Adipocyte, Immune Cells, and miRNA Crosstalk: A Novel Regulator of Metabolic Dysfunction and Obesity
Abstract
1. Introduction
2. MicroRNA—An Exciting Discovery
3. Role of miRs in AT Inflammation
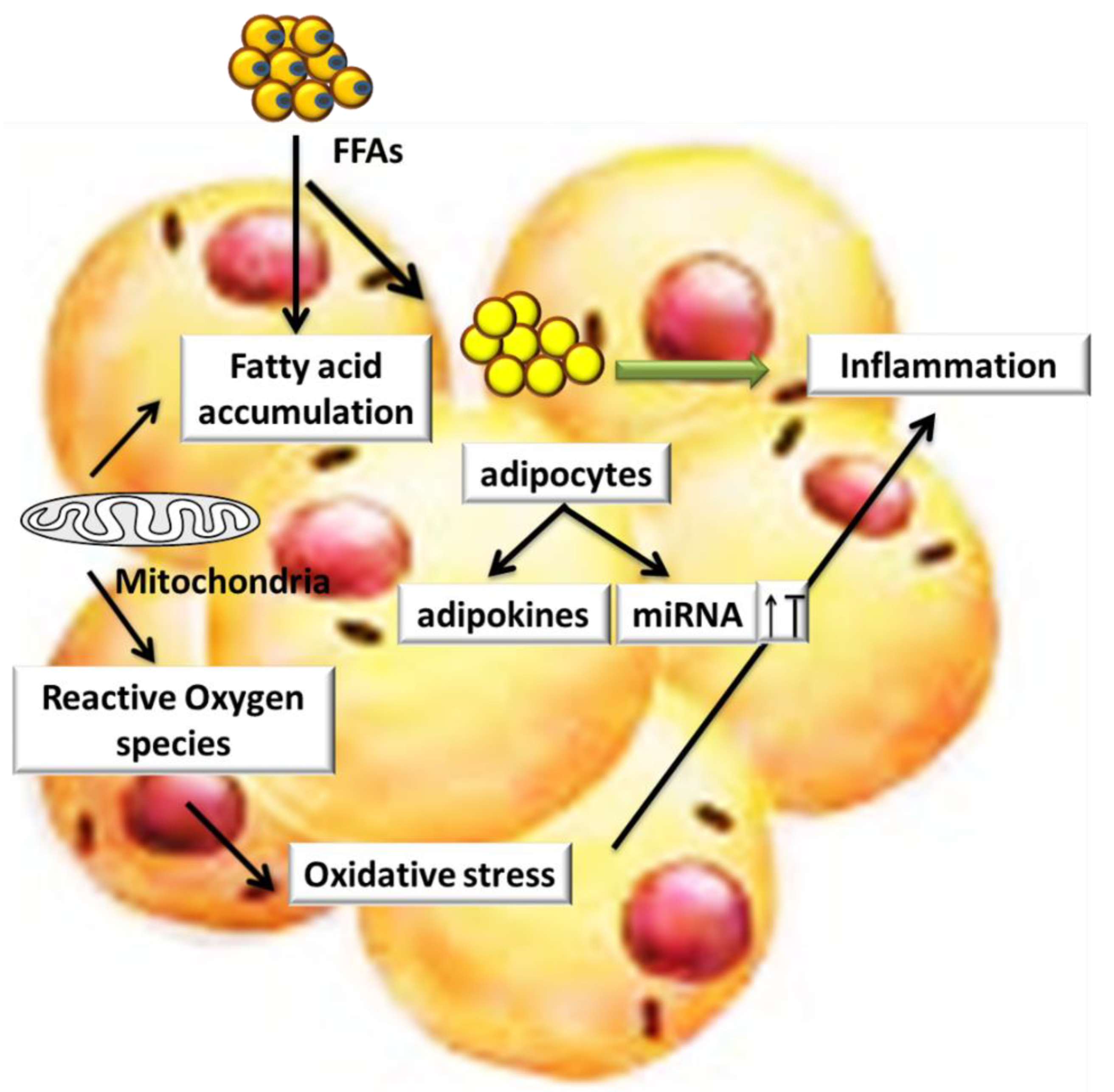
4. Role of miRs and Oxidative Stress in Obesity and Its Associated Diseases
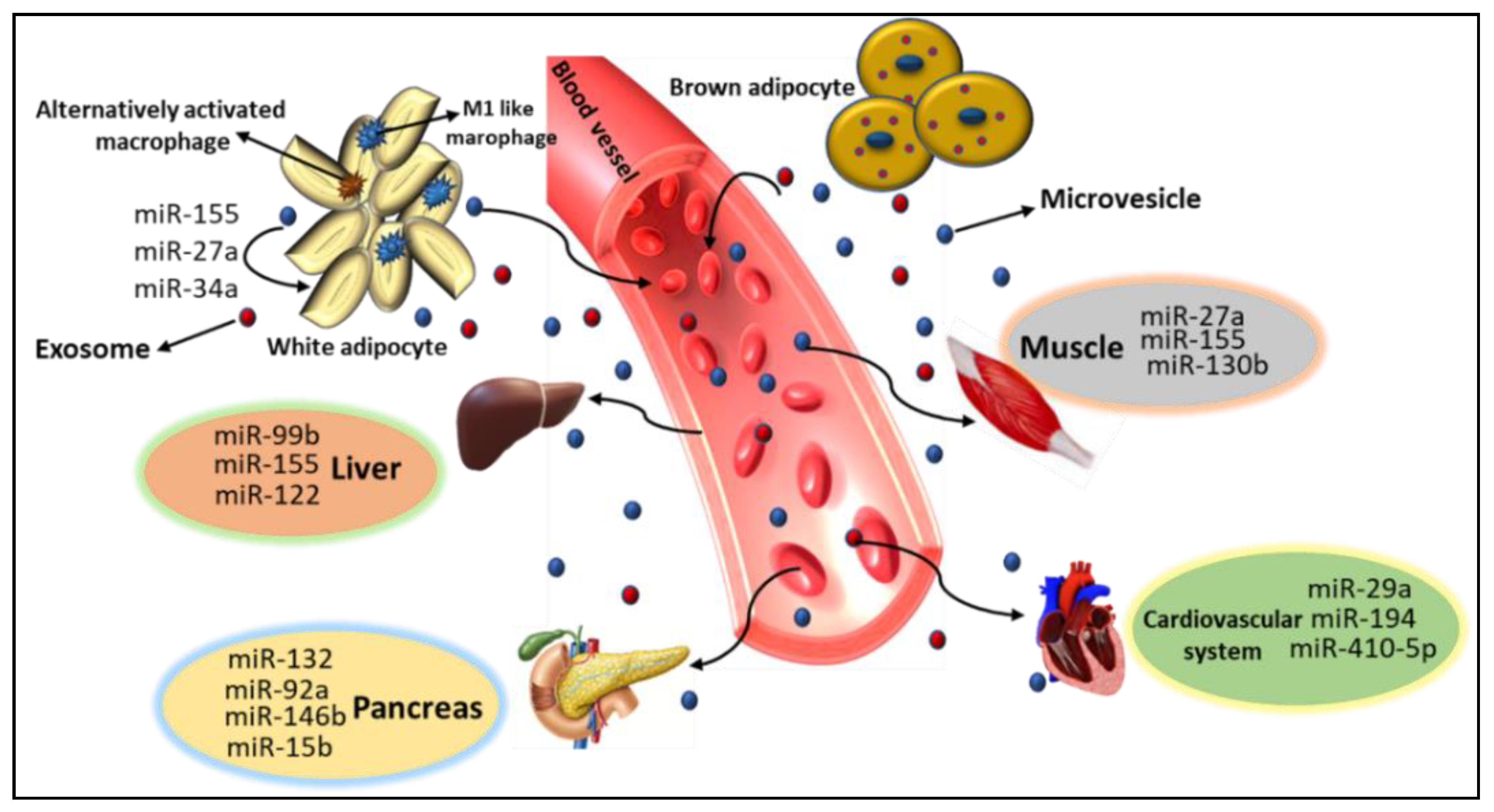
5. Role of miRs in Metabolic Organ Cross-Talk
6. AT Derived miRs
7. miRs as A Bridge between Adipocytes and AT Macrophages
8. AT and Skeletal Muscle miRs
9. miRs in AT–Pancreas Crosstalk
10. AT-Cardiovascular System miRs
11. Endocrine Function in the Liver by miRs
12. Extracellular miRs as Disease Biomarkers
13. miRs-Based Therapeutics
14. Future Prospective and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Polyzos, S.A.; Kountouras, J.; Mantzoros, C.S. Obesity and nonalcoholic fatty liver disease: From pathophysiology to therapeutics. Metabolism 2019, 92, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Cai, J.; Gonzalez, F.J. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2021, 1–13. [Google Scholar] [CrossRef]
- Rahman, I.; Athar, T.; Islam, M. Type 2 Diabetes, Obesity, and Cancer Share Some Common and Critical Pathways. Front. Oncol. 2021, 10, 600824. [Google Scholar] [CrossRef] [PubMed]
- Passos, G.R.; Ghezzi, A.C.; Antunes, E.; de Oliveira, M.G.; Mónica, F.Z. The Role of Periprostatic Adipose Tissue on Prostate Function in Vascular-Related Disorders. Front. Pharmacol. 2021, 12, 626155. [Google Scholar] [CrossRef]
- Jung, U.J.; Choi, M.-S. Obesity and Its Metabolic Complications: The Role of Adipokines and the Relationship between Obesity, Inflammation, Insulin Resistance, Dyslipidemia and Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2014, 15, 6184–6223. [Google Scholar] [CrossRef]
- Kawai, T.; Autieri, M.V.; Scalia, R. Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol. Physiol. 2021, 320, C375–C391. [Google Scholar] [CrossRef]
- Del Cornò, M.; Conti, L.; Gessani, S. Innate Lymphocytes in Adipose Tissue Homeostasis and Their Alterations in Obesity and Colorectal Cancer. Front. Immunol. 2018, 9, 2556. [Google Scholar] [CrossRef]
- Bradley, C.A. Specialized macrophages contribute to obesity. Nat. Rev. Endocrinol. 2017, 13, 690. [Google Scholar] [CrossRef]
- Blücher, C.; Stadler, S.C. Obesity and Breast Cancer: Current Insights on the Role of Fatty Acids and Lipid Metabolism in Promoting Breast Cancer Growth and Progression. Front. Endocrinol. 2017, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Fall, C.H.; Sachdev, H.S.; Osmond, C.; Lakshmy, R.; Biswas, S.D.; Prabhakaran, D.; Tandon, N.; Ramji, S.; Reddy, K.S.; Barker, D.J.; et al. Adult Metabolic Syndrome and Impaired Glucose Tolerance Are Associated With Different Patterns of BMI Gain During Infancy: Data from the New Delhi birth cohort. Diabetes Care 2008, 31, 2349–2356. [Google Scholar] [CrossRef] [PubMed]
- Moro, K.; Yamada, T.; Tanabe, M.; Takeuchi, T.; Ikawa, T.; Kawamoto, H.; Furusawa, J.i.; Ohtani, M.; Fujii, H.; Koyasu, S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature 2010, 463, 540–544. [Google Scholar] [CrossRef]
- Brestoff, J.R.; Kim, B.S.; Saenz, S.A.; Stine, R.R.; Monticelli, L.A.; Sonnenberg, G.F.; Thome, J.J.; Farber, D.L.; Lutfy, K.; Seale, P.; et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nat. Cell Biol. 2015, 519, 242–246. [Google Scholar] [CrossRef]
- Molofsky, A.B.; Nussbaum, J.C.; Liang, H.-E.; Van Dyken, S.J.; Cheng, L.E.; Mohapatra, A.; Chawla, A.; Locksley, R.M. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 2013, 210, 535–549. [Google Scholar] [CrossRef]
- Miller, A.M.; Asquith, D.L.; Hueber, A.J.; Anderson, L.A.; Holmes, W.M.; McKenzie, A.N.; Xu, D.; Sattar, N.; McInnes, I.B.; Liew, F.Y. Interleukin-33 Induces Protective Effects in Adipose Tissue Inflammation During Obesity in Mice. Circ. Res. 2010, 107, 650–658. [Google Scholar] [CrossRef]
- O’Sullivan, T.E.; Rapp, M.; Fan, X.; Weizman, O.-E.; Bhardwaj, P.; Adams, N.M.; Walzer, T.; Dannenberg, A.J.; Sun, J.C. Adipose-Resident Group 1 Innate Lymphoid Cells Promote Obesity-Associated Insulin Resistance. Immunity 2016, 45, 428–441. [Google Scholar] [CrossRef]
- Covarrubias, A.J.; Horng, T. IL-6 Strikes a Balance in Metabolic Inflammation. Cell Metab. 2014, 19, 898–899. [Google Scholar] [CrossRef]
- Martin, S.S.; Qasim, A.; Reilly, M.P. Leptin Resistance: A Possible Interface of Inflammation and Metabolism in Obesity-Related Cardiovascular Disease. J. Am. Coll. Cardiol. 2008, 52, 1201–1210. [Google Scholar] [CrossRef]
- Cawthorn, W.P.; Sethi, J.K. TNF-α and adipocyte biology. FEBS Lett. 2007, 582, 117–131. [Google Scholar] [CrossRef]
- Jamaluddin, M.S.; Weakley, S.M.; Yao, Q.; Chen, C. Resistin: Functional roles and therapeutic considerations for cardiovascular disease. Br. J. Pharmacol. 2012, 165, 622–632. [Google Scholar] [CrossRef]
- Panee, J. Monocyte Chemoattractant Protein 1 (MCP-1) in obesity and diabetes. Cytokine 2012, 60, 1–12. [Google Scholar] [CrossRef]
- Wu, H.; Ghosh, S.; Perrard, X.D.; Feng, L.; Garcia, G.E.; Perrard, J.L.; Sweeney, J.F.; Peterson, L.E.; Chan, L.; Smith, C.W.; et al. T-Cell Accumulation and Regulated on Activation, Normal T Cell Expressed and Secreted Upregulation in Adipose Tissue in Obesity. Circulation 2007, 115, 1029–1038. [Google Scholar] [CrossRef]
- Fagiolo, U.; Cossarizza, A.; Scala, E.; Fanales-Belasio, E.; Ortolani, C.; Cozzi, E.; Monti, D.; Franceschi, C.; Paganelli, R. Increased cytokine production in mononuclear cells of healthy elderly people. Eur. J. Immunol. 1993, 23, 2375–2378. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Wei, Y.; Zhu, M.; Schober, A. Macrophage MicroRNAs as Therapeutic Targets for Atherosclerosis, Metabolic Syndrome, and Cancer. Int. J. Mol. Sci. 2018, 19, 1756. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.; Yang, X.; Bam, M.; Murphy, E.A.; Nagarkatti, P.S.; Nagarkatti, M. MicroRNA-30 modulates metabolic inflammation by regulating Notch signaling in adipose tissue macrophages. Int. J. Obes. 2018, 42, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, K.; Sivasankar, V. MicroRNAs—Biology and clinical applications. J. Oral Maxillofac. Pathol. JOMFP 2014, 18, 229–234. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nat. Cell Biol. 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. et Biophys. Acta (BBA) Bioenerg. 2010, 1803, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Valdmanis, P.N.; Kim, H.K.; Chu, K.; Zhang, F.; Xu, J.; Munding, E.M.; Shen, J.; Kay, M.A. miR-122 removal in the liver activates imprinted microRNAs and enables more effective microRNA-mediated gene repression. Nat. Commun. 2018, 9, 5321. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Chen, J.-F.; Wang, D.-Z. Transgenic overexpression of miR-133a in skeletal muscle. BMC Musculoskelet. Disord. 2011, 12, 115. [Google Scholar] [CrossRef] [PubMed]
- Iannone, F.; Montesanto, A.; Cione, E.; Crocco, P.; Caroleo, M.C.; Dato, S.; Rose, G.; Passarino, G. Expression Patterns of Muscle-Specific miR-133b and miR-206 Correlate with Nutritional Status and Sarcopenia. Nutrients 2020, 12, 297. [Google Scholar] [CrossRef]
- van Rooij, E.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J., Jr.; Olson, E.N. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef]
- Nachtigall, P.G.; Dias, M.C.; Carvalho, R.F.; Martins, C.; Pinhal, D. MicroRNA-499 Expression Distinctively Correlates to Target Genes sox6 and rod1 Profiles to Resolve the Skeletal Muscle Phenotype in Nile Tilapia. PLoS ONE 2015, 10, e0119804. [Google Scholar] [CrossRef]
- Aoi, W.; Ichikawa, H.; Mune, K.; Tanimura, Y.; Mizushima, K.; Naito, Y.; Yoshikawa, T. Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front. Physiol. 2013, 4, 80. [Google Scholar] [CrossRef]
- Chen, J.-F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The Role of MicroRNA-1 and MicroRNA-133 in Skeletal Muscle Proliferation and Differentiation. Nat. Genet. 2005, 38, 228–233. [Google Scholar] [CrossRef]
- Boštjančič, E.; Zidar, N.; Štajer, D.; Glavač, D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 Are Dysregulated in Human Myocardial Infarction. Cardiology 2010, 115, 163–169. [Google Scholar] [CrossRef]
- Sun, Y.; Luo, Z.-M.; Guo, X.-M.; Su, D.-F.; Liu, X. An updated role of microRNA-124 in central nervous system disorders: A review. Front. Cell. Neurosci. 2015, 9, 193. [Google Scholar] [CrossRef]
- Ong, S.-G.; Lee, W.H.; Kodo, K.; Wu, J.C. MicroRNA-mediated regulation of differentiation and trans-differentiation in stem cells. Adv. Drug Deliv. Rev. 2015, 88, 3–15. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Penfornis, P.; Vallabhaneni, K.C.; Whitt, J.; Pochampally, R. Extracellular vesicles as carriers of microRNA, proteins and lipids in tumor microenvironment. Int. J. Cancer 2016, 138, 14–21. [Google Scholar] [CrossRef]
- Perron, M.P.; Provost, P. Protein Components of the microRNA Pathway and Human Diseases. In Methods in Molecular Biology; Springer Science and Business Media LLC: Clifton, NJ, USA, 2009; Volume 487, pp. 1–17. [Google Scholar]
- Belleannée, C. Extracellular microRNAs from the epididymis as potential mediators of cell-to-cell communication. Asian J. Androl. 2015, 17, 730–736. [Google Scholar] [CrossRef]
- Chartoumpekis, D.V.; Zaravinos, A.; Ziros, P.G.; Iskrenova, R.P.; Psyrogiannis, A.I.; Kyriazopoulou, V.E.; Habeos, I.G. Differential Expression of MicroRNAs in Adipose Tissue after Long-Term High-Fat Diet-Induced Obesity in Mice. PLoS ONE 2012, 7, e34872. [Google Scholar] [CrossRef]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Pardo, G.; Sabater, M.; Hummel, M.; Ferrer, A.; Rodriguez-Hermosa, J.I.; Ruiz, B.; Ricart, W.; Peral, B.; et al. MiRNA Expression Profile of Human Subcutaneous Adipose and during Adipocyte Differentiation. PLoS ONE 2010, 5, e9022. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Miller, N.; McAnena, O.J.; O’Brien, T.; Kerin, M.J. Differential miRNA Expression in Omental Adipose Tissue and in the Circulation of Obese Patients Identifies Novel Metabolic Biomarkers. J. Clin. Endocrinol. Metab. 2011, 96, E846–E850. [Google Scholar] [CrossRef]
- Arner, E.; Mejhert, N.; Kulyté, A.; Balwierz, P.J.; Pachkov, M.; Cormont, M.; Lorente-Cebrián, S.; Ehrlund, A.; Laurencikiene, J.; Hedén, P.; et al. Adipose Tissue MicroRNAs as Regulators of CCL2 Production in Human Obesity. Diabetes 2012, 61, 1986–1993. [Google Scholar] [CrossRef]
- Parra, P.; Serra, F.; Palou, A. Expression of Adipose MicroRNAs Is Sensitive to Dietary Conjugated Linoleic Acid Treatment in Mice. PLoS ONE 2010, 5, e13005. [Google Scholar] [CrossRef]
- Strum, J.C.; Johnson, J.H.; Ward, J.; Xie, H.; Feild, J.; Hester, A.; Alford, A.; Waters, K.M. MicroRNA 132 Regulates Nutritional Stress-Induced Chemokine Production through Repression of SirT. Mol. Endocrinol. 2009, 23, 1876–1884. [Google Scholar] [CrossRef]
- Ge, Q.; Gérard, J.; Noel, L.; Scroyen, I.; Brichard, S.M. MicroRNAs Regulated by Adiponectin as Novel Targets for Controlling Adipose Tissue Inflammation. Endocrinology 2012, 153, 5285–5296. [Google Scholar] [CrossRef]
- Jindra, P.T.; Bagley, J.; Godwin, J.G.; Iacomini, J. Costimulation-Dependent Expression of MicroRNA-214 Increases the Ability of T Cells To Proliferate by Targeting Pten. J. Immunol. 2010, 185, 990–997. [Google Scholar] [CrossRef]
- Neilson, J.R.; Zheng, G.X.; Burge, C.B.; Sharp, P.A. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007, 21, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Vega-Cárdenas, M.; Uresti-Rivera, E.E.; Cortés-García, J.D.; Briones-Espinoza, M.; Ruíz-Rodríguez, V.M.; Reynaga-Hernández, E.; Mendez-Mancilla, A.; Portales-Pérez, D.P. Increased levels of adipose tissue-resident Th17 cells in obesity associated with miR-326. Immunol. Lett. 2019, 211, 60–67. [Google Scholar] [CrossRef]
- Ying, W.; Tseng, A.; Chang, R.C.-A.; Wang, H.; Lin, Y.-L.; Kanameni, S.; Brehm, T.; Morin, A.; Jones, B.; Splawn, T.; et al. miR-150 regulates obesity-associated insulin resistance by controlling B cell functions. Sci. Rep. 2016, 6, 20176. [Google Scholar] [CrossRef] [PubMed]
- Metere, A.; Graves, C.E.; Pietraforte, D.; Casella, G. The Effect of Sleeve Gastrectomy on Oxidative Stress in Obesity. Biomedicines 2020, 8, 168. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Raad, H.; Paciet, M.-H.; Boussetta, T.; Kroviarski, Y.; Morel, F.; Quinn, M.T.; Gougerot-Pocidalo, M.-A.; Dang, P.M.-C.; El-Benna, J. Regulation of the phagocyte NADPH oxidase activity: Phosphorylation of gp91 phox /NOX2 by protein kinase C enhances its diaphorase activity and binding to Rac2, p67 phox, and p47 phox. FASEB J. 2008, 23, 1011–1022. [Google Scholar] [CrossRef]
- Dong, L.G.; Lu, F.-F.; Zu, J.; Zhang, W.; Xu, C.-Y.; Jin, G.-L.; Yang, X.-X.; Xiao, Q.-H.; Cui, C.-C.; Xu, R.; et al. MiR-133b inhibits MPP+-induced apoptosis in Parkinson’s disease model by inhibiting the ERK1/2 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11192–11198. [Google Scholar]
- Choi, S.-E.; Fu, T.; Seok, S.; Kim, D.-H.; Yu, E.; Lee, K.-W.; Kang, Y.; Li, X.; Kemper, B.; Kemper, J.K. Elevated microRNA-34a in obesity reduces NAD+levels and SIRT1 activity by directly targeting NAMPT. Aging Cell 2013, 12, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Menghini, R.; Casagrande, V.; Cardellini, M.; Martelli, E.; Terrinoni, A.; Amati, F.; Vasa-Nicotera, M.; Ippoliti, A.; Novelli, G.; Melino, G.; et al. MicroRNA 217 Modulates Endothelial Cell Senescence via Silent Information Regulator. Circulation 2009, 120, 1524–1532. [Google Scholar] [CrossRef]
- Xia, N.; Strand, S.; Schlufter, F.; Siuda, D.; Reifenberg, G.; Kleinert, H.; Förstermann, U.; Li, H. Role of SIRT1 and FOXO factors in eNOS transcriptional activation by resveratrol. Nitric Oxide 2013, 32, 29–35. [Google Scholar] [CrossRef]
- Sun, H.-X.; Zeng, D.-Y.; Li, R.-T.; Pang, R.-P.; Yang, H.; Hu, Y.-L.; Zhang, Q.; Jiang, Y.; Huang, L.-Y.; Tang, Y.-B.; et al. Essential Role of MicroRNA-155 in Regulating Endothelium-Dependent Vasorelaxation by Targeting Endothelial Nitric Oxide Synthase. Hypertens. 2012, 60, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Y.; Chen, W.; Xie, L.; Zhao, Z.-A.; Yang, J.; Chen, Y.; Lei, W.; Shen, Z. MicroRNA-133 overexpression promotes the therapeutic efficacy of mesenchymal stem cells on acute myocardial infarction. Stem Cell Res. Ther. 2017, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I.; et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 2004, 114, 1752–1761. [Google Scholar] [CrossRef]
- Löffler, D.; Brocke-Heidrich, K.; Pfeifer, G.; Stocsits, C.; Hackermüller, J.; Kretzschmar, A.K.; Burger, R.; Gramatzki, M.; Blumert, C.; Bauer, K.; et al. Interleukin-6–dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007, 110, 1330–1333. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 Activation of miR-21 and miR-181b-1 via PTEN and CYLD Are Part of the Epigenetic Switch Linking Inflammation to Cancer. Mol. Cell 2010, 39, 493–506. [Google Scholar] [CrossRef]
- Oh, Y.S.; Bae, G.D.; Park, E.-Y.; Jun, H.-S. MicroRNA-181c Inhibits Interleukin-6-mediated Beta Cell Apoptosis by Targeting TNF-α Expression. Molecules 2019, 24, 1410. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Hu, J.; Lu, P.; Cao, H.; Yu, C.; Li, X.; Qian, X.; Yang, X.; Yang, Y.; Han, N.; et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Birsoy, K.; Festuccia, W.T.; Laplante, M. A comparative perspective on lipid storage in animals. J. Cell Sci. 2013, 126, 1541–1552. [Google Scholar] [CrossRef]
- Saely, C.H.; Geiger, K.; Drexel, H. Brown versus White Adipose Tissue: A Mini-Review. Gerontology 2012, 58, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tan, C. miRNAs in Adipocyte-Derived Extracellular Vesicles: Multiple Roles in Development of Obesity-Associated Disease. Front. Mol. Biosci. 2020, 7, 171. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-Derived Circulating miRNAs Regulate Gene Expression in Other Tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Brandão, B.B.; Madsen, S.; Rabiee, A.; Oliverio, M.; Ruiz, G.P.; Ferrucci, D.L.; Branquinho, J.L.; Razolli, D.; Pinto, S.; Nielsen, T.S.; et al. Dynamic changes in DICER levels in adipose tissue control metabolic adaptations to exercise. Proc. Natl. Acad. Sci. USA 2020, 117, 23932–23941. [Google Scholar] [CrossRef]
- Mudhasani, R.R.; Puri, V.; Hoover, K.; Czech, M.P.; Imbalzano, A.N.; Jones, S.N. Dicer is required for the formation of white but not brown adipose tissue. J. Cell. Physiol. 2010, 226, 1399–1406. [Google Scholar] [CrossRef]
- Hubal, M.J.; Nadler, E.P.; Ferrante, S.C.; Barberio, M.D.; Suh, J.-H.; Wang, J.; Dohm, G.L.; Pories, W.J.; Mietus-Snyder, M.; Freishtat, R.J. Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obesity 2017, 25, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Serbulea, V.; Upchurch, C.M.; Schappe, M.S.; Voigt, P.; Deweese, D.E.; Desai, B.N.; Meher, A.K.; Leitinger, N. Macrophage phenotype and bioenergetics are controlled by oxidized phospholipids identified in lean and obese adipose tissue. Proc. Natl. Acad. Sci. USA 2018, 115, E6254–E6263. [Google Scholar] [CrossRef]
- Ieronymaki, E.; Theodorakis, E.M.; Lyroni, K.; Vergadi, E.; Lagoudaki, E.; Al-Qahtani, A.; Aznaourova, M.; Neofotistou-Themeli, E.; Eliopoulos, A.G.; Vaporidi, K.; et al. Insulin Resistance in Macrophages Alters Their Metabolism and Promotes an M2-Like Phenotype. J. Immunol. 2019, 202, 1786–1797. [Google Scholar] [CrossRef]
- Ricardo-Gonzalez, R.R.; Eagle, A.R.; Odegaard, J.I.; Jouihan, H.; Morel, C.R.; Heredia, J.E.; Mukundan, L.; Wu, D.; Locksley, R.M.; Chawla, A. IL-4/STAT6 immune axis regulates peripheral nutrient metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. USA 2010, 107, 22617–22622. [Google Scholar] [CrossRef] [PubMed]
- Kern, L.; Mittenbühler, M.J.; Vesting, A.J.; Ostermann, A.L.; Wunderlich, C.M.; Wunderlich, F.T. Obesity-Induced TNFα and IL-6 Signaling: The Missing Link between Obesity and Inflammation—Driven Liver and Colorectal Cancers. Cancers 2018, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xu, W.; Feng, X.; He, Y.; Liu, X.; Gao, Y.; Yang, S.; Shao, Z.; Yang, C.; Ye, Z. TNF-a mediated inflammatory macrophage polarization contributes to the pathogenesis of steroid-induced osteonecrosis in mice. Int. J. Immunopathol. Pharmacol. 2015, 28, 351–361. [Google Scholar] [CrossRef]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003, 112, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384. [Google Scholar] [CrossRef]
- Cho, Y.K.; Son, Y.; Kim, S.-N.; Song, H.-D.; Kim, M.; Park, J.-H.; Jung, Y.-S.; Ahn, S.-Y.; Saha, A.; Granneman, J.G.; et al. MicroRNA-10a-5p regulates macrophage polarization and promotes therapeutic adipose tissue remodeling. Mol. Metab. 2019, 29, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.-D.; Cheng, P.; Liu, T.; Wang, Z. BMSC-Derived Exosomal miR-29a Promotes Angiogenesis and Osteogenesis. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Leung, S.W. Identification of microRNA biomarkers in type 2 diabetes: A meta-analysis of controlled profiling studies. Diabetologia 2015, 58, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.; Mehrpouya-Bahrami, P.; Nagarkatti, P.S.; Nagarkatti, M. Cannabinoid Receptor 1 Blockade Attenuates Obesity and Adipose Tissue Type 1 Inflammation Through miR-30e-5p Regulation of Delta-Like-4 in Macrophages and Consequently Downregulation of Th1 Cells. Front. Immunol. 2019, 10, 1049. [Google Scholar] [CrossRef]
- Jaiswal, A.; Reddy, S.S.; Maurya, M.; Maurya, P.; Barthwal, M.K. MicroRNA-99a mimics inhibit M1 macrophage phenotype and adipose tissue inflammation by targeting TNFα. Cell. Mol. Immunol. 2018, 16, 495–507. [Google Scholar] [CrossRef]
- Tryggestad, J.B.; Teague, A.M.; Sparling, D.P.; Jiang, S.; Chernausek, S.D. Macrophage-Derived microRNA-155 Increases in Obesity and Influences Adipocyte Metabolism by Targeting Peroxisome Proliferator-Activated Receptor Gamma. Obesity 2019, 27, 1856–1864. [Google Scholar] [CrossRef]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Investig. 2019, 129, 834–849. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, Y.; Liu, Y.; Zhu, D.; Yu, J.; Li, G.; Sun, Z.; Wang, W.; Jiang, H.; Hong, Z. MiR-27a promotes insulin resistance and mediates glucose metabolism by targeting PPAR-γ-mediated PI3K/AKT signaling. Aging 2019, 11, 7510–7524. [Google Scholar] [CrossRef] [PubMed]
- Collares, R.V.A.; Salgado, W., Jr.; da Cunda Tirapelli, D.P.; dos Santos, J.S. The expression of LEP, LEPR, IGF1 and IL10 in obesity and the relationship with microRNAs. PLoS ONE 2014, 9, e93512. [Google Scholar]
- Kim, H.I.; Ahn, Y.H. Role of peroxisome proliferator-activated receptor-gamma in the glucose-sensing apparatus of liver and beta-cells. Diabetes 2004, 53 (Suppl. 1), S60–S65. [Google Scholar] [CrossRef]
- Ma, S.; Liu, M.; Xu, Z.; Li, Y.; Guo, H.; Ge, Y.; Liu, Y.; Zheng, D.; Shi, J. A double feedback loop mediated by microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage polarization and thus regulates cancer progression. Oncotarget 2016, 7, 13502–13519. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-Derived Exosomal MiR-27a Induces Insulin Resistance in Skeletal Muscle Through Repression of PPARγ. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-F.; Ku, H.-C.; Lin, H. PGC-1α as a Pivotal Factor in Lipid and Metabolic Regulation. Int. J. Mol. Sci. 2018, 19, 3447. [Google Scholar] [CrossRef] [PubMed]
- LaPierre, M.P.; Stoffel, M. MicroRNAs as stress regulators in pancreatic beta cells and diabetes. Mol. Metab. 2017, 6, 1010–1023. [Google Scholar] [CrossRef]
- Mziaut, H.; Henniger, G.; Ganss, K.; Hempel, S.; Wolk, S.; Mcchord, J.; Chowdhury, K.; Ravassard, P.; Knoch, K.-P.; Krautz, C.; et al. MiR-132 controls pancreatic beta cell proliferation and survival through Pten/Akt/Foxo3 signaling. Mol. Metab. 2020, 31, 150–162. [Google Scholar] [CrossRef]
- Cui, X.; You, L.; Zhu, L.; Wang, X.; Zhou, Y.; Li, Y.; Wen, J.; Xia, Y.; Wang, X.; Ji, C.; et al. Change in circulating microRNA profile of obese children indicates future risk of adult diabetes. Metabolism 2018, 78, 95–105. [Google Scholar] [CrossRef]
- Min, P.-K.; Chan, S.Y. The biology of circulating microRNAs in cardiovascular disease. Eur. J. Clin. Investig. 2015, 45, 860–874. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, X.; Liu, X.; Du, H.; Sun, C.; Shao, X.; Tian, J.; Gu, X.; Wang, H.; Tian, J.; et al. Adipose-Derived Exosomes Exert Proatherogenic Effects by Regulating Macrophage Foam Cell Formation and Polarization. J. Am. Heart Assoc. 2018, 7, e007442. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, P.; Liu, J.; Xie, X. Exosomal microRNA-122 mediates obesity-related cardiomyopathy through suppressing mitochondrial ADP-ribosylation factor-like. Clin. Sci. 2019, 133, 1871–1881. [Google Scholar] [CrossRef]
- McMullen, J.R.; Bernardo, B.C. Inhibition of miR-29 protects against cardiac hypertrophy and fibrosis: New insight for the role of miR-29 in the heart. Non-Coding RNA Investig. 2018, 2, 14. [Google Scholar] [CrossRef]
- Zou, T.; Zhu, M.; Ma, Y.-C.; Xiao, F.; Yu, X.; Xu, L.; Ma, L.-Q.; Yang, J.; Dong, J.-Z. MicroRNA-410-5p exacerbates high-fat diet-induced cardiac remodeling in mice in an endocrine fashion. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Jopling, C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012, 9, 137–142. [Google Scholar] [CrossRef]
- Burchard, J.; Zhang, C.; Liu, A.M.; Poon, R.T.P.; Lee, N.P.Y.; Wong, K.; Sham, P.C.; Lam, B.Y.; Ferguson, M.D.; Tokiwa, G.; et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol. Syst. Biol. 2010, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Miyaaki, H.; Ichikawa, T.; Kamo, Y.; Taura, N.; Honda, T.; Shibata, H.; Milazzo, M.; Fornari, F.; Gramantieri, L.; Bolondi, L.; et al. Significance of serum and hepatic microRNA-122 levels in patients with non-alcoholic fatty liver disease. Liver Int. 2014, 34, e302–e307. [Google Scholar] [CrossRef]
- Li, P.; Fan, C.; Cai, Y.; Fang, S.; Zeng, Y.; Zhang, Y.; Lin, X.; Zhang, H.; Xue, Y.; Guan, M. Transplantation of brown adipose tissue up-regulates miR-99a to ameliorate liver metabolic disorders in diabetic mice by targeting NOX4. Adipocyte 2020, 9, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Eunji, L.; Jung, J.; Lee, J.W.; Kim, H.J.; Kim, J.; Yoo, H.j.; Lee, H.J.; Chae, S.Y.; Jeon, S.M.; et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene 2018, 37, 2982–2991. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Dezso, Z.; MacKenzie, C.; Oestreicher, J.; Agoulnik, S.; Byrne, M.; Bernier, F.; Yanagimachi, M.; Aoshima, K.; Oda, Y. Circulating miRNA Biomarkers for Alzheimer’s Disease. PLoS ONE 2013, 8, e69807. [Google Scholar] [CrossRef]
- Fu, Y.; Hu, X.; Zheng, C.; Sun, G.; Xu, J.; Luo, S.; Cao, P. Intrahippocampal miR-342-3p inhibition reduces β-amyloid plaques and ameliorates learning and memory in Alzheimer’s disease. Metab. Brain Dis. 2019, 34, 1355–1363. [Google Scholar] [CrossRef]
- Hong, H.; Li, Y.; Su, B. Identification of Circulating miR-125b as a Potential Biomarker of Alzheimer’s Disease in APP/PS1 Transgenic Mouse. J. Alzheimers Dis. 2017, 59, 1449–1458. [Google Scholar] [CrossRef]
- Alipoor, B.; Ghaedi, H.; Meshkani, R.; Torkamandi, S.; Saffari, S.; Iranpour, M.; Omrani, M.D. Association of MiR-146a Expression and Type 2 Diabetes Mellitus: A Meta-Analysis. Int. J. Mol. Cell. Med. 2017, 6, 156–163. [Google Scholar]
- Parrizas, M.; Mundet, X.; Castaño, C.; Canivell, S.; Cos, X.; Brugnara, L.; Giráldez-García, C.; Regidor, E.; Mata-Cases, M.; Franch-Nadal, J.; et al. miR-10b and miR-223-3p in serum microvesicles signal progression from prediabetes to type 2 diabetes. J. Endocrinol. Investig. 2019, 43, 451–459. [Google Scholar] [CrossRef]
- Zhang, T.; Ji, C.; Shi, R. miR-142-3p promotes pancreatic β cell survival through targeting FOXO1 in gestational diabetes mellitus. Int. J. Clin. Exp. Pathol. 2019, 12, 1529–1538. [Google Scholar] [PubMed]
- Liu, Y.; Gao, G.; Yang, C.; Zhou, K.; Shen, B.; Liang, H.; Jiang, X. The Role of Circulating MicroRNA-126 (miR-126): A Novel Biomarker for Screening Prediabetes and Newly Diagnosed Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2014, 15, 10567–10577. [Google Scholar] [CrossRef] [PubMed]
- Li, X. miR-375, a microRNA related to diabetes. Gene 2014, 533, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Corral-Fernández, N.E.; Salgado-Bustamante, M.; Martínez-Leija, M.E.; Cortez-Espinosa, N.; García-Hernández, M.H.; Reynaga-Hernández, E.; Quezada-Calvillo, R.; Portales-Pérez, D.P. Dysregulated miR-155 expression in peripheral blood mononuclear cells from patients with type 2 diabetes. Exp. Clin. Endocrinol. Diabetes 2013, 121, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lin, L.; Huang, Y.; Sun, J.; Wang, X.; Wang, P. MicroRNA-138 Suppresses Adipogenic Differentiation in Human Adipose Tissue-Derived Mesenchymal Stem Cells by Targeting Lipoprotein Lipase. Yonsei Med. J. 2019, 60, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- González-Arce, L.M.; Lara-Riegos, J.C.; Pérez-Mendoza, G.J.; Rubí-Castellanos, R.; Vega-Marcín, M.; Valencia-Pacheco, G.; Torres-Romero, J.C.; González-Herrera, L. High expression levels of circulating microRNA -122 and microRNA -222 are associated with obesity in children with Mayan ethnicity. Am. J. Hum. Biol. 2020, e23540. [Google Scholar] [CrossRef]
- Wei, Z.; Qin, X.; Kang, X.; Zhou, H.; Wang, S.; Wei, D. MiR-142-3p inhibits adipogenic differentiation and autophagy in obesity through targeting KLF9. Mol. Cell. Endocrinol. 2020, 518, 111028. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Chen, J.; Peng, W.; Yuan, B.; Bi, Q.; Xu, Y. Exosomes from adipose-derived stem cells promote chondrogenesis and suppress inflammation by upregulating miR-145 and miR-221. Mol. Med. Rep. 2020, 21, 1881–1889. [Google Scholar] [CrossRef] [PubMed]
- Abplanalp, W.T.; Fischer, A.; John, D.; Zeiher, A.M.; Gosgnach, W.; Darville, H.; Montgomery, R.; Pestano, L.; Allée, G.; Paty, I.; et al. Efficiency and Target Derepression of Anti-miR-92a: Results of a First in Human Study. Nucleic Acid Ther. 2020, 30, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-W.; Shen, Y.-J.; Shi, J.; Yu, J.-G. MiR-223-3p in Cardiovascular Diseases: A Biomarker and Potential Therapeutic Target. Front. Cardiovasc. Med. 2021, 7, 610561. [Google Scholar] [CrossRef]
- Mishra, S.; Rizvi, A.; Pradhan, A.; Perrone, M.A.; Ali, W. Circulating microRNA-126 &122 in patients with coronary artery disease: Correlation with small dense LDL. Prostaglandins Other Lipid Mediat. 2021, 153, 106536. [Google Scholar] [CrossRef] [PubMed]
- Zhelankin, A.V.; Vasiliev, S.V.; Stonogina, D.A.; Babalyan, K.A.; Sharova, E.I.; Doludin, Y.V.; Shchekochikhin, D.Y.; Generozov, E.V.; Akselrod, A.S. Elevated Plasma Levels of Circulating Extracellular miR-320a-3p in Patients with Paroxysmal Atrial Fibrillation. Int. J. Mol. Sci. 2020, 21, 3485. [Google Scholar] [CrossRef]
- Bai, X.; Tang, Y.; Yu, M.; Wu, L.; Liu, F.; Ni, J.; Wang, Z.; Wang, J.; Fei, J.; Wang, W.; et al. Downregulation of blood serum microRNA 29 family in patients with Parkinson’s disease. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Cao, X.-Y.; Lu, J.-M.; Zhao, Z.-Q.; Li, M.-C.; Lu, T.; An, X.-S.; Xue, L.-J. MicroRNA biomarkers of Parkinson’s disease in serum exosome-like microvesicles. Neurosci. Lett. 2017, 644, 94–99. [Google Scholar] [CrossRef]
- Zhou, S.; Zhang, D.; Guo, J.; Zhang, J.; Chen, Y. Knockdown of SNHG14 Alleviates MPP+-Induced Injury in the Cell Model of Parkinson’s Disease by Targeting the miR-214-3p/KLF4 Axis. Front. Neurosci. 2020, 14, 930. [Google Scholar] [CrossRef]
- Zhu, G.; Cao, B.; Liang, X.; Li, L.; Hao, Y.; Meng, W.; He, C.; Wang, L.; Li, L. Small extracellular vesicles containing miR-192/215 mediate hypoxia-induced cancer-associated fibroblast development in head and neck squamous cell carcinoma. Cancer Lett. 2021, 506, 11–22. [Google Scholar] [CrossRef]
- Yu, H.; Qin, L.; Peng, Y.; Bai, W.; Wang, Z. Exosomes Derived From Hypertrophic Cardiomyocytes Induce Inflammation in Macrophages via miR-155 Mediated MAPK Pathway. Front. Immunol. 2021, 11, 606045. [Google Scholar] [CrossRef]
- Jia, Y.-C.; Ding, Y.-X.; Mei, W.-T.; Wang, Y.-T.; Zheng, Z.; Qu, Y.-X.; Liang, K.; Li, J.; Cao, F.; Li, F. Extracellular vesicles and pancreatitis: Mechanisms, status and perspectives. Int. J. Biol. Sci. 2021, 17, 549–561. [Google Scholar] [CrossRef]
- Hong, L.Z.; Zhou, L.; Zou, R.; Khoo, C.M.; Chew, A.L.S.; Chin, C.-L.; Shih, S.-J. Systematic evaluation of multiple qPCR platforms, NanoString and miRNA-Seq for microRNA biomarker discovery in human biofluids. Sci. Rep. 2021, 11, 4435. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, R.; Nardelli, C.; Pilone, V.; Buonomo, T.; Liguori, R.; Castanò, I.; Buono, P.; Masone, S.; Persico, G.; Forestieri, P.; et al. miR-519d Overexpression Is Associated With Human Obesity. Obesity 2010, 18, 2170–2176. [Google Scholar] [CrossRef] [PubMed]
- Belgardt, B.-F.; Ahmed, K.; Spranger, M.; Latreille, M.; Denzler, R.; Kondratiuk, N.; Von Meyenn, F.; Villena, F.N.; Herrmanns, K.; Bosco, D.; et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat. Med. 2015, 21, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Asl, E.R.; Rasmi, Y.; Baradaran, B. MicroRNA-124-3p suppresses PD-L1 expression and inhibits tumorigenesis of colorectal cancer cells via modulating STAT3 signaling. J. Cell. Physiol. 2021. [Google Scholar] [CrossRef]
- Sayed, R.; Fernández-Ortiz, M.; Fernández-Martínez, J.; Martínez, P.A.; Guerra-Librero, A.; Rodríguez-Santana, C.; de Haro, T.; Escames, G.; Acuña-Castroviejo, D.; Rusanova, I. The Impact of Melatonin and NLRP3 Inflammasome on the Expression of microRNAs in Aged Muscle. Antioxidants 2021, 10, 524. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Wang, C.; Chen, L.; Wong, G. Dysregulation of MicroRNAs and PIWI-Interacting RNAs in a Caenorhabditis elegans Parkinson’s Disease Model Overexpressing Human α-Synuclein and Influence of tdp-1. Front. Neurosci. 2021, 15, 600462. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, S.; Hug, C.; Todorov, P.; Moret, N.; Boswell, S.A.; Evans, K.; Zhou, G.; Johnson, N.T.; Hyman, B.T.; Sorger, P.K.; et al. Machine learning identifies candidates for drug repurposing in Alzheimer’s disease. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yang, X.; Liu, M.; Zhang, Z.; Xing, E. Roles of miRNA dysregulation in the pathogenesis of multiple myeloma. Cancer Gene Ther. 2021, 1–13. [Google Scholar] [CrossRef]
- Rocic, P. Can microRNAs be Biomarkers or Targets for Therapy of Ischemic Coronary Artery Disease in Metabolic Syndrome? Curr. Drug Targets 2017, 18, 1722–1732. [Google Scholar] [CrossRef] [PubMed]
- Casciaro, M.; Di Salvo, E.; Brizzi, T.; Rodolico, C.; Gangemi, S. Involvement of miR-126 in autoimmune disorders. Clin. Mol. Allergy 2018, 16, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Alevizos, I.; Illei, G.G. MicroRNAs as biomarkers in rheumatic diseases. Nat. Rev. Rheumatol. 2010, 6, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef]
- Schmidt, M.F. miRNA Targeting Drugs: The Next Blockbusters? Methods Mol. Biol. 2017, 1517, 3–22. [Google Scholar]
- Jafari, N.; Abediankenari, S. MicroRNA-34 dysregulation in gastric cancer and gastric cancer stem cell. Tumor Biol. 2017, 39, 1010428317701652. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Cai, W.; Liang, Y.; Yao, J.; Wang, X.; Shen, J. In situ self-assembly of Au-antimiR-155 nanocomplexes mediates TLR3-dependent apoptosis in hepatocellular carcinoma cells. Aging 2020, 13, 241–261. [Google Scholar] [CrossRef] [PubMed]
- Lucas, T.; Schäfer, F.; Müller, P.; Eming, S.A.; Heckel, A.; Dimmeler, S. Light-inducible antimiR-92a as a therapeutic strategy to promote skin repair in healing-impaired diabetic mice. Nat. Commun. 2017, 8, 15162. [Google Scholar] [CrossRef]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501. [Google Scholar] [CrossRef]
- Gaudet, A.D.; Fonken, L.K.; Gushchina, L.V.; Aubrecht, T.G.; Maurya, S.K.; Periasamy, M.; Nelson, R.J.; Popovich, P.G. miR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 2016, 6, 22862. [Google Scholar] [CrossRef]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Xihua, L.; Shengjie, T.; Weiwei, G.; Matro, E.; Tingting, T.; Lin, L.; Fang, W.; Jiaqiang, Z.; Fenping, Z.; Hong, L. Circulating miR-143-3p inhibition protects against insulin resistance in Metabolic Syndrome via targeting of the insulin-like growth factor 2 receptor. Transl. Res. 2019, 205, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Querfeld, C.; Foss, F.M.; Kim, Y.H.; Pinter-Brown, L.; William, B.M.; Porcu, P.; Pacheco, T.; Haverkos, B.M.; DeSimone, J.; Guitart, J.; et al. Phase 1 Trial of Cobomarsen, an Inhibitor of Mir-155, in Cutaneous T Cell Lymphoma. Blood 2018, 132, 2903. [Google Scholar] [CrossRef]
- Ottosen, S.; Parsley, T.B.; Yang, L.; Zeh, K.; van Doorn, L.; van der Veer, E.; Raney, A.K.; Hodges, M.R.; Patick, A.K. In Vitro Antiviral Activity and Preclinical and Clinical Resistance Profile of Miravirsen, a Novel Anti-Hepatitis C Virus Therapeutic Targeting the Human Factor miR-122. Antimicrob. Agents Chemother. 2015, 59, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Kang, Y.-K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.-L.; Kim, T.-Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Reid, G.; Kao, S.C.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; Van Zandwijk, N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 2016, 8, 1079–1085. [Google Scholar] [CrossRef]
- Lee, J.J.; Pope, J.E. Emerging drugs and therapeutics for systemic sclerosis. Expert Opin. Emerg. Drugs 2016, 21, 421–430. [Google Scholar] [CrossRef]

| Sr. No | Disease | miRs | References |
|---|---|---|---|
| 1. | Alzheimer’s disease (AD) | let-7d | [111] |
| miR-342 | [112] | ||
| miR-125b | [113] | ||
| 2. | T2DM | miR-146a | [114,115] |
| miR-223 | [116] | ||
| miR-142-3p | [117] | ||
| miR-126 | [118] | ||
| miR-375 | [119] | ||
| miR-155 | |||
| 3. | Obesity | miR-138 | |
| miR-122 | [120] | ||
| miR-142-3p | [121] | ||
| miR-221 | [122] | ||
| miR-145 | [123] | ||
| 4. | Cardiac disorders | miR-92a | [124] |
| miR-223 | [125] | ||
| miR-126 | [126] | ||
| miR-320a | [127] | ||
| 5. | Parkinson’s disease (PD) | miR-29a/c | [128] |
| miR-19b | [129] | ||
| miR-214 | [130] | ||
| miR-133b | [60] |
| Sr. No. | miRNAs | Diseases | Drug | Ref. |
|---|---|---|---|---|
| 1 | anti-miR-155 | Cutaneous T and B-cell lymphoma | MRG-106 | [154] |
| 2 | anti-miR-122 | Hepatitis C virus infection | Mirvirasen, RG-101 | [155] |
| 3 | anti-miR-103/107 | T2DM with nonalcoholic fatty liver disease | RG-125/AZD4076 | |
| 4 | miR-34 mimic | solid tumors | MRX34 | [156] |
| 5 | miR-16 mimic | Malignant pleural mesothelioma, non-small-cell lung cancer | MesomiR-1 | [157] |
| 6 | miR-29 mimic | Scleroderma | MRG-201 | [158] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiran, S.; Kumar, V.; Kumar, S.; Price, R.L.; Singh, U.P. Adipocyte, Immune Cells, and miRNA Crosstalk: A Novel Regulator of Metabolic Dysfunction and Obesity. Cells 2021, 10, 1004. https://doi.org/10.3390/cells10051004
Kiran S, Kumar V, Kumar S, Price RL, Singh UP. Adipocyte, Immune Cells, and miRNA Crosstalk: A Novel Regulator of Metabolic Dysfunction and Obesity. Cells. 2021; 10(5):1004. https://doi.org/10.3390/cells10051004
Chicago/Turabian StyleKiran, Sonia, Vijay Kumar, Santosh Kumar, Robert L Price, and Udai P. Singh. 2021. "Adipocyte, Immune Cells, and miRNA Crosstalk: A Novel Regulator of Metabolic Dysfunction and Obesity" Cells 10, no. 5: 1004. https://doi.org/10.3390/cells10051004
APA StyleKiran, S., Kumar, V., Kumar, S., Price, R. L., & Singh, U. P. (2021). Adipocyte, Immune Cells, and miRNA Crosstalk: A Novel Regulator of Metabolic Dysfunction and Obesity. Cells, 10(5), 1004. https://doi.org/10.3390/cells10051004








