The p23 of Citrus Tristeza Virus Interacts with Host FKBP-Type Peptidyl-Prolylcis-Trans Isomerase 17-2 and Is Involved in the Intracellular Movement of the Viral Coat Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Source and Plant Material
2.2. Gene Cloning
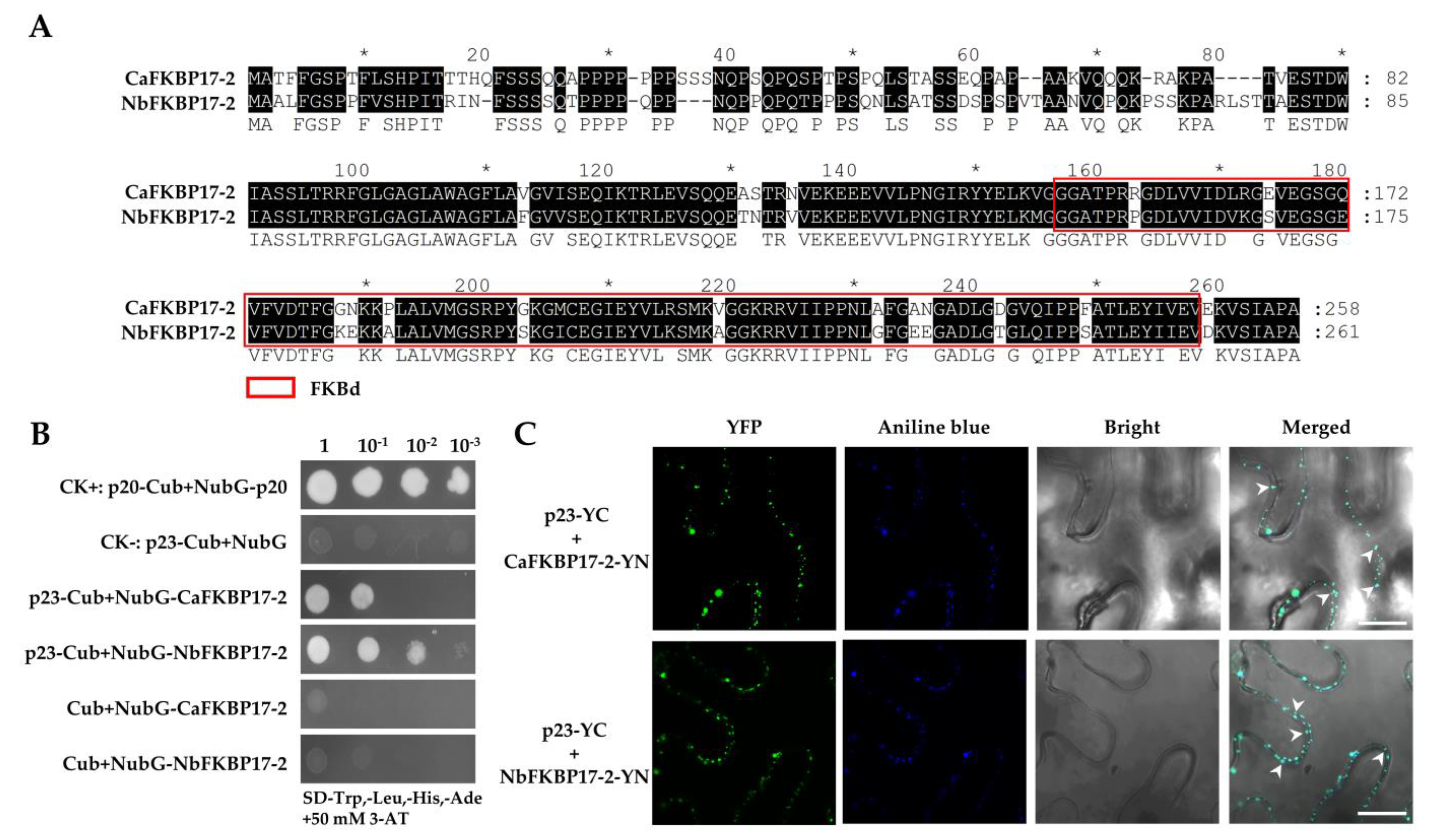
2.3. DUALMembrane Yeast Two-Hybrid Assay
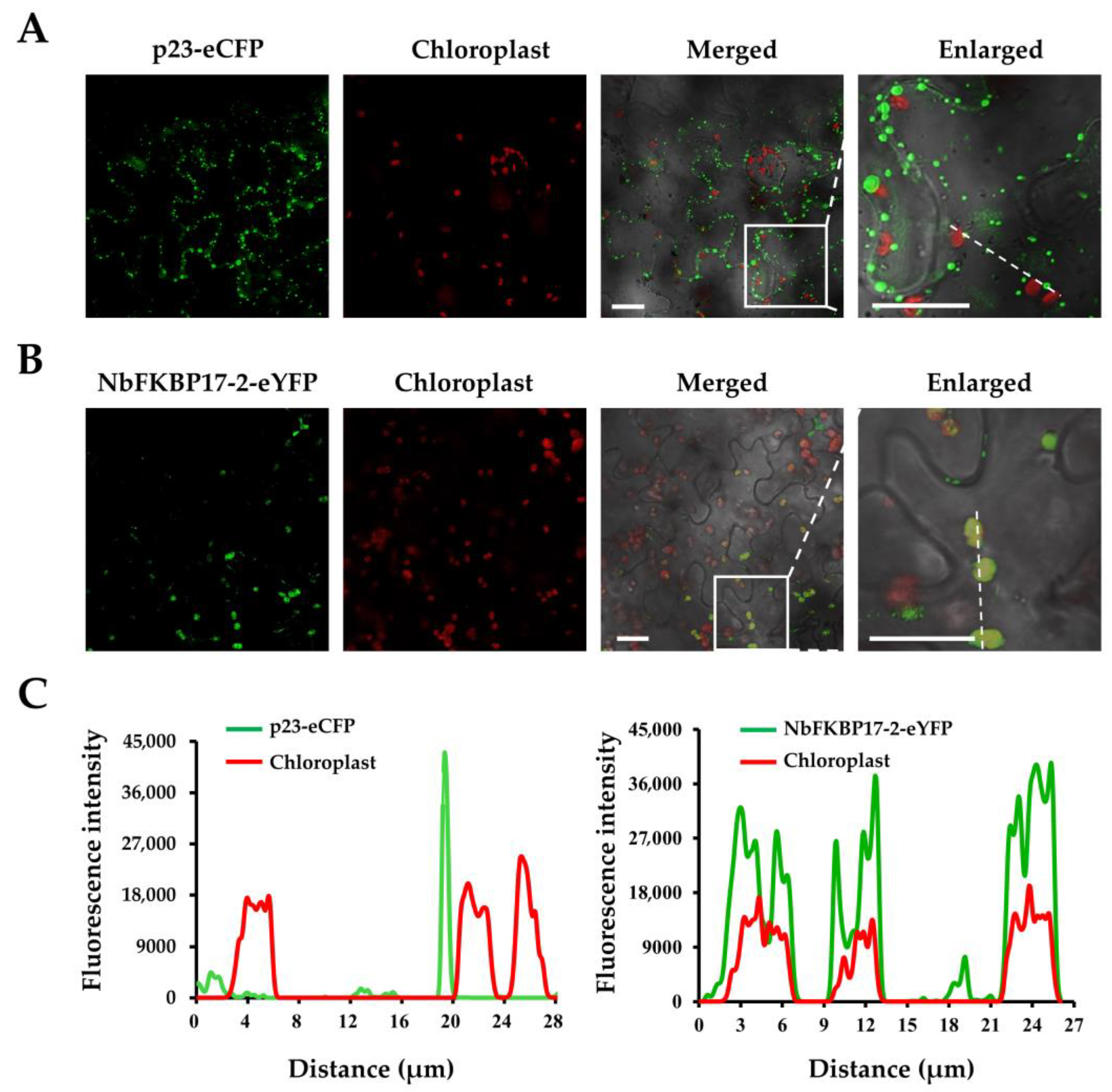
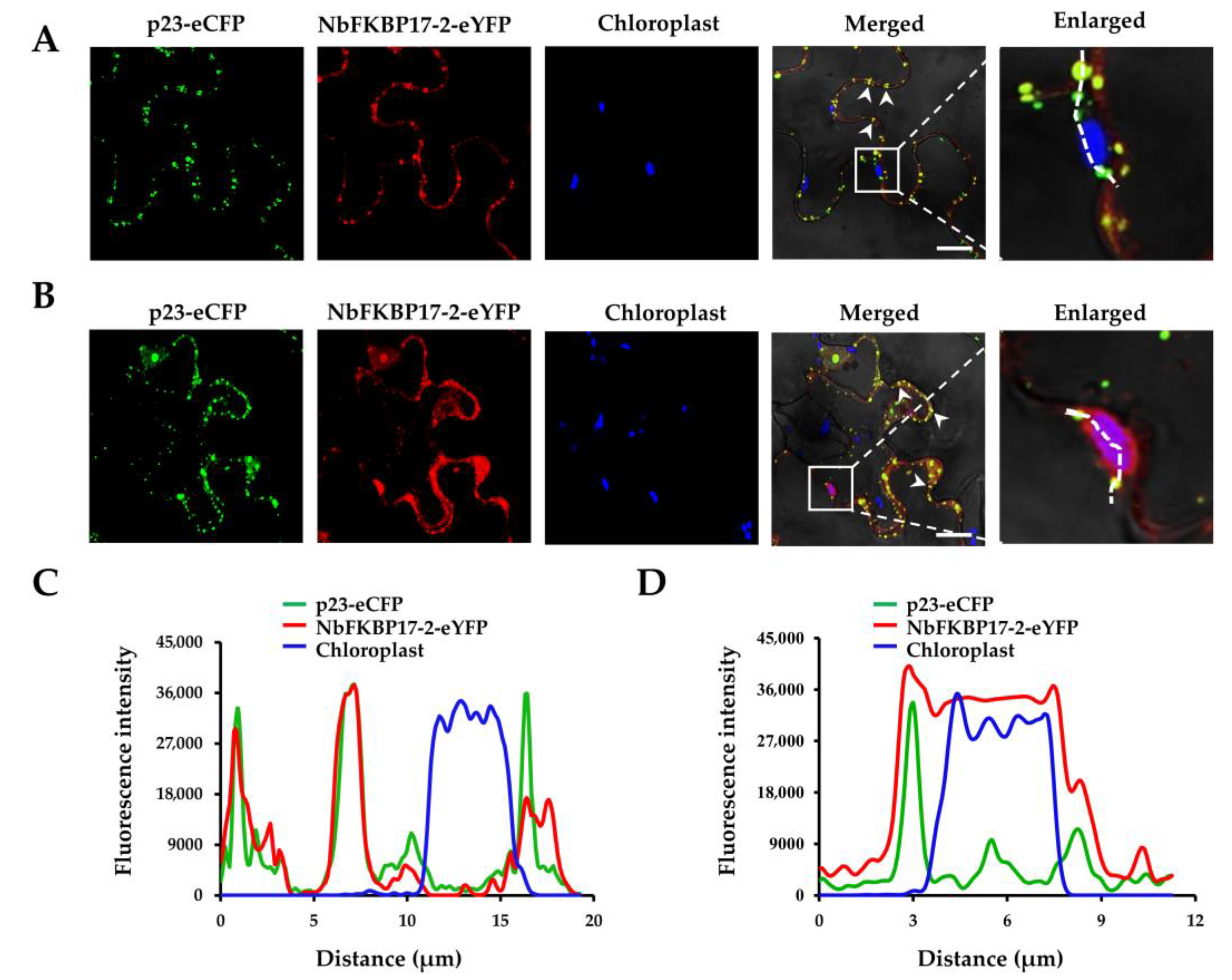
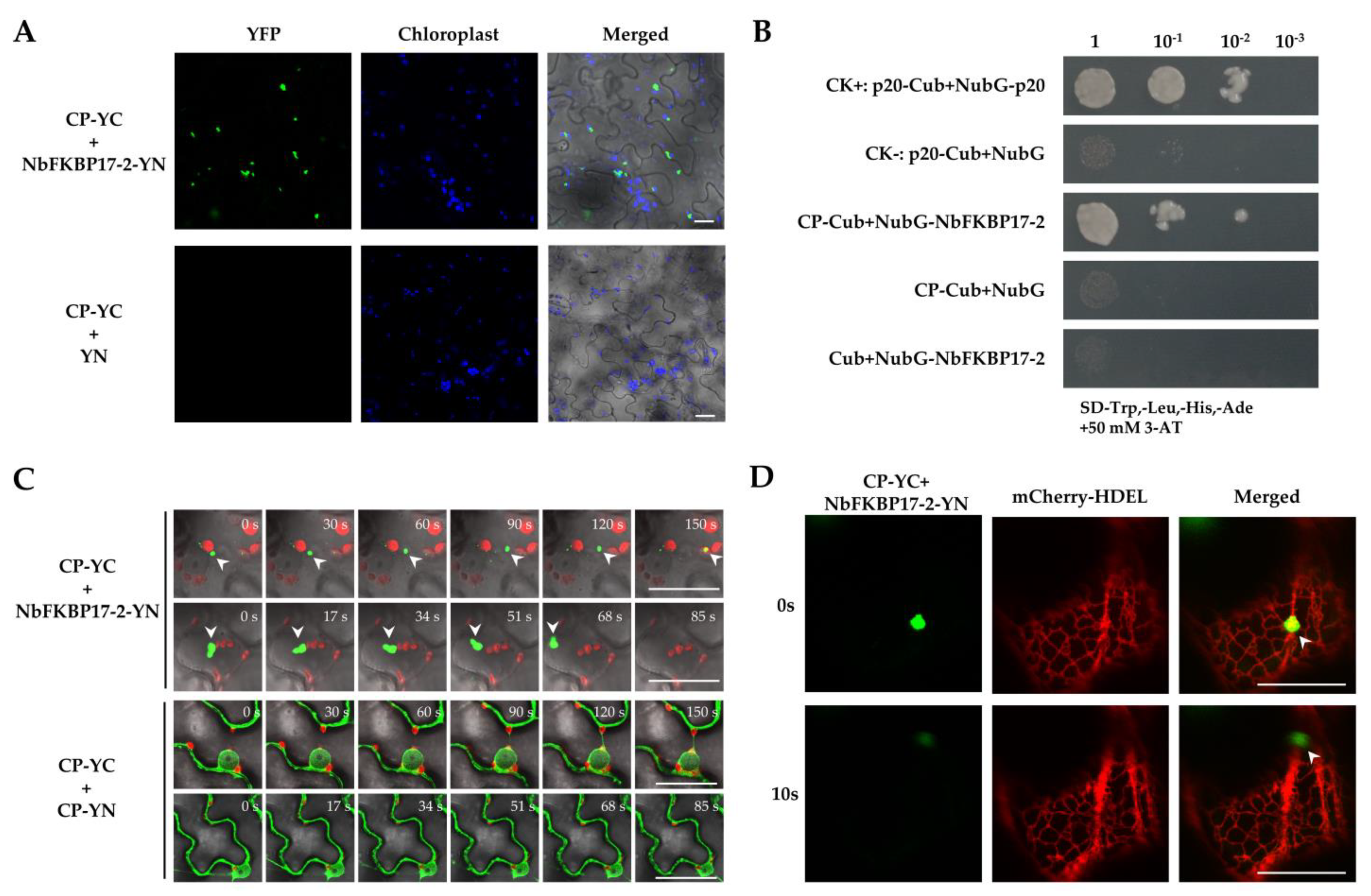
2.4. BiFC and Subcellular Localization Assays
2.5. Virus-Induced Gene Silencing (VIGS) and CTV Inoculation
2.6. Real-Time RT-PCR
2.7. Sequence Analyses
3. Results
3.1. CTV p23 Interacts with FKBP17-2 In Vivo and In Vitro
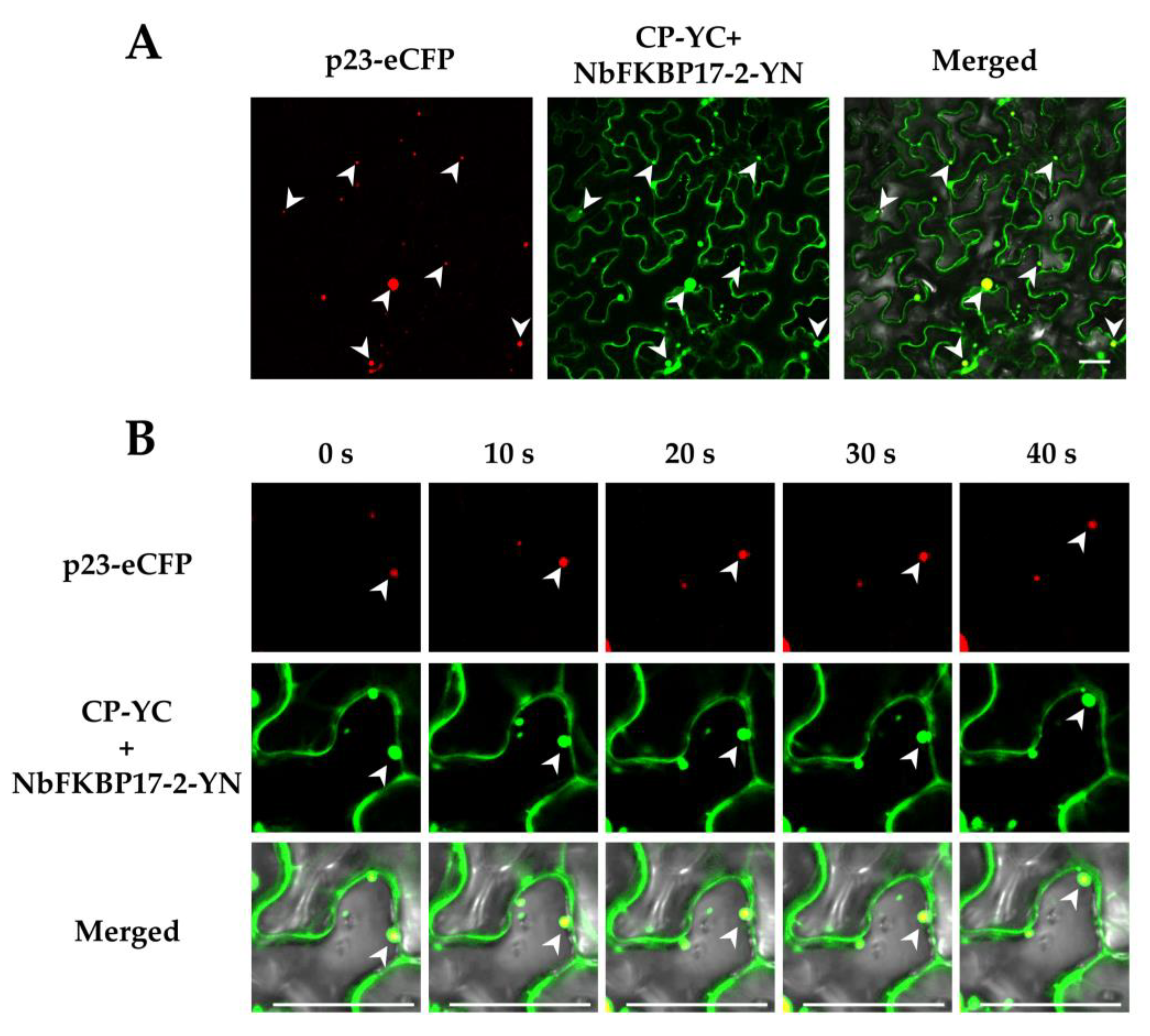
3.2. Subcellular Localization of p23 and NbFKBP17-2 in N. benthamiana Leaf Cells
3.3. NbFKBP17-2 and p23 Were Involved in CTV CP Movement
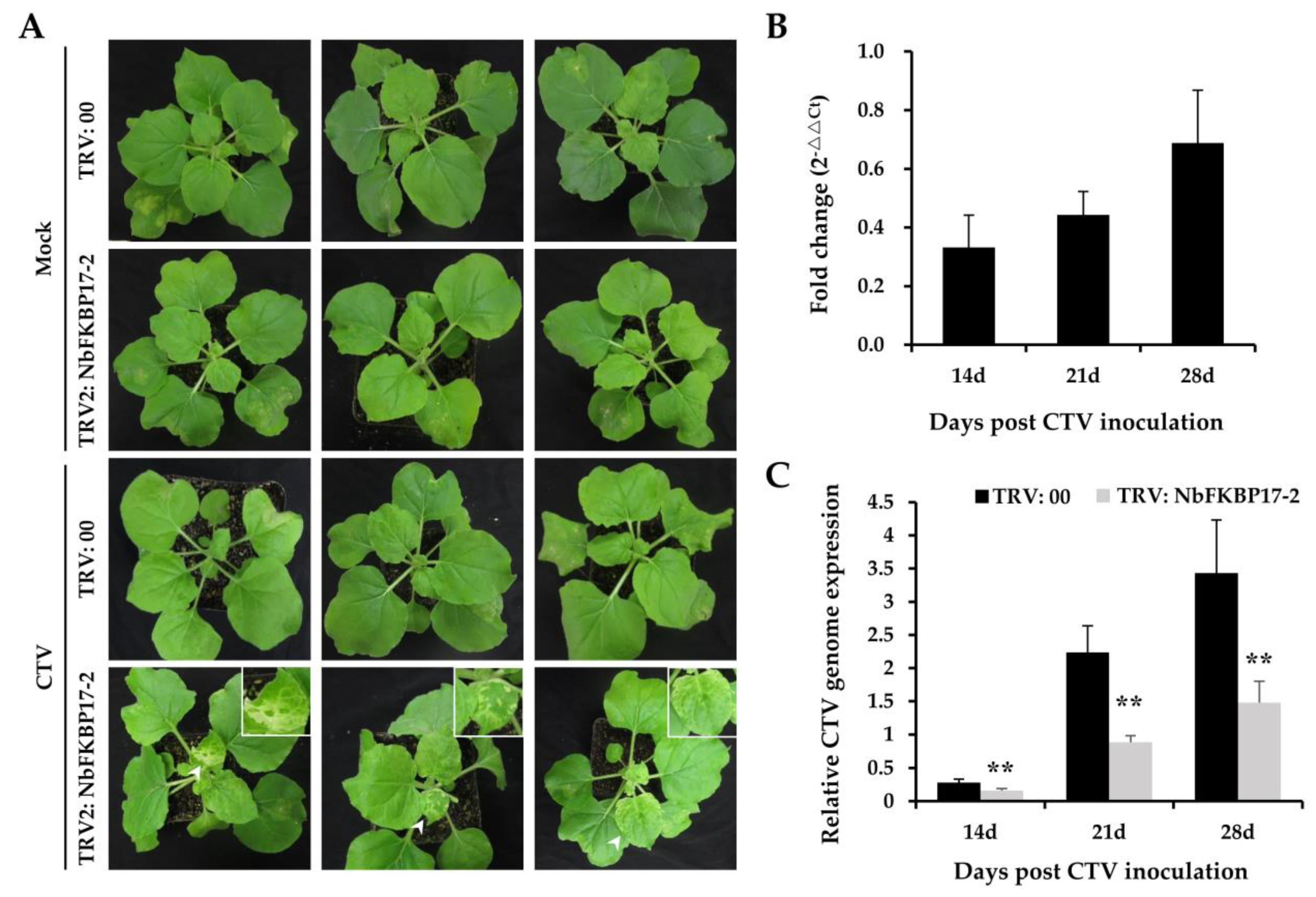
3.4. Knock-Down Expression of NbFKBP17-2 in N. benthamiana Led to Decreased CTV Accumulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karasev, A.; Boyko, V.; Gowda, S.; Nikolaeva, O.; Hilf, M.; Koonin, E.; Niblett, C.; Cline, K.; Gumpf, D.; Lee, R.J.V. Complete sequence of the citrus tristeza virus RNA genome. Virology 1995, 208, 511–520. [Google Scholar] [CrossRef]
- Dawson, W.O.; Bar-Joseph, M.; Garnsey, S.M.; Moreno, P. Citrus tristeza virus: Making an ally from an enemy. Annu. Rev. Phytopathol. 2015, 53, 137–155. [Google Scholar] [CrossRef]
- Mawassi, M.; Mietkiewska, E.; Gofman, R.; Yang, G.; Bar-Joseph, M. Unusual sequence relationships between two isolates of citrus tristeza virus. J. Gen. Virol. 1996, 77, 2359–2364. [Google Scholar] [CrossRef]
- Harper, S.J. Citrus tristeza virus: Evolution of complex and varied genotypic groups. Front. Microbiol. 2013, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Hilf, M.E.; Karasev, A.V.; Pappu, H.R.; Gumpf, D.J.; Niblett, C.L.; Garnsey, S.M. Characterization of citrus tristeza virus subgenomic RNAs in infected tissue. Virology 1995, 208, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J.P.T. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef]
- Febres, V.J.; Ashoulin, L.; Mawassi, M.; Frank, A.; Bar-Joseph, M.; Manjunath, K.L.; Niblett, C.L. The p27 protein is present at one end of Citrus tristeza virus particles. Phytopathology 1996, 86, 1331–1335. [Google Scholar] [CrossRef]
- Gowda, S.; Tatineni, S.; Folimonova, S.Y.; Hilf, M.E.; Dawson, W.O. Accumulation of a 5′ proximal subgenomic RNA of Citrus tristeza virus is correlated with encapsidation by the minor coat protein. Virology 2009, 389, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, T.; Gowda, S.; Mawassi, M.; Albiach-Martí, M.R.; Ayllón, M.A.; Robertson, C.; Garnsey, S.M.; Dawson, W.O. Closterovirus Encoded HSP70 Homolog and p61 in Addition to Both Coat Proteins Function in Efficient Virion Assembly. Virology 2000, 278, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, T.; Gowda, S.; Ayllon, M.A.; Dawson, W.O. Closterovirus bipolar virion: Evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5’ region. Proc. Natl. Acad. Sci. USA 2004, 101, 799–804. [Google Scholar] [CrossRef]
- Tatineni, S.; Robertson, C.J.; Garnsey, S.M.; Bar-Joseph, M.; Gowda, S.; Dawson, W.O. Three genes of Citrus tristeza virus are dispensable for infection and movement throughout some varieties of citrus trees. Virology 2008, 376, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, S.; Gowda, S.; Dawson, W.O. Heterologous minor coat proteins of Citrus tristeza virus strains affect encapsidation, but the coexpression of HSP70h and p61 restores encapsidation to wild-type levels. Virology 2010, 402, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.X.; Falk, B.W.; Dawson, W.O.; Ding, S.W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef]
- Flores, R.; Ruiz-Ruiz, S.; Soler, N.; Sánchez-Navarro, J.; Fagoaga, C.; López, C.; Navarro, L.; Moreno, P.; Peña, L. Citrus tristeza virus p23: A unique protein mediating key virus–host interactions. Front. Microbiol. 2013, 4, 98. [Google Scholar] [CrossRef] [PubMed]
- López, C.; Navas-Castillo, J.; Gowda, S.; Moreno, P.; Flores, R. The 23-kDa protein coded by the 3′-terminal gene of citrus tristeza virus is an RNA-binding protein. Virology 2000, 269, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, T.; Gowda, S.; Ayllón, M.A.; Albiach-Martí, M.R.; Rabindran, S.; Dawson, W.O. The p23 protein of Citrus tristeza virus controls asymmetrical RNA accumulation. J. Virol. 2002, 76, 473–483. [Google Scholar] [CrossRef]
- Ruiz-Ruiz, S.; Soler, N.; Sánchez-Navarro, J.; Fagoaga, C.; López, C.; Navarro, L.; Moreno, P.; Peña, L.; Flores, R. Citrus tristeza virus p23: Determinants for nucleolar localization and their influence on suppression of RNA silencing and pathogenesis. Mol. Plant Microbe Interact. 2013, 26, 306–318. [Google Scholar] [CrossRef]
- Ghorbel, R.; López, C.; Fagoaga, C.; Moreno, P.; Navarro, L.; Flores, R.; Peña, L. Transgenic citrus plants expressing the citrus tristeza virus p23 protein exhibit viral-like symptoms. Mol. Plant Pathol. 2001, 2, 27–36. [Google Scholar] [CrossRef]
- Fagoaga, C.; López, C.; Moreno, P.; Navarro, L.; Flores, R.; Peña, L. Viral-like symptoms induced by the ectopic expression of the p23 gene of Citrus tristeza virus are citrus specific and do not correlate with the pathogenicity of the virus strain. Mol. Plant Microbe Interact. 2005, 18, 435–445. [Google Scholar] [CrossRef]
- Weber, P.H.; Bujarski, J. Multiple functions of capsid proteins in (+) stranded RNA viruses during plant-virus interactions. Virus Res. 2015, 22, 140–149. [Google Scholar] [CrossRef]
- Levy, A.; El-Mochtar, C.; Wang, C.X.; Goodin, M.; Orbovic, V. A new toolset for protein expression and subcellular localization studies in citrus and its application to citrus tristeza virus proteins. Plant Methods 2018, 14, 2. [Google Scholar] [CrossRef]
- Whitham, S.A.; Wang, Y. Roles for host factors in plant viral pathogenicity. Curr. Opin. Plant Biol. 2004, 7, 365–371. [Google Scholar] [CrossRef]
- Satyanarayana, T.; Gowda, S.; Boyko, V.P.; Albiach-Marti, M.R.; Mawassi, M.; Navas-Castillo, J.; Karasev, A.V.; Dolja, V.; Hilf, M.E.; Lewandowski, D.J.; et al. An Engineered Closterovirus RNA Replicon and Analysis of Heterologous Terminal Sequences for Replication. Proc. Natl. Acad. Sci. USA 1999, 96, 7433–7438. [Google Scholar] [CrossRef]
- Sun, Y.D.; Folimonova, S.Y. The p33 protein of Citrus tristeza virus affects viral pathogenicity by modulating a host immune response. New Phytol. 2019, 221, 2039–2053. [Google Scholar] [CrossRef]
- Sun, Y.D.; Zhang, L.; Folimonova, S.Y. Citrus miraculin-like protein hijacks a viral movement-related p33 protein and induces cellular oxidative stress in defence against Citrus tristeza virus. Plant Biotechnol. J. 2020. [Google Scholar] [CrossRef]
- Ambrós, S.; El-Mohtar, C.; Ruiz-Ruiz, S.; Peña, L.; Guerri, J.; Dawson, W.O.; Moreno, P. Agroinoculation of Citrus tristeza virus causes systemic infection and symptoms in the presumed nonhost Nicotiana benthamiana. Mol. Plant Microbe Interact. 2011, 24, 1119–1131. [Google Scholar] [CrossRef] [PubMed]
- Bak, A.; Folimonova, S.Y. The conundrum of a unique protein encoded by citrus tristeza virus that is dispensable for infection of most hosts yet shows characteristics of a viral movement protein. Virology 2015, 485, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Bergua, M.; Kang, S.H.; Folimonova, S.Y. Understanding superinfection exclusion by complex populations of Citrus tristeza virus. Virology 2016, 499, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Sun, Y.D.; Atallah, O.O.; Huguet-Tapia, J.C.; Noble, J.D.; Folimonova, S.Y. A Long Non-Coding RNA of Citrus tristeza virus: Role in the Virus Interplay with the Host Immunity. Viruses 2019, 11, 436. [Google Scholar] [CrossRef]
- Ruiz-Ruiz, S.; Spanò, R.; Navarro, L.; Moreno, P.; Peña, L.; Flores, R. Citrus tristeza virus co-opts glyceraldehyde 3-phosphate dehydrogenase for its infectious cycle by interacting with the viral-encoded protein p23. Plant Mol. Biol. 2018, 98, 363–373. [Google Scholar] [CrossRef]
- Harding, M.W.; Galat, A.; Uehling, D.E.; Schreiber, S.L. A receptor for the immunosup-pressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 1989, 341, 758–760. [Google Scholar] [CrossRef] [PubMed]
- Siekierka, J.J.; Staruch, M.J.; Hung, S.H.; Sigal, N.H. FK506, a potent novel immunosup-pressive agent, binds to a cytosolic protein which is distinct from the cyclosporin A-binding protein, cyclophilin. J. Immunol. 1989, 143, 1580–1583. [Google Scholar]
- Gollan, P.J.; Bhave, M.; Aro, E.M. The FKBP families of higher plants: Exploring the structures and functions of protein interaction specialists. FEBS Lett. 2012, 586, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Petersson, U.A.; Haas, B.J.; Funk, C.; Schröder, W.P.; Kieselbach, T. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 8354–8365. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Y.; Li, L.G.; Luan, S. Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 2004, 134, 1248–1267. [Google Scholar] [CrossRef]
- Yang, F. The Effects of a Severe CTV Isolate on Citrus Gene Expression and the Identification of Proteins Involved in Virus-Host Interaction. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2013. [Google Scholar]
- Moreno, P.; Ambrós, S.; Albiach-Martí, M.R.; Guerri, J.; Pena, L. Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 2008, 9, 251–268. [Google Scholar] [CrossRef]
- Yang, F.; Wang, G.P.; Jiang, B.; Liu, Y.H.; Liu, Y.; Wu, G.W.; Hong, N. Differentially expressed genes and temporal and spatial expression of genes during interactions between Mexican lime (Citrus aurantifolia) and a severe Citrus tristeza virus isolate. Physiol. Mol. Plant Pathol. 2013, 83, 17–24. [Google Scholar] [CrossRef]
- Lu, Q.; Tang, X.R.; Tian, G.; Wang, F.; Liu, K.D.; Nguyen, V.; Kohalmi, S.E.; Keller, W.A.; Tsang, E.W.; Harada, J.J.; et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: Components and anchoring nucleoporin. Plant J. 2010, 61, 259–270. [Google Scholar] [CrossRef]
- Yang, G.G.; Tang, L.G.; Gong, Y.D.; Xie, J.T.; Fu, Y.P.; Jiang, D.H.; Li, G.Q.; Collinge, D.B.; Chen, W.D.; Cheng, J.S. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 2018, 217, 739–755. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Rehman, A.U.; Li, Z.R.; Yang, Z.K.; Waqas, M.; Wang, G.P.; Xu, W.X.; Li, F.; Hong, N. The Coat Protein of Citrus Yellow Vein Clearing Virus Interacts with Viral Movement Proteins and Serves as an RNA Silencing Suppressor. Viruses 2019, 11, 329. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Diaz, T.; Zhang, D.; Fan, P.; Wang, L.; Ding, X.; Jiang, Y.; Jimenez-Gongora, T.; Medina-Puche, L.; Zhao, X.; Feng, Z.; et al. A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl. Acad. Sci. USA 2018, 115, 1388–1393. [Google Scholar] [CrossRef]
- Liu, Y.L.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef]
- Ruiz-Ruiz, S.; Moreno, P.; Guerri, J.; Ambrós, S. A real-time RT-PCR assay for detection and absolute quantitation of Citrus tristeza virus in different plant tissues. J. Virol. Methods 2007, 145, 96–105. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, G.P.; Wang, Z.Q.; Yang, F.; Wu, G.W.; Hong, N. Identification of differentially expressed genes in response to infection of a mild Citrus tristeza virus isolate in Citrus aurantifolia by suppression subtractive hybridization. Sci. Hortic. 2012, 134, 144–149. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Albiach-Marti, M.R.; Robertson, C.; Gowda, S.; Tatineni, S.; Belliure, B.; Garnsey, S.M.; Folimonova, S.Y.; Moreno, P.; Dawson, W.O. The pathogenicity determinant of Citrus tristeza virus causing the seedling yellows syndrome maps at the 3’-terminal region of the viral genome. Mol. Plant Pathol. 2010, 11, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Roy, G.; Sudarshana, M.R.; Ullman, D.E.; Ding, S.W.; Dandekar, A.M.; Falk, B.W. Chimeric cDNA sequences from Citrus tristeza virus confer RNA silencing-mediated resistance in transgenic Nicotiana benthamiana plants. Phytopathology 2006, 96, 819–827. [Google Scholar] [CrossRef]
- Heinlein, M. Plasmodesmata: Channels for viruses on the move. Methods Mol. Biol. 2015, 1217, 25–52. [Google Scholar] [CrossRef] [PubMed]
- Tilsner, J.; Linnik, O.; Louveaux, M.; Roberts, I.M.; Chapman, S.N.; Oparka, K.J. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 2013, 201, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Lucas, W.J. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 2006, 344, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Tilsner, J.; Oparka, K.J. Missing links?—The connection between replication and movement of plant RNA viruses. Curr. Opin. Virol. 2012, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Peña, E.J.; Heinlein, M. RNA transport during TMV cell-to-cell movement. Front. Plant Sci. 2012, 28, 193. [Google Scholar] [CrossRef] [PubMed]
- Harries, P.A.; Park, J.W.; Sasaki, N.; Ballard, K.D.; Maule, A.J.; Nelson, R.S. Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc. Natl. Acad. Sci. USA 2009, 106, 17594–17599. [Google Scholar] [CrossRef]
- Cui, X.Y.; Wei, T.Y.; Chowda-Reddy, R.V.; Sun, G.Y.; Wang, A.M. The Tobacco etch virus P3 protein forms mobile inclusions via the early secretory pathway and traffics along actin microfilaments. Virology 2010, 397, 56–63. [Google Scholar] [CrossRef]
- Wei, T.Y.; Zhang, C.W.; Hong, J.; Xiong, R.Y.; Kasschau, K.D.; Zhou, X.P.; Carrington, J.C.; Wang, A.M. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 2010, 6, e1000962. [Google Scholar] [CrossRef]
- Harries, P.A.; Palanichelvam, K.; Yu, W.C.; Schoelz, J.E.; Nelson, R.S. The Cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol. 2009, 149, 1005–1016. [Google Scholar] [CrossRef]
- Feng, Z.K.; Chen, X.J.; Bao, Y.Q.; Dong, J.H.; Zhang, Z.K.; Tao, X.R. Nucleocapsid of Tomato spotted wilt tospovirus forms mobile particles that traffic on an actin/endoplasmic reticulum network driven by myosin XI-K. New Phytol. 2013, 200, 1212–1224. [Google Scholar] [CrossRef]
- Heinlein, M. Plant virus replication and movement. Virology 2015, 479–480, 657–671. [Google Scholar] [CrossRef]
- Scholthof,, H.B. Plant virus transport: Motions of functional equivalence. Trends Plant Sci. 2005, 10, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Alzhanova, D.V.; Hagiwara, Y.; Peremyslov, V.V.; Dolja, V.V. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology 2000, 268, 192–200. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, A.Y.S.; Peng, J.H.C.; Polek, M.; Tian, T.; Ludman, M.; Fátyol, K.; Ng, J.C.K. Comparative analysis identifies amino acids critical for citrus tristeza virus (T36CA) encoded proteins involved in suppression of RNA silencing and differential systemic infection in two plant species. Mol. Plant Pathol. 2021, 22, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Fagoaga, C.; Pensabene-Bellavia, G.; Moreno, P.; Navarro, L.; Flores, R.; Peña, L. Ectopic expression of the p23 silencing suppressor of Citrus tristeza virus differentially modifies viral accumulation and tropism in two transgenic woody hosts. Mol. Plant Pathol. 2011, 12, 898–910. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Zhang, Y.; Wang, G.; Wen, S.; Wang, Y.; Li, L.; Xiao, F.; Hong, N. The p23 of Citrus Tristeza Virus Interacts with Host FKBP-Type Peptidyl-Prolylcis-Trans Isomerase 17-2 and Is Involved in the Intracellular Movement of the Viral Coat Protein. Cells 2021, 10, 934. https://doi.org/10.3390/cells10040934
Yang Z, Zhang Y, Wang G, Wen S, Wang Y, Li L, Xiao F, Hong N. The p23 of Citrus Tristeza Virus Interacts with Host FKBP-Type Peptidyl-Prolylcis-Trans Isomerase 17-2 and Is Involved in the Intracellular Movement of the Viral Coat Protein. Cells. 2021; 10(4):934. https://doi.org/10.3390/cells10040934
Chicago/Turabian StyleYang, Zuokun, Yongle Zhang, Guoping Wang, Shaohua Wen, Yanxiang Wang, Liu Li, Feng Xiao, and Ni Hong. 2021. "The p23 of Citrus Tristeza Virus Interacts with Host FKBP-Type Peptidyl-Prolylcis-Trans Isomerase 17-2 and Is Involved in the Intracellular Movement of the Viral Coat Protein" Cells 10, no. 4: 934. https://doi.org/10.3390/cells10040934
APA StyleYang, Z., Zhang, Y., Wang, G., Wen, S., Wang, Y., Li, L., Xiao, F., & Hong, N. (2021). The p23 of Citrus Tristeza Virus Interacts with Host FKBP-Type Peptidyl-Prolylcis-Trans Isomerase 17-2 and Is Involved in the Intracellular Movement of the Viral Coat Protein. Cells, 10(4), 934. https://doi.org/10.3390/cells10040934




