The Contribution of Lysosomes to DNA Replication
Abstract
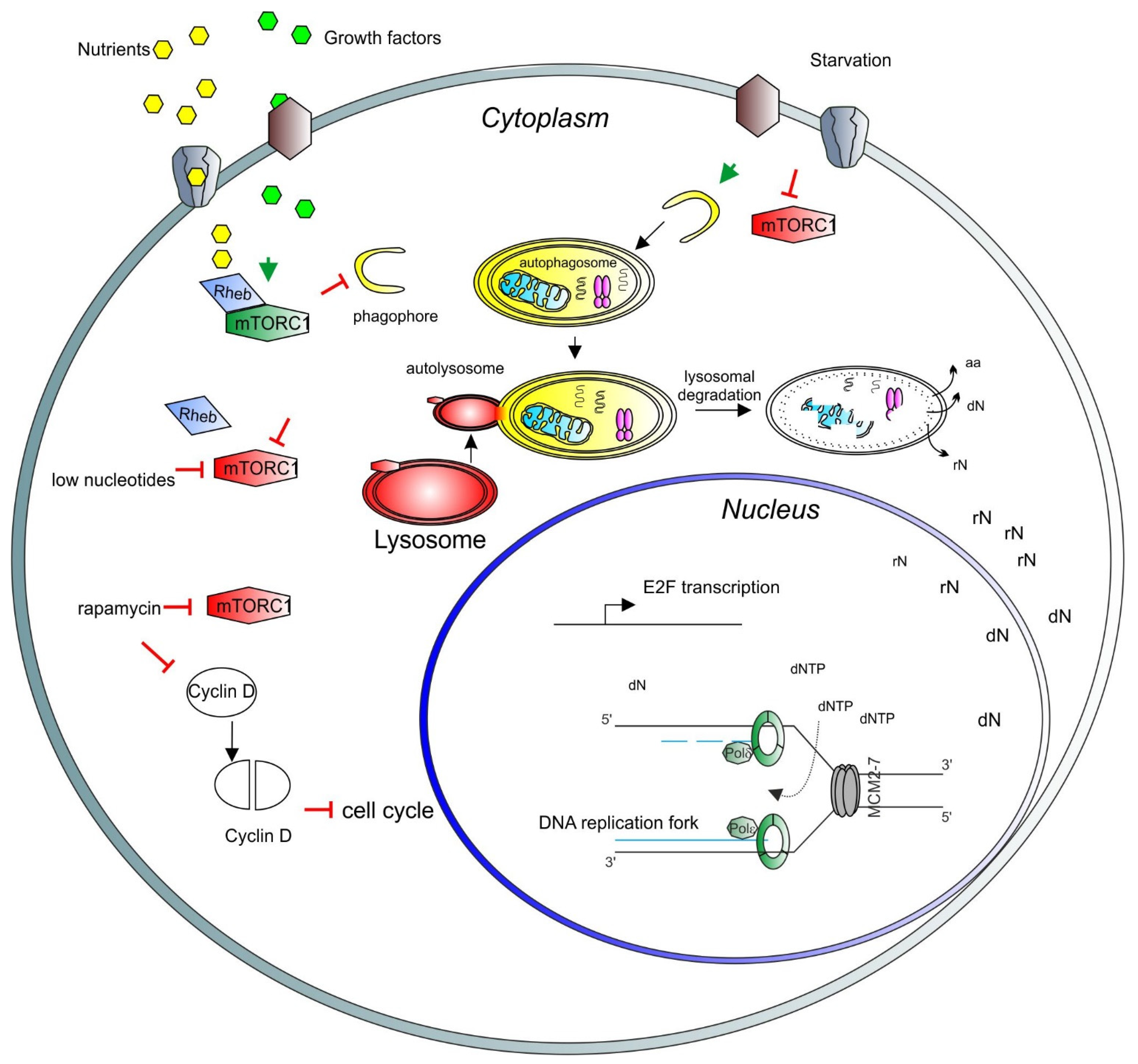
1. Introduction
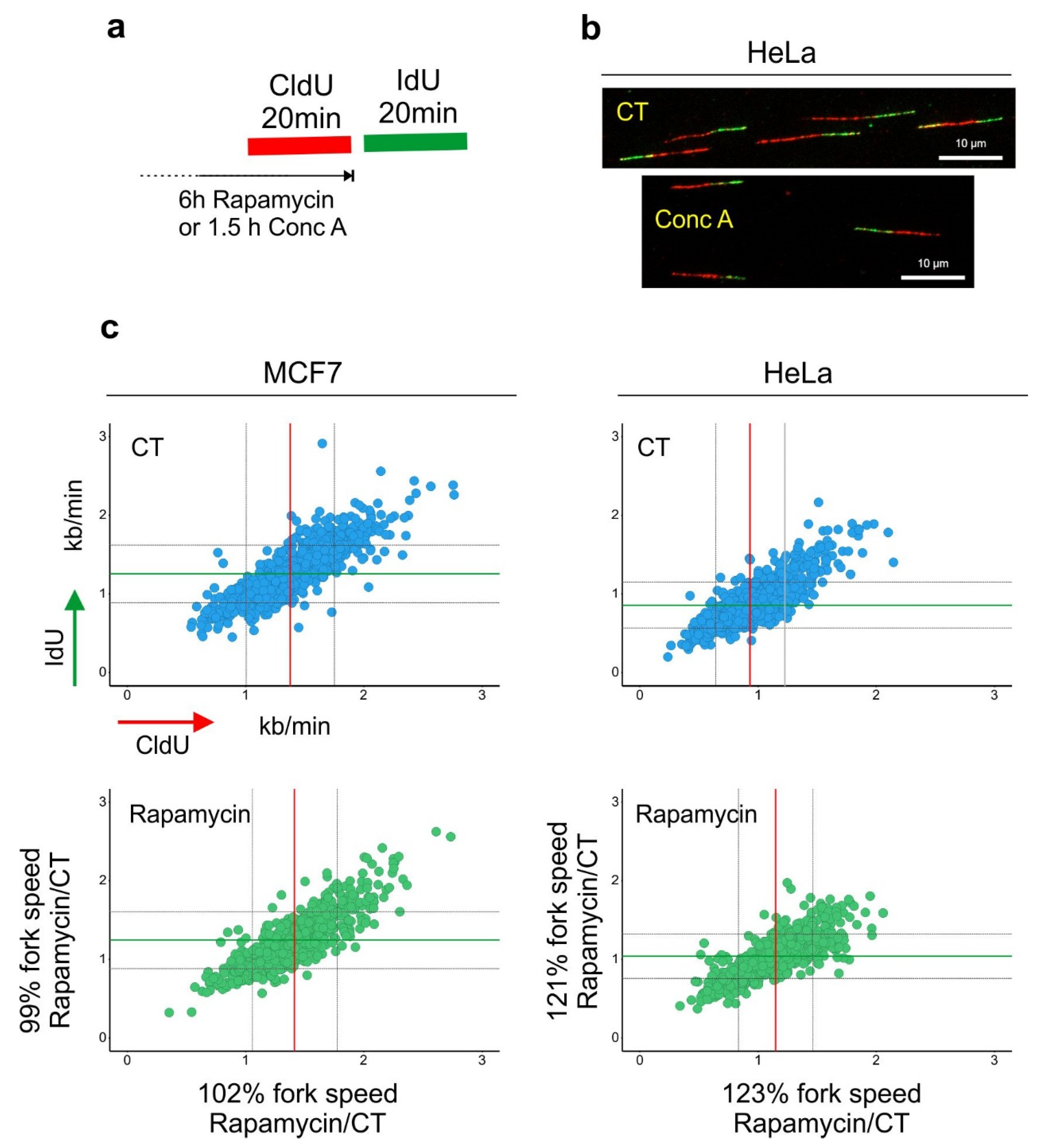
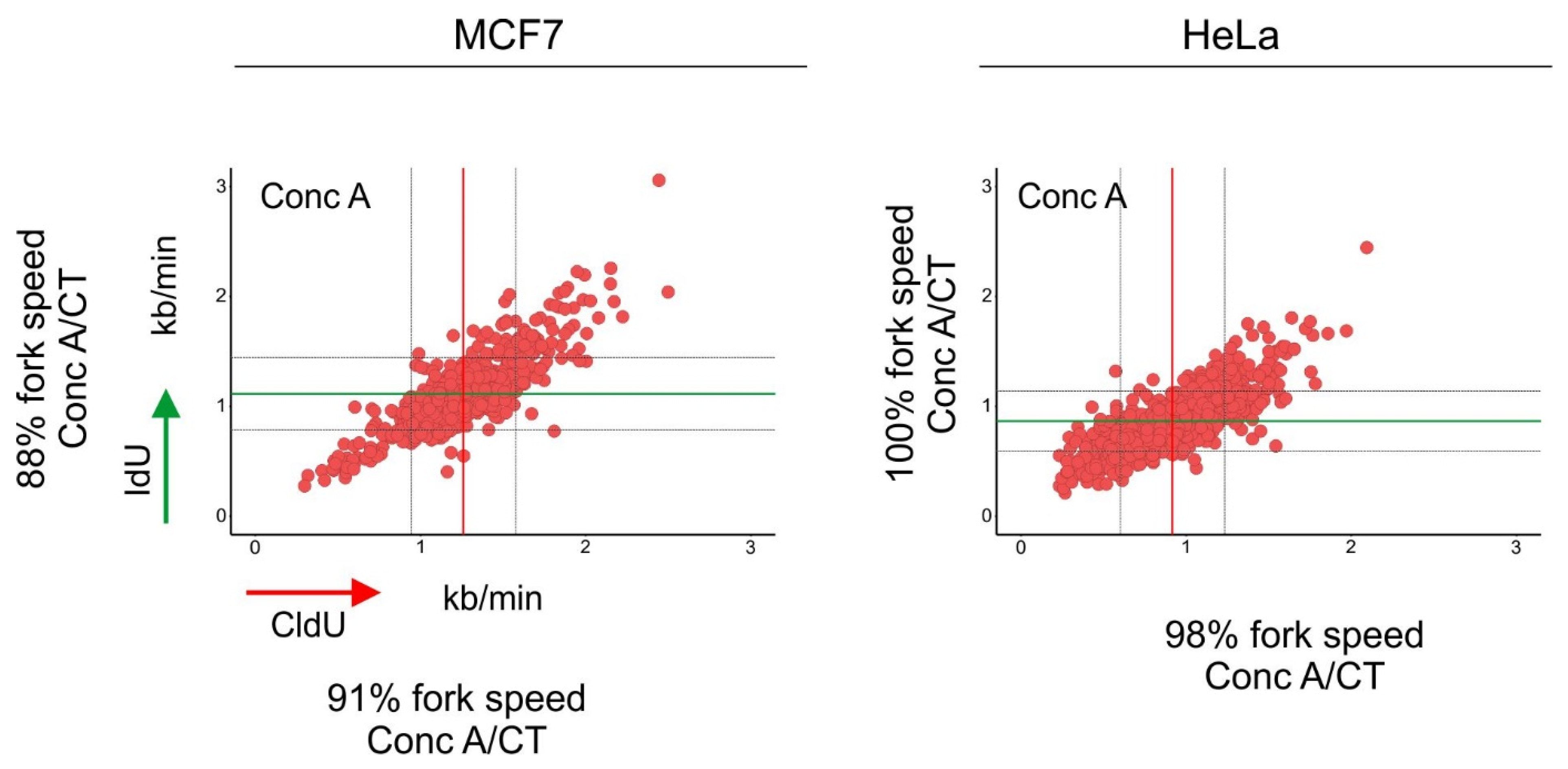
2. mTORC1 Activity and DNA Synthesis
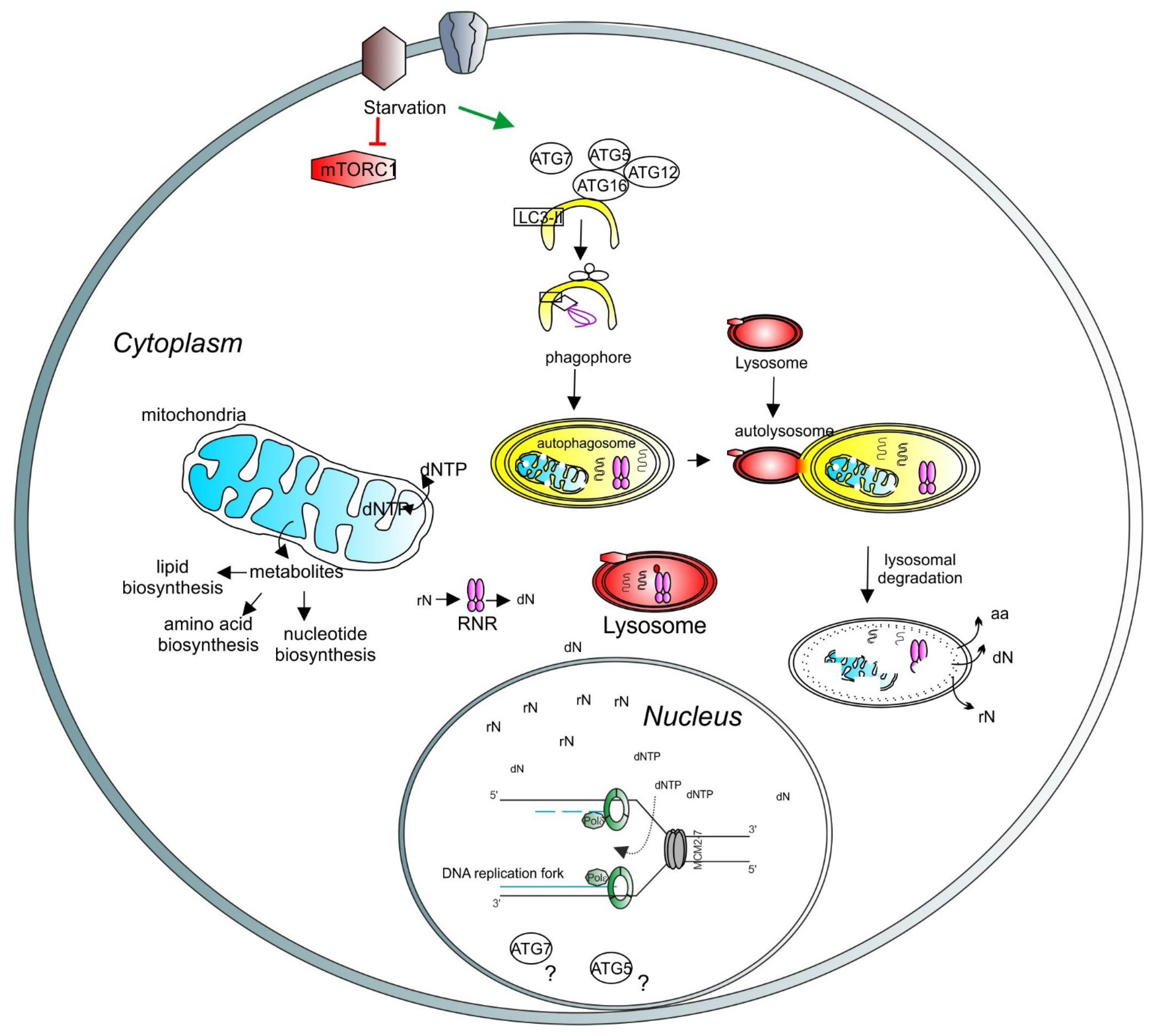
3. Autophagy and DNA Synthesis
4. Recycling of Nucleic Acids
5. Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davidson, S.M.; Vander Heiden, M.G. Critical functions of the lysosome in cancer biology. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 481–507. [Google Scholar] [CrossRef] [PubMed]
- Luzio, J.P.; Pryor, P.R.; Bright, N.A. Lysosomes: Fusion and function. Nat. Rev. Mol. Cell Biol. 2007, 8, 622–632. [Google Scholar] [CrossRef]
- Winchester, B.G. Lysosomal membrane proteins. Eur. J. Paediatr. Neurol. 2001, 5 (Suppl. A), 11–19. [Google Scholar] [CrossRef] [PubMed]
- Woychik, N.A.; Cardelli, J.A.; Dimond, R.L. A conformationally altered precursor to the lysosomal enzyme alpha-mannosidase accumulates in the endoplasmic reticulum in a mutant strain of Dictyostelium discoideum. J. Biol. Chem. 1986, 261, 9595–9602. [Google Scholar] [CrossRef]
- Galluzzi, L.; Pietrocola, F.; Bravo-San Pedro, J.M.; Amaravadi, R.K.; Baehrecke, E.H.; Cecconi, F.; Codogno, P.; Debnath, J.; Gewirtz, D.A.; Karantza, V.; et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015, 34, 856–880. [Google Scholar] [CrossRef]
- Diao, J.; Liu, R.; Rong, Y.; Zhao, M.; Zhang, J.; Lai, Y.; Zhou, Q.; Wilz, L.M.; Li, J.; Vivona, S.; et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015, 520, 563–566. [Google Scholar] [CrossRef]
- Audano, M.; Schneider, A.; Mitro, N. Mitochondria, lysosomes, and dysfunction: Their meaning in neurodegeneration. J. Neurochem. 2018, 147, 291–309. [Google Scholar] [CrossRef]
- Rothman, J.H.; Yamashiro, C.T.; Raymond, C.K.; Kane, P.M.; Stevens, T.H. Acidification of the lysosome-like vacuole and the vacuolar H+-ATPase are deficient in two yeast mutants that fail to sort vacuolar proteins. J. Cell Biol. 1989, 109, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Bastos, A.L.; Moura Nunes, J.F.; Vigario, J.D.; Nunes Petisca, J.L.; Terrinha, A.M. The effect of lysosome permeability changes on DNA synthesis and mitosis of cells growing in vitro. Z. Krebsforsch. 1967, 69, 1–9. [Google Scholar] [CrossRef]
- Comings, O.E.; Kovacs, B.W.; Avelino, E.; Harris, D.C. Mechanisms of chromosome banding. V. Quinacrine banding. Chromosoma 1975, 50, 111–114. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tsun, Z.Y.; Wolfson, R.L.; Shen, K.; Wyant, G.A.; Plovanich, M.E.; Yuan, E.D.; Jones, T.D.; Chantranupong, L.; Comb, W.; et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015, 347, 188–194. [Google Scholar] [CrossRef]
- Kim, E.; Goraksha-Hicks, P.; Li, L.; Neufeld, T.P.; Guan, K.L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008, 10, 935–945. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef]
- Kalender, A.; Selvaraj, A.; Kim, S.Y.; Gulati, P.; Brule, S.; Viollet, B.; Kemp, B.E.; Bardeesy, N.; Dennis, P.; Schlager, J.J.; et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010, 11, 390–401. [Google Scholar] [CrossRef]
- Lin, S.C.; Hardie, D.G. AMPK: Sensing glucose as well as cellular energy status. Cell Metab. 2018, 27, 299–313. [Google Scholar] [CrossRef]
- Chavez, J.A.; Roach, W.G.; Keller, S.R.; Lane, W.S.; Lienhard, G.E. Inhibition of GLUT4 translocation by Tbc1d1, a Rab GTPase-activating protein abundant in skeletal muscle, is partially relieved by AMP-activated protein kinase activation. J. Biol. Chem. 2008, 283, 9187–9195. [Google Scholar] [CrossRef]
- Ben-Sahra, I.; Hoxhaj, G.; Ricoult, S.J.H.; Asara, J.M.; Manning, B.D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 2016, 351, 728–733. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Hughes-Hallett, J.; Timson, R.C.; Ilagan, E.; Yuan, M.; Asara, J.M.; Ben-Sahra, I.; Manning, B.D. The mTORC1 Signaling Network Senses Changes in Cellular Purine Nucleotide Levels. Cell Rep. 2017, 21, 1331–1346. [Google Scholar] [CrossRef]
- Emmanuel, N.; Ragunathan, S.; Shan, Q.; Wang, F.; Giannakou, A.; Huser, N.; Jin, G.; Myers, J.; Abraham, R.T.; Unsal-Kacmaz, K. Purine Nucleotide Availability Regulates mTORC1 Activity through the Rheb GTPase. Cell Rep. 2017, 19, 2665–2680. [Google Scholar] [CrossRef]
- French, J.B.; Jones, S.A.; Deng, H.; Pedley, A.M.; Kim, D.; Chan, C.Y.; Hu, H.; Pugh, R.J.; Zhao, H.; Zhang, Y.; et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science 2016, 351, 733–737. [Google Scholar] [CrossRef]
- Ben-Sahra, I.; Howell, J.J.; Asara, J.M.; Manning, B.D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 2013, 339, 1323–1328. [Google Scholar] [CrossRef]
- Martel, R.R.; Klicius, J.; Galet, S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can. J. Physiol. Pharmacol. 1977, 55, 48–51. [Google Scholar] [CrossRef]
- Kahan, B.D.; Chang, J.Y.; Sehgal, S.N. Preclinical evaluation of a new potent immunosuppressive agent, rapamycin. Transplantation 1991, 52, 185–191. [Google Scholar] [CrossRef]
- Ilagan, E.; Manning, B.D. Emerging role of mTOR in the response to cancer therapeutics. Trends Cancer 2016, 2, 241–251. [Google Scholar] [CrossRef]
- Pollizzi, K.N.; Powell, J.D. Regulation of T cells by mTOR: The known knowns and the known unknowns. Trends Immunol. 2015, 36, 13–20. [Google Scholar] [CrossRef]
- Vanzo, R.; Bartkova, J.; Merchut-Maya, J.M.; Hall, A.; Bouchal, J.; Dyrskjot, L.; Frankel, L.B.; Gorgoulis, V.; Maya-Mendoza, A.; Jaattela, M.; et al. Autophagy role(s) in response to oncogenes and DNA replication stress. Cell Death Differ. 2020, 27, 1134–1153. [Google Scholar] [CrossRef]
- Matsushime, H.; Quelle, D.E.; Shurtleff, S.A.; Shibuya, M.; Sherr, C.J.; Kato, J.Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 1994, 14, 2066–2076. [Google Scholar] [CrossRef] [PubMed]
- Peters, G. The D-type cyclins and their role in tumorigenesis. J. Cell Sci. Suppl. 1994, 18, 89–96. [Google Scholar] [CrossRef]
- Weinberg, R.A. E2F and cell proliferation: A world turned upside down. Cell 1996, 85, 457–459. [Google Scholar] [CrossRef]
- Hashemolhosseini, S.; Nagamine, Y.; Morley, S.J.; Desrivieres, S.; Mercep, L.; Ferrari, S. Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J. Biol. Chem. 1998, 273, 14424–14429. [Google Scholar] [CrossRef]
- Diffley, J.F. Regulation of early events in chromosome replication. Curr. Biol. 2004, 14, R778–R786. [Google Scholar] [CrossRef]
- Siddiqui, K.; On, K.F.; Diffley, J.F. Regulating DNA replication in eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef]
- Akerman, I.; Kasaai, B.; Bazarova, A.; Sang, P.B.; Peiffer, I.; Artufel, M.; Derelle, R.; Smith, G.; Rodriguez-Martinez, M.; Romano, M.; et al. A predictable conserved DNA base composition signature defines human core DNA replication origins. Nat. Commun. 2020, 11, 4826. [Google Scholar] [CrossRef]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Maya-Mendoza, A.; Moudry, P.; Merchut-Maya, J.M.; Lee, M.; Strauss, R.; Bartek, J. High speed of fork progression induces DNA replication stress and genomic instability. Nature 2018, 559, 279–284. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 2012, 22, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Stricher, F.; Macri, C.; Ruff, M.; Muller, S. HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 2013, 9, 1937–1954. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Lee, J.S.; Kim, M.S.; Lee, Y.H.; Jung, J.H.; Hagiwara, A.; Zhou, B.; Lee, J.S.; Jeong, C.B. Genome-wide identification of 99 autophagy-related (Atg) genes in the monogonont rotifer Brachionus spp. and transcriptional modulation in response to cadmium. Aquat. Toxicol. 2018, 201, 73–82. [Google Scholar] [CrossRef]
- Karanasios, E.; Stapleton, E.; Manifava, M.; Kaizuka, T.; Mizushima, N.; Walker, S.A.; Ktistakis, N.T. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J. Cell Sci. 2013, 126, 5224–5238. [Google Scholar] [CrossRef]
- Dooley, H.C.; Razi, M.; Polson, H.E.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell 2014, 55, 238–252. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Kuma, A.; Murakami, M.; Kishi, C.; Yamamoto, A.; Mizushima, N. Autophagy is essential for preimplantation development of mouse embryos. Science 2008, 321, 117–120. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Ueno, T.; Iwata, J.; Murata, S.; Tanida, I.; Ezaki, J.; Mizushima, N.; Ohsumi, Y.; Uchiyama, Y.; et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005, 169, 425–434. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Poole, L.P.; Macleod, K.F. Mitophagy in tumorigenesis and metastasis. Cell. Mol. Life Sci. 2021. [Google Scholar] [CrossRef]
- Liu, L.; Tian, Z.; Zhang, Y.; Liu, P.; Xin, Z.; Zhao, Y.; Li, Y.; Miao, S.; Shi, J.; Chen, Z.; et al. Ligand-based discovery of small molecules suppressing cancer cell proliferation via autophagic flux inhibition. J. Mol. Med. 2020, 98, 1573–1589. [Google Scholar] [CrossRef] [PubMed]
- Fitzwalter, B.E.; Thorburn, A. Recent insights into cell death and autophagy. FEBS J. 2015, 282, 4279–4288. [Google Scholar] [CrossRef] [PubMed]
- Karantza-Wadsworth, V.; Patel, S.; Kravchuk, O.; Chen, G.; Mathew, R.; Jin, S.; White, E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007, 21, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Young, A.R.; Narita, M.; Ferreira, M.; Kirschner, K.; Sadaie, M.; Darot, J.F.; Tavare, S.; Arakawa, S.; Shimizu, S.; Watt, F.M.; et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009, 23, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Galanos, P.; Vougas, K.; Walter, D.; Polyzos, A.; Maya-Mendoza, A.; Haagensen, E.J.; Kokkalis, A.; Roumelioti, F.M.; Gagos, S.; Tzetis, M.; et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat. Cell Biol. 2016, 18, 777–789. [Google Scholar] [CrossRef]
- Filippi-Chiela, E.C.; Bueno e Silva, M.M.; Thome, M.P.; Lenz, G. Single-cell analysis challenges the connection between autophagy and senescence induced by DNA damage. Autophagy 2015, 11, 1099–1113. [Google Scholar] [CrossRef]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Gillespie, D.A.; Ryan, K.M. Autophagy is critically required for DNA repair by homologous recombination. Mol. Cell. Oncol. 2016, 3, e1030538. [Google Scholar] [CrossRef]
- Dyavaiah, M.; Rooney, J.P.; Chittur, S.V.; Lin, Q.; Begley, T.J. Autophagy-dependent regulation of the DNA damage response protein ribonucleotide reductase 1. Mol. Cancer Res. 2011, 9, 462–475. [Google Scholar] [CrossRef]
- Bester, A.C.; Roniger, M.; Oren, Y.S.; Im, M.M.; Sarni, D.; Chaoat, M.; Bensimon, A.; Zamir, G.; Shewach, D.S.; Kerem, B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 2011, 145, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative stress and autophagy: The clash between damage and metabolic needs. Cell Death. Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Gui, D.Y.; Hosios, A.M.; Bush, L.N.; Freinkman, E.; Vander Heiden, M.G. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 2015, 162, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.; Moshitch-Moshkovitz, S.; Kvam, E.; O’Toole, E.; Winey, M.; Goldfarb, D.S. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 129–141. [Google Scholar] [CrossRef]
- Dou, Z.; Xu, C.; Donahue, G.; Shimi, T.; Pan, J.A.; Zhu, J.; Ivanov, A.; Capell, B.C.; Drake, A.M.; Shah, P.P.; et al. Autophagy mediates degradation of nuclear lamina. Nature 2015, 527, 105–109. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Huna, A.; Salmina, K.; Jackson, T.R.; Cragg, M.S. Macroautophagy-aided elimination of chromatin: Sorting of waste, sorting of fate? Autophagy 2012, 8, 1877–1881. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, T.; Olziersky, A.M.; Harry, D.; De Sousa, F.; Vassal, H.; Eskat, A.; Meraldi, P. Mild replication stress causes chromosome mis-segregation via premature centriole disengagement. Nat. Commun. 2019, 10, 3585. [Google Scholar] [CrossRef]
- Krupina, K.; Goginashvili, A.; Cleveland, D.W. Causes and consequences of micronuclei. Curr. Opin. Cell Biol. 2021, 70, 91–99. [Google Scholar] [CrossRef]
- Rello-Varona, S.; Lissa, D.; Shen, S.; Niso-Santano, M.; Senovilla, L.; Marino, G.; Vitale, I.; Jemaa, M.; Harper, F.; Pierron, G.; et al. Autophagic removal of micronuclei. Cell Cycle 2012, 11, 170–176. [Google Scholar] [CrossRef]
- Iadevaia, V.; Liu, R.; Proud, C.G. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin. Cell Dev. Biol. 2014, 36, 113–120. [Google Scholar] [CrossRef]
- Deshpande, R.A.; Shankar, V. Ribonucleases from T2 family. Crit. Rev. Microbiol. 2002, 28, 79–122. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.J.; Aguilera, R.J. DNase II: Genes, enzymes and function. Gene 2003, 322, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hase, K.; Fujiwara, Y.; Kikuchi, H.; Aizawa, S.; Hakuno, F.; Takahashi, S.; Wada, K.; Kabuta, T. RNautophagy/DNautophagy possesses selectivity for RNA/DNA substrates. Nucleic Acids Res. 2015, 43, 6439–6449. [Google Scholar] [CrossRef]
- Huang, H.; Kawamata, T.; Horie, T.; Tsugawa, H.; Nakayama, Y.; Ohsumi, Y.; Fukusaki, E. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 2015, 34, 154–168. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Kikuchi, H.; Aizawa, S.; Furuta, A.; Hatanaka, Y.; Konya, C.; Uchida, K.; Wada, K.; Kabuta, T. Direct uptake and degradation of DNA by lysosomes. Autophagy 2013, 9, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.Y.; Londono, D.; Bouley, R.; Rooney, M.S.; Hacohen, N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 2014, 9, 180–192. [Google Scholar] [CrossRef]
- Coquel, F.; Neumayer, C.; Lin, Y.L.; Pasero, P. SAMHD1 and the innate immune response to cytosolic DNA during DNA replication. Curr. Opin. Immunol. 2019, 56, 24–30. [Google Scholar] [CrossRef]
- Zhao, Z.; Qin, P.; Huang, Y.W. Lysosomal ion channels involved in cellular entry and uncoating of enveloped viruses: Implications for therapeutic strategies against SARS-CoV-2. Cell Calcium 2021, 94, 102360. [Google Scholar] [CrossRef] [PubMed]
- Miao, G.; Zhao, H.; Li, Y.; Ji, M.; Chen, Y.; Shi, Y.; Bi, Y.; Wang, P.; Zhang, H. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell 2021, 56, 427–442.e425. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ko, B.; Hensley, C.T.; Jiang, L.; Wasti, A.T.; Kim, J.; Sudderth, J.; Calvaruso, M.A.; Lumata, L.; Mitsche, M.; et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 2014, 56, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Watts, C. The endosome-lysosome pathway and information generation in the immune system. Biochim. Biophys. Acta 2012, 1824, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug. Discov. 2019, 18, 923–948. [Google Scholar] [CrossRef]
- Ferlinz, K.; Hurwitz, R.; Weiler, M.; Suzuki, K.; Sandhoff, K.; Vanier, M.T. Molecular analysis of the acid sphingomyelinase deficiency in a family with an intermediate form of Niemann-Pick disease. Am. J. Hum. Genet. 1995, 56, 1343–1349. [Google Scholar]
- Torres, S.; Balboa, E.; Zanlungo, S.; Enrich, C.; Garcia-Ruiz, C.; Fernandez-Checa, J.C. Lysosomal and Mitochondrial Liaisons in Niemann-Pick Disease. Front. Physiol. 2017, 8, 982. [Google Scholar] [CrossRef]
- Kirkegaard, T.; Roth, A.G.; Petersen, N.H.; Mahalka, A.K.; Olsen, O.D.; Moilanen, I.; Zylicz, A.; Knudsen, J.; Sandhoff, K.; Arenz, C.; et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature 2010, 463, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Mikami, T.; Takahashi, T.; Ishida, A.; Minamiya, Y.; Ida, H.; Takada, G. Signaling pathway for radiation-induced apoptosis in the lymphoblasts from neuronopathic (type A) and non-neuronopathic (type B) forms of Niemann-Pick disease. J. Neurol. Sci. 2002, 199, 39–43. [Google Scholar] [CrossRef]
- Merchut-Maya, J.M.; Bartek, J.; Maya-Mendoza, A. Regulation of replication fork speed: Mechanisms and impact on genomic stability. DNA Repair 2019, 81, 102654. [Google Scholar] [CrossRef]
- Kapuscinski, J.; Darzynkiewicz, Z. Condensation of nucleic acids by intercalating aromatic cations. Proc. Natl. Acad. Sci. 1984, 81, 7368–7372. [Google Scholar] [CrossRef]
- Kusuzaki, K.; Murata, H.; Takeshita, H.; Hashiguchi, S.; Nozaki, T.; Emoto, K.; Ashihara, T.; Hirasawa, Y. Intracellular binding sites of acridine orange in living osteosarcoma cells. Anticancer Res. 2000, 20, 971–975. [Google Scholar]
- Ellegaard, A.M.; Dehlendorff, C.; Vind, A.C.; Anand, A.; Cederkvist, L.; Petersen, N.H.T.; Nylandsted, J.; Stenvang, J.; Mellemgaard, A.; Osterlind, K.; et al. Repurposing cationic amphiphilic antihistamines for cancer treatment. EBioMedicine 2016, 9, 130–139. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merchut-Maya, J.M.; Maya-Mendoza, A. The Contribution of Lysosomes to DNA Replication. Cells 2021, 10, 1068. https://doi.org/10.3390/cells10051068
Merchut-Maya JM, Maya-Mendoza A. The Contribution of Lysosomes to DNA Replication. Cells. 2021; 10(5):1068. https://doi.org/10.3390/cells10051068
Chicago/Turabian StyleMerchut-Maya, Joanna Maria, and Apolinar Maya-Mendoza. 2021. "The Contribution of Lysosomes to DNA Replication" Cells 10, no. 5: 1068. https://doi.org/10.3390/cells10051068
APA StyleMerchut-Maya, J. M., & Maya-Mendoza, A. (2021). The Contribution of Lysosomes to DNA Replication. Cells, 10(5), 1068. https://doi.org/10.3390/cells10051068




