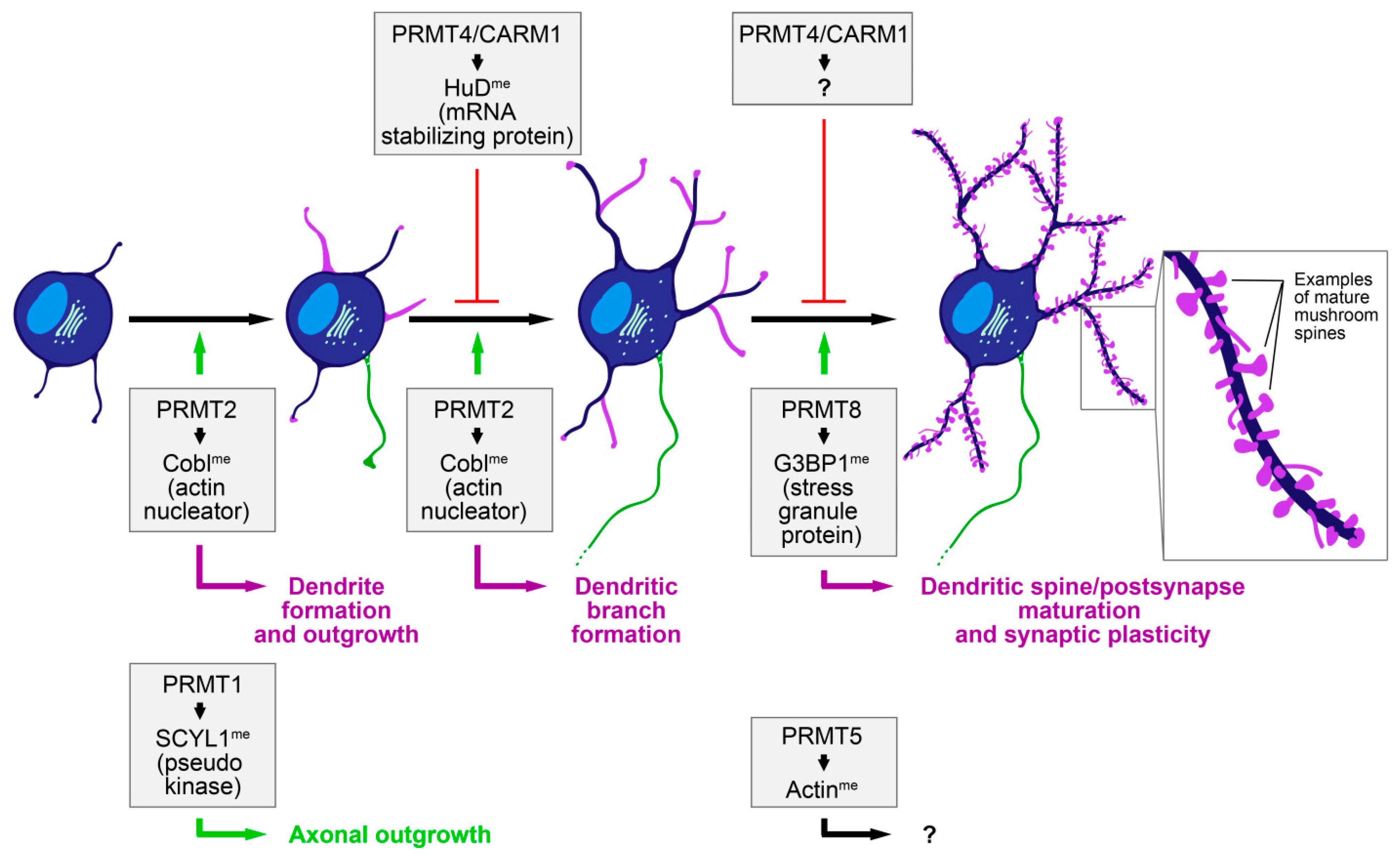
2. Neuromorphogenesis Controlled by Protein Arginine Methylation
3. A Role of Protein Arginine Methylation in Dendritic Spine Formation and Maturation—Signaling to Actin Cytoskeletal Factors?
4. Direct Modulation of Actin by Arginine Methylation
5. Arginine Methylation of Actin Cytoskeletal Effectors Controls Neuromorphogenesis
6. Additional Cytoskeletal Targets for Neuronal Arginine Protein Methylation
7. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Blanc, R.S.; Richard, S. Arginine methylation: The coming of age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef]
- Wolf, S.S. The protein arginine methyltransferase family: An update about function, new perspectives and the physiological role in humans. Cell. Mol. Life Sci. 2009, 66, 2109–2121. [Google Scholar] [CrossRef] [PubMed]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef]
- Fuhrmann, J.; Clancy, K.W.; Thompson, P.R. Chemical biology of protein arginine modifications in epigenetic regulation. Chem. Rev. 2015, 115, 5413–5461. [Google Scholar] [CrossRef] [PubMed]
- Kakimoto, Y.; Matsuoka, Y.; Miyake, M.; Konishi, H. Methylated amino acid residues of proteins of brain and other organs. J. Neurochem. 1975, 24, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Murata, K.; Ishida, J.; Kanou, A.; Kasuya, Y.; Fukamizu, A. Severe hypomyelination and developmental defects are caused in mice lacking protein arginine methyltransferase 1 (PRMT1) in the central nervous system. J. Biol. Chem. 2016, 291, 2237–2245. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Fukamizu, A.; Nakagawa, T.; Kizuka, Y. Roles of protein arginine methyltransferase 1 (PRMT1) in brain development and disease. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129776. [Google Scholar] [CrossRef]
- Couto, E.; Silva, A.; Wu, C.Y.; Citadin, C.T.; Clemons, G.A.; Possoit, H.E.; Grames, M.S.; Lien, C.F.; Minagar, A.; Lee, R.H.; et al. Protein arginine methyltransferases in cardiovascular and neuronal function. Mol. Neurobiol. 2020, 57, 1716–1732. [Google Scholar] [CrossRef] [PubMed]
- Miranda, T.B.; Miranda, M.; Frankel, A.; Clarke, S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem. 2004, 279, 22902–22907. [Google Scholar] [CrossRef] [PubMed]
- Dotti, C.G.; Sullivan, C.A.; Banker, G.A. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 1988, 8, 1454–1468. [Google Scholar] [CrossRef] [PubMed]
- Kessels, M.M.; Schwintzer, L.; Schlobinski, D.; Qualmann, B. Controlling actin cytoskeletal organization and dynamics during neuronal morphogenesis. Eur. J. Cell Biol. 2011, 90, 926–933. [Google Scholar] [CrossRef]
- Pollard, T.D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 451–477. [Google Scholar] [CrossRef] [PubMed]
- Qualmann, B.; Kessels, M.M. New players in actin polymerization—WH2-domain-containing actin nucleators. Trends Cell Biol. 2009, 19, 276–285. [Google Scholar] [CrossRef]
- Williams, D.S.; Linberg, K.A.; Vaughan, D.K.; Fariss, R.N.; Fisher, S.K. Disruption of microfilament organization and deregulation of disk membrane morphogenesis by cytochalasin D in rod and cone photoreceptors. J. Comp. Neurol. 1988, 272, 161–176. [Google Scholar] [CrossRef]
- Edson, K.; Weisshaar, B.; Matus, A. Actin depolymerisation induces process formation on MAP2-transfected non-neuronal cells. Development 1993, 117, 689–700. [Google Scholar] [CrossRef]
- Kwiatkowski, A.V.; Rubinson, D.A.; Dent, E.W.; Van Veen, J.E.; Leslie, J.D.; Zhang, J.; Mebane, L.M.; Philippar, U.; Pinheiro, E.M.; Burds, A.A.; et al. Ena/VASP is required for neuritogenesis in the developing cortex. Neuron 2007, 56, 441–455. [Google Scholar] [CrossRef]
- Dent, E.W.; Kwiatkowski, A.V.; Mebane, L.M.; Philippar, U.; Barzik, M.; Rubinson, D.A.; Gupton, S.; Van Veen, J.E.; Furman, C.; Zhang, J.; et al. Filopodia are required for cortical neurite initiation. Nat. Cell Biol. 2007, 9, 1347–1359. [Google Scholar] [CrossRef]
- Chesarone, M.A.; Goode, B.L. Actin nucleation and elongation factors: Mechanisms and interplay. Curr. Opin. Cell Biol. 2009, 21, 28–37. [Google Scholar] [CrossRef]
- Campellone, K.G.; Welch, M.D. A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010, 11, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Strasser, G.A.; Rahim, N.A.; VanderWaal, K.E.; Gertler, F.B.; Lanier, L.M. Arp2/3 is a negative regulator of growth cone translocation. Neuron 2004, 43, 81–94. [Google Scholar] [CrossRef]
- Dharmalingam, E.; Haeckel, A.; Pinyol, R.; Schwintzer, L.; Koch, D.; Kessels, M.M.; Qualmann, B. F-BAR proteins of the syndapin family shape the plasma membrane and are crucial for neuromorphogenesis. J. Neurosci. 2009, 29, 13715–13727. [Google Scholar] [CrossRef]
- Pinyol, R.; Haeckel, A.; Ritter, A.; Qualmann, B.; Kessels, M.M. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS ONE 2007, 2, e400. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, R.; Pinyol, R.; Reichenbach, N.; Custer, L.; Klingensmith, J.; Kessels, M.M.; Qualmann, B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell 2007, 131, 337–350. [Google Scholar] [CrossRef]
- Quinlan, M.E.; Heuser, J.E.; Kerkhoff, E.; Mullins, R.D. Drosophila Spire is an actin nucleation factor. Nature 2005, 433, 382–388. [Google Scholar] [CrossRef]
- Chereau, D.; Boczkowska, M.; Skwarek-Maruszewska, A.; Fujiwara, I.; Hayes, D.B.; Rebowski, G.; Lappalainen, P.; Pollard, T.D.; Dominguez, R. Leiomodin is an actin filament nucleator in muscle cells. Science 2008, 320, 239–243. [Google Scholar] [CrossRef]
- Zuchero, J.B.; Coutts, A.S.; Quinlan, M.E.; Thangue, N.B.; Mullins, R.D. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 2009, 11, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Ziv, N.E.; Smith, S.J. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 1996, 17, 91–102. [Google Scholar] [CrossRef]
- Hering, H.; Sheng, M. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J. Neurosci. 2003, 23, 11759–11769. [Google Scholar] [CrossRef]
- Bourne, J.; Harris, K.M. Do thin spines learn to be mushroom spines that remember? Curr. Opin. Neurobiol. 2007, 17, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.M.; Jensen, F.E.; Tsao, B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: Implications for the maturation of synaptic physiology and long-term potentiation. J. Neurosci. 1992, 12, 2685–2705. [Google Scholar] [CrossRef] [PubMed]
- Hotulainen, P.; Hoogenraad, C.C. Actin in dendritic spines: Connecting dynamics to function. J. Cell Biol. 2010, 189, 619–629. [Google Scholar] [CrossRef]
- Bertling, E.; Hotulainen, P. New waves in dendritic spine actin cytoskeleton: From branches and bundles to rings, from actin binding proteins to post-translational modifications. Mol. Cell. Neurosci. 2017, 84, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Cimato, T.R.; Ettinger, M.J.; Zhou, X.; Aletta, J.M. Nerve growth factor-specific regulation of protein methylation during neuronal differentiation of PC12 cells. J. Cell Biol. 1997, 138, 1089–1103. [Google Scholar] [CrossRef]
- Cheng, D.; Yadav, N.; King, R.W.; Swanson, M.S.; Weinstein, E.J.; Bedford, M.T. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 2004, 279, 23892–23899. [Google Scholar] [CrossRef]
- Bonham, K.; Hemmers, S.; Lim, Y.H.; Hill, D.M.; Finn, M.G.; Mowen, K.A. Effects of a novel arginine methyltransferase inhibitor on T-helper cell cytokine production. FEBS J. 2010, 277, 2096–2108. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Qian, K.; Ho, M.C.; Zheng, Y.G. Small molecule inhibitors of protein arginine methyltransferases. Expert Opin. Investig. Drugs 2016, 25, 335–358. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.S.; Alkon, D.L. Protein kinase C stimulates HuD-mediated mRNA stability and protein expression of neurotrophic factors and enhances dendritic maturation of hippocampal neurons in culture. Hippocampus 2012, 22, 2303–2319. [Google Scholar] [CrossRef]
- Aggarwal, P.; Vaites, L.P.; Kim, J.K.; Mellert, H.; Gurung, B.; Nakagawa, H.; Herlyn, M.; Hua, X.; Rustgi, A.K.; McMahon, S.B.; et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 2010, 18, 329–340. [Google Scholar] [CrossRef]
- Maggipinto, M.; Rabiner, C.; Kidd, G.J.; Hawkins, A.J.; Smith, R.; Barbarese, E. Increased expression of the MBP mRNA binding protein HnRNP A2 during oligodendrocyte differentiation. J. Neurosci. Res. 2004, 75, 614–623. [Google Scholar] [CrossRef]
- Hong, S.; Heo, J.; Lee, S.; Heo, S.; Kim, S.S.; Lee, Y.D.; Kwon, M.; Hong, S. Methyltransferase-inhibition interferes with neuronal differentiation of P19 embryonal carcinoma cells. Biochem. Biophys. Res. Commun. 2008, 377, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Nemitz, S.; Schopper, S.; Nielsen, M.L.; Kessels, M.M.; Qualmann, B. Arginine methylation by PRMT2 controls the functions of the actin nucleator Cobl. Dev. Cell 2018, 45, 262–275. [Google Scholar] [CrossRef]
- Amano, G.; Matsuzaki, S.; Mori, Y.; Miyoshi, K.; Han, S.; Shikada, S.; Takamura, H.; Yoshimura, T.; Katayama, T. SCYL1 arginine methylation by PRMT1 is essential for neurite outgrowth via Golgi morphogenesis. Mol. Biol. Cell 2020, 31, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.S.; Alkon, D.L. Inhibition of coactivator-associated arginine methyltransferase 1 modulates dendritic arborization and spine maturation of cultured hippocampal neurons. J. Biol. Chem. 2017, 292, 6402–6413. [Google Scholar] [CrossRef]
- Fujiwara, T.; Mori, Y.; Chu, D.L.; Koyama, Y.; Miyata, S.; Tanaka, H.; Yachi, K.; Kubo, T.; Yoshikawa, H.; Tohyama, M. CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol. Cell. Biol. 2006, 26, 2273–2285. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Higashimoto, K.; Kuhn, P.; Desai, D.; Cheng, X.; Xu, W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc. Natl. Acad. Sci. USA 2007, 104, 12318–12323. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.; Mori, Y.; Tohyama, M. PRMT1 and Btg2 regulates neurite outgrowth of Neuro2a cells. Neurosci. Lett. 2008, 445, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.J.; Gary, J.D.; Yang, M.C.; Clarke, S.; Herschman, H.R. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 1996, 271, 15034–15044. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.; Mori, Y.; Tohyama, M. PRMT3 is essential for dendritic spine maturation in rat hippocampal neurons. Brain Res. 2010, 1352, 11–20. [Google Scholar] [CrossRef]
- Liu, H.; Jiang, J.; Zhao, L. Protein arginine methyltransferase-1 deficiency restrains depression-like behavior of mice by inhibiting inflammation and oxidative stress via Nrf-2. Biochem. Biophys. Res. Commun. 2019, 518, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Sayegh, J.; Daniel, J.; Clarke, S.; Bedford, M.T. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 2005, 280, 32890–32896. [Google Scholar] [CrossRef] [PubMed]
- Taneda, T.; Miyata, S.; Kousaka, A.; Inoue, K.; Koyama, Y.; Mori, Y.; Tohyama, M. Specific regional distribution of protein arginine methyltransferase 8 (PRMT8) in the mouse brain. Brain Res. 2007, 1155, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kousaka, A.; Mori, Y.; Koyama, Y.; Taneda, T.; Miyata, S.; Tohyama, M. The distribution and characterization of endogenous protein arginine N-methyltransferase 8 in mouse CNS. Neuroscience 2009, 163, 1146–1157. [Google Scholar] [CrossRef]
- Kim, J.D.; Park, K.E.; Ishida, J.; Kako, K.; Hamada, J.; Kani, S.; Takeuchi, M.; Namiki, K.; Fukui, H.; Fukuhara, S.; et al. PRMT8 as a phospholipase regulates Purkinje cell dendritic arborization and motor coordination. Sci. Adv. 2015, 1, e1500615. [Google Scholar] [CrossRef]
- Klein, J. Functions and pathophysiological roles of phospholipase D in the brain. J. Neurochem. 2005, 94, 1473–1487. [Google Scholar] [CrossRef]
- Lin, Y.L.; Tsai, Y.J.; Liu, Y.F.; Cheng, Y.C.; Hung, C.M.; Lee, Y.J.; Pan, H.; Li, C. The critical role of protein arginine methyltransferase prmt8 in zebrafish embryonic and neural development is non-redundant with its paralogue prmt1. PLoS ONE 2013, 8, e55221. [Google Scholar] [CrossRef]
- Lee, P.K.; Goh, W.W.; Sng, J.C. Network-based characterization of the synaptic proteome reveals that removal of epigenetic regulator Prmt8 restricts proteins associated with synaptic maturation. J. Neurochem. 2017, 140, 613–628. [Google Scholar] [CrossRef]
- Penney, J.; Seo, J.; Kritskiy, O.; Elmsaouri, S.; Gao, F.; Pao, P.C.; Su, S.C.; Tsai, L.H. Loss of protein arginine methyltransferase 8 alters synapse composition and function, resulting in behavioral defects. J. Neurosci. 2017, 37, 8655–8666. [Google Scholar] [CrossRef] [PubMed]
- Lo, L.H.; Dong, R.; Lyu, Q.; Lai, K.O. The protein arginine methyltransferase PRMT8 and substrate G3BP1 control Rac1-PAK1 signaling and actin cytoskeleton for dendritic spine maturation. Cell Rep. 2020, 31, 107744. [Google Scholar] [CrossRef] [PubMed]
- Hotulainen, P.; Llano, O.; Smirnov, S.; Tanhuanpää, K.; Faix, J.; Rivera, C.; Lappalainen, P. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J. Cell Biol. 2009, 185, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Pyronneau, A.; He, Q.; Hwang, J.Y.; Porch, M.; Contractor, A.; Zukin, R.S. Aberrant Rac1-cofilin signaling mediates defects in dendritic spines, synaptic function, and sensory perception in fragile X syndrome. Sci. Signal. 2017, 10, eaan0852. [Google Scholar] [CrossRef]
- Bikkavilli, R.K.; Malbon, C.C. Arginine methylation of G3BP1 in response to Wnt3a regulates β-catenin mRNA. J. Cell Sci. 2011, 124, 2310–2320. [Google Scholar] [CrossRef]
- Guo, A.; Gu, H.; Zhou, J.; Mulhern, D.; Wang, Y.; Lee, K.A.; Yang, V.; Aguiar, M.; Kornhauser, J.; Jia, X.; et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 2014, 13, 372–387. [Google Scholar] [CrossRef]
- Tsai, W.C.; Gayatri, S.; Reineke, L.C.; Sbardella, G.; Bedford, M.T.; Lloyd, R.E. Arginine demethylation of G3BP1 promotes stress granule assembly. J. Biol. Chem. 2016, 291, 22671–22685. [Google Scholar] [CrossRef]
- Martin, S.; Zekri, L.; Metz, A.; Maurice, T.; Chebli, K.; Vignes, M.; Tazi, J. Deficiency of G3BP1, the stress granules assembly factor, results in abnormal synaptic plasticity and calcium homeostasis in neurons. J. Neurochem. 2013, 125, 175–184. [Google Scholar] [CrossRef]
- Terman, J.R.; Kashina, A. Post-translational modification and regulation of actin. Curr. Opin. Cell Biol. 2013, 25, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Bertling, E.; Englund, J.; Minkeviciene, R.; Koskinen, M.; Segerstråle, M.; Castrén, E.; Taira, T.; Hotulainen, P. Actin tyrosine-53-phosphorylation in neuronal maturation and synaptic plasticity. J. Neurosci. 2016, 36, 5299–5313. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Zhong, Y.; Albrecht, A.; Sang, P.B.; Maples, A.; Liu, Z.; Vinayachandran, V.; Reja, R.; Lee, C.F.; Kumar, A.; et al. Actin R256 mono-methylation is a conserved post-translational modification involved in transcription. Cell Rep. 2020, 32, 108172. [Google Scholar] [CrossRef]
- Rubenstein, P.A.; Wen, K.K. Insights into the effects of disease-causing mutations in human actins. Cytoskeleton (Hoboken) 2014, 71, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.C.; Pannu, H.; Tran-Fadulu, V.; Papke, C.L.; Yu, R.K.; Avidan, N.; Bourgeois, S.; Estrera, A.L.; Safi, H.J.; Sparks, E.; et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat. Genet. 2007, 39, 1488–1493. [Google Scholar] [CrossRef]
- Guo, D.C.; Papke, C.L.; Tran-Fadulu, V.; Regalado, E.S.; Avidan, N.; Johnson, R.J.; Kim, D.H.; Pannu, H.; Willing, M.C.; Sparks, E.; et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease. Am. J. Hum. Genet. 2009, 84, 617–627. [Google Scholar] [CrossRef]
- Schwintzer, L.; Koch, N.; Ahuja, R.; Grimm, J.; Kessels, M.M.; Qualmann, B. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 2011, 30, 3147–3159. [Google Scholar] [CrossRef]
- Haag, N.; Schwintzer, L.; Ahuja, R.; Koch, N.; Grimm, J.; Heuer, H.; Qualmann, B.; Kessels, M.M. The actin nucleator Cobl is crucial for Purkinje cell development and works in close conjunction with the F-actin binding protein Abp1. J. Neurosci. 2012, 32, 17842–17856. [Google Scholar] [CrossRef] [PubMed]
- Schüler, S.; Hauptmann, J.; Perner, B.; Kessels, M.M.; Englert, C.; Qualmann, B. Ciliated sensory hair cell formation and function require the F-BAR protein syndapin I and the WH2 domain-based actin nucleator Cobl. J. Cell Sci. 2013, 126, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Izadi, M.; Nemitz, S.; Haag, N.; Kessels, M.M.; Qualmann, B. The actin nucleator Cobl is controlled by calcium and Calmodulin. PLoS Biol. 2015, 13, e1002233. [Google Scholar] [CrossRef] [PubMed]
- Haag, N.; Schüler, S.; Nietzsche, S.; Hübner, C.A.; Strenzke, N.; Qualmann, B.; Kessels, M.M. The actin nucleator Cobl is critical for centriolar positioning, postnatal planar cell polarity refinement, and function of the cochlea. Cell Rep. 2018, 24, 2418–2431.e6. [Google Scholar] [CrossRef] [PubMed]
- Beer, A.J.; González Delgado, J.; Steiniger, F.; Qualmann, B.; Kessels, M.M. The actin nucleator Cobl organises the terminal web of enterocytes. Sci. Rep. 2020, 10, 11156. [Google Scholar] [CrossRef]
- Hertzog, M.; van Heijenoort, C.; Didry, D.; Gaudier, M.; Coutant, J.; Gigant, B.; Didelot, G.; Préat, T.; Knossow, M.; Guittet, E.; et al. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 2004, 117, 611–623. [Google Scholar] [CrossRef]
- Jaworski, J.; Kapitein, L.C.; Gouveia, S.M.; Dortland, B.R.; Wulf, P.S.; Grigoriev, I.; Camera, P.; Spangler, S.A.; Di Stefano, P.; Demmers, J.; et al. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 2009, 61, 85–100. [Google Scholar] [CrossRef] [PubMed]
- McVicker, D.P.; Awe, A.M.; Richters, K.E.; Wilson, R.L.; Cowdrey, D.A.; Hu, X.; Chapman, E.R.; Dent, E.W. Transport of a kinesin-cargo pair along microtubules into dendritic spines undergoing synaptic plasticity. Nat. Comm. 2016, 7, 12741. [Google Scholar] [CrossRef]
- Zhao, J.; Fok, A.H.K.; Fan, R.; Kwan, P.Y.; Chan, H.L.; Lo, L.H.; Chan, Y.S.; Yung, W.H.; Huang, J.; Lai, C.S.W.; et al. Specific depletion of the motor protein KIF5B leads to deficits in dendritic transport, synaptic plasticity and memory. Elife 2020, 9, e53456. [Google Scholar] [CrossRef]
- Yang, Y.; Bedford, M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 2013, 13, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Walport, L.J.; Hopkinson, R.J.; Chowdhury, R.; Schiller, R.; Ge, W.; Kawamura, A.; Schofield, C.J. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Comm. 2016, 7, 11974. [Google Scholar] [CrossRef] [PubMed]
- Wesche, J.; Kühn, S.; Kessler, B.M.; Salton, M.; Wolf, A. Protein arginine methylation: A prominent modification and its demethylation. Cell. Mol. Life Sci. 2017, 74, 3305–3315. [Google Scholar] [CrossRef]
- Zhang, J.; Jing, L.; Li, M.; He, L.; Guo, Z. Regulation of histone arginine methylation/demethylation by methylase and demethylase. Mol. Med. Rep. 2019, 19, 3963–3971. [Google Scholar] [CrossRef] [PubMed]
- Basso, M.; Pennuto, M. Serine phosphorylation and arginine methylation at the crossroads to neurodegeneration. Exp. Neurol. 2015, 271, 77–83. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qualmann, B.; Kessels, M.M. The Role of Protein Arginine Methylation as Post-Translational Modification on Actin Cytoskeletal Components in Neuronal Structure and Function. Cells 2021, 10, 1079. https://doi.org/10.3390/cells10051079
Qualmann B, Kessels MM. The Role of Protein Arginine Methylation as Post-Translational Modification on Actin Cytoskeletal Components in Neuronal Structure and Function. Cells. 2021; 10(5):1079. https://doi.org/10.3390/cells10051079
Chicago/Turabian StyleQualmann, Britta, and Michael M. Kessels. 2021. "The Role of Protein Arginine Methylation as Post-Translational Modification on Actin Cytoskeletal Components in Neuronal Structure and Function" Cells 10, no. 5: 1079. https://doi.org/10.3390/cells10051079
APA StyleQualmann, B., & Kessels, M. M. (2021). The Role of Protein Arginine Methylation as Post-Translational Modification on Actin Cytoskeletal Components in Neuronal Structure and Function. Cells, 10(5), 1079. https://doi.org/10.3390/cells10051079





