Hypertrophy-Reduced Autophagy Causes Cardiac Dysfunction by Directly Impacting Cardiomyocyte Contractility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Echocardiography
2.3. Blood-Pressure Measurement
2.4. Immunohistochemistry
2.5. Lysosomal Activity (Cathepsin D&E Activity)
2.6. Immunoblot Analyses
2.7. Cardiomyocyte Culture and Treatment
2.8. Real-Time qPCR
2.9. Atg5 Silencing
2.10. Measurement of Cardiomyocyte Contractility
2.11. Statistics
3. Results
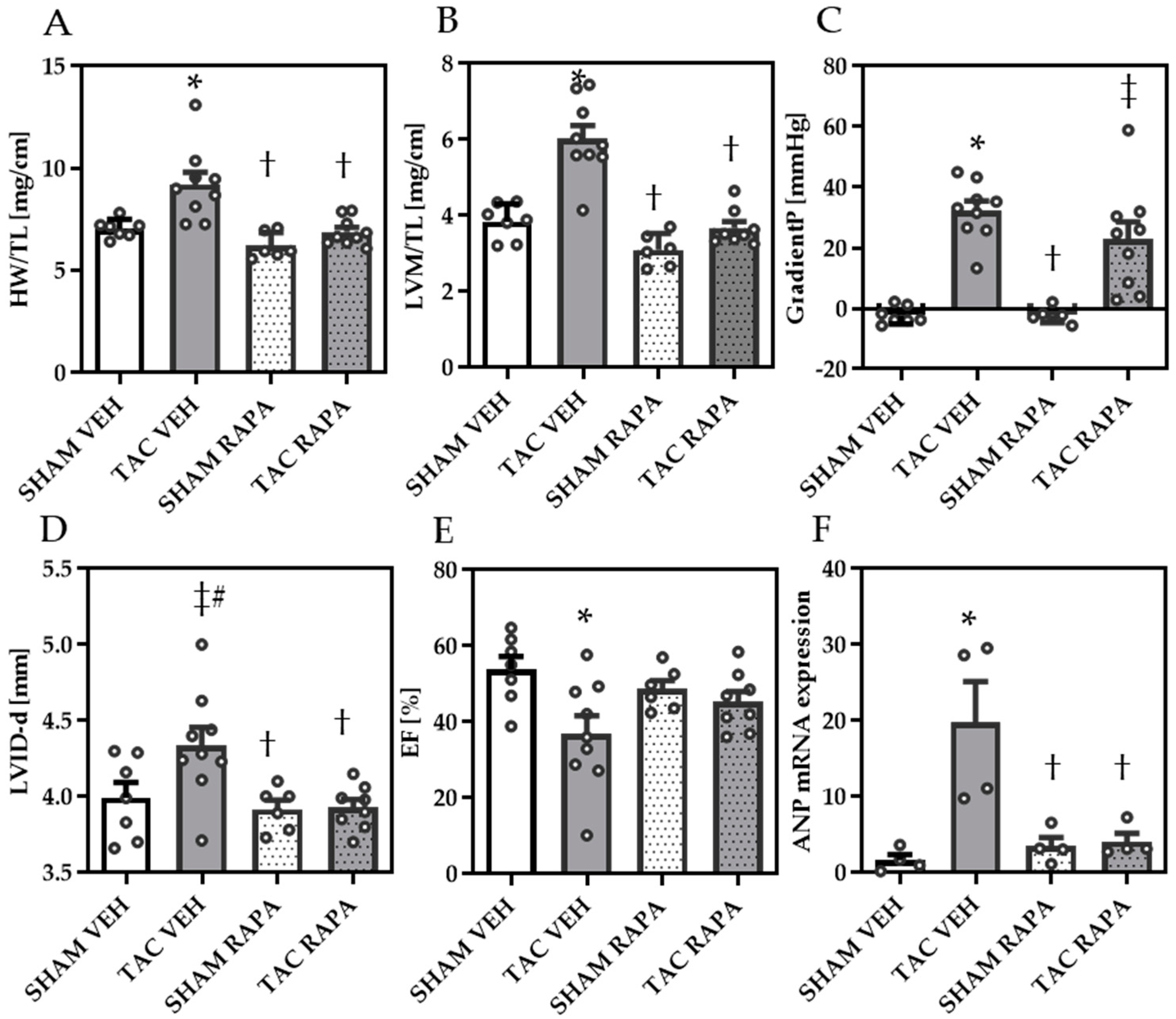
3.1. RAPA-Treatment Improves LV Dysfunction in TAC Mice
3.2. RAPA Attenuated Cardiac Remodeling in Pressure-Overloaded Mice
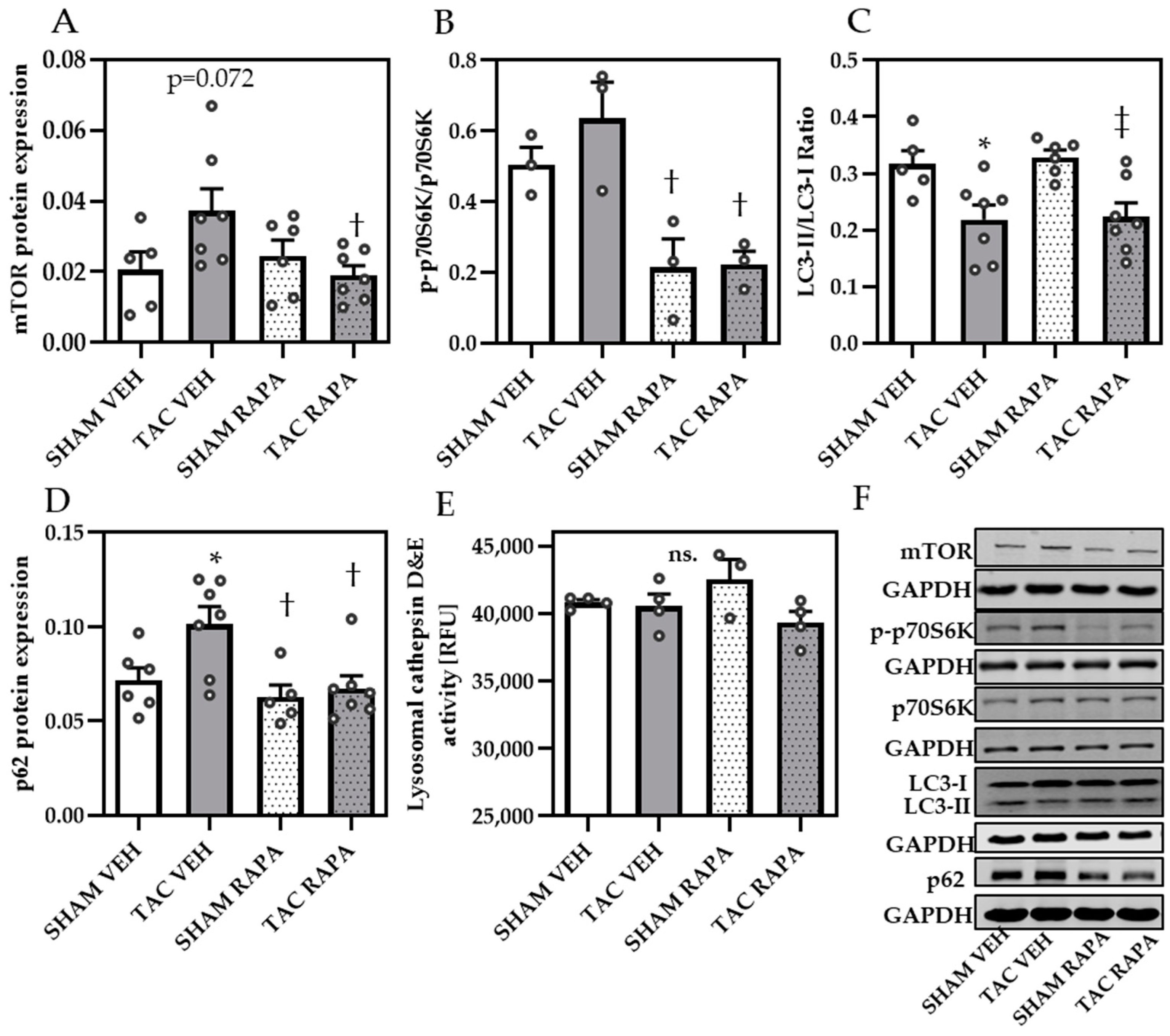
3.3. RAPA Treatment Leads to Improved Autophagy in Hypertrophic Conditions
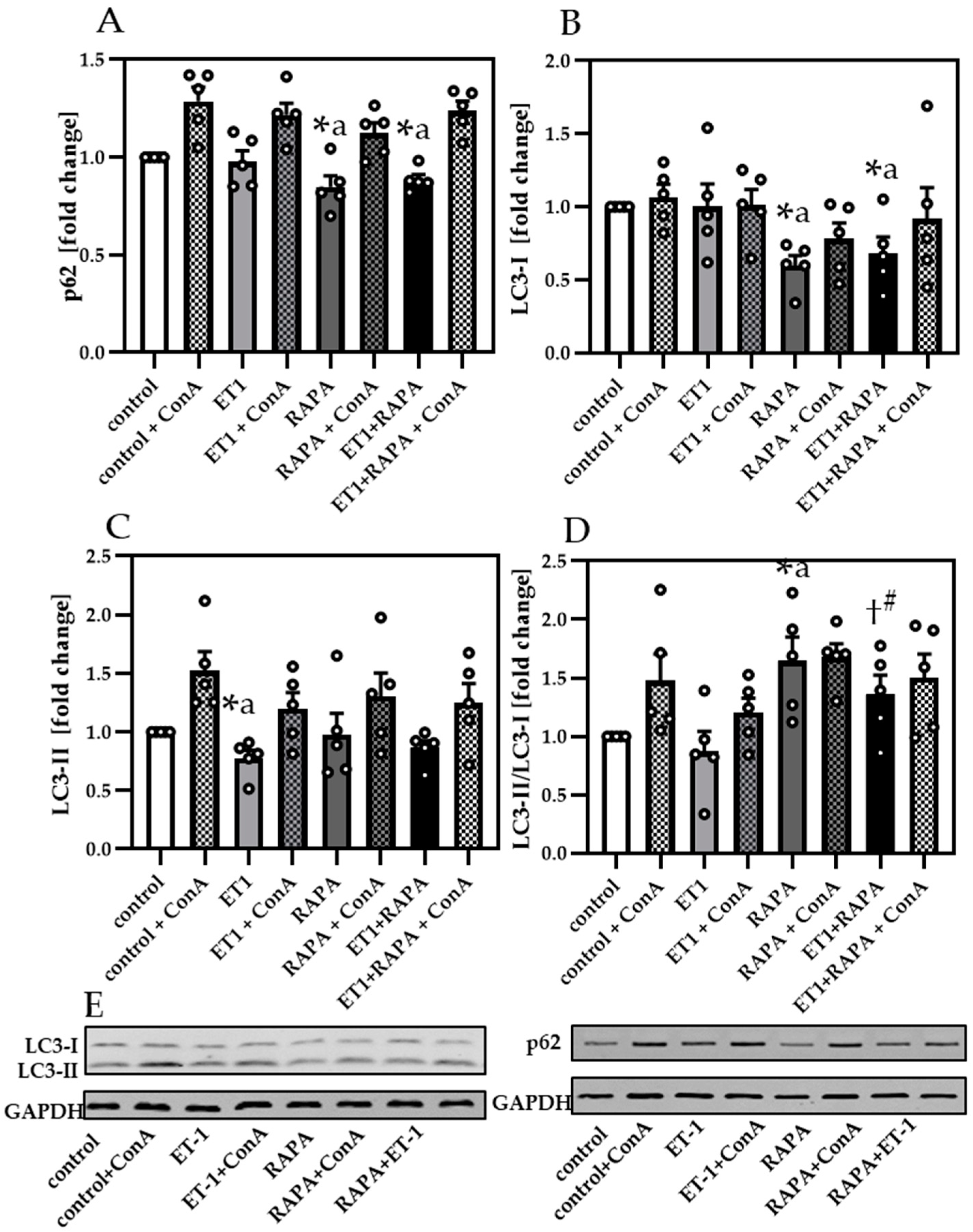
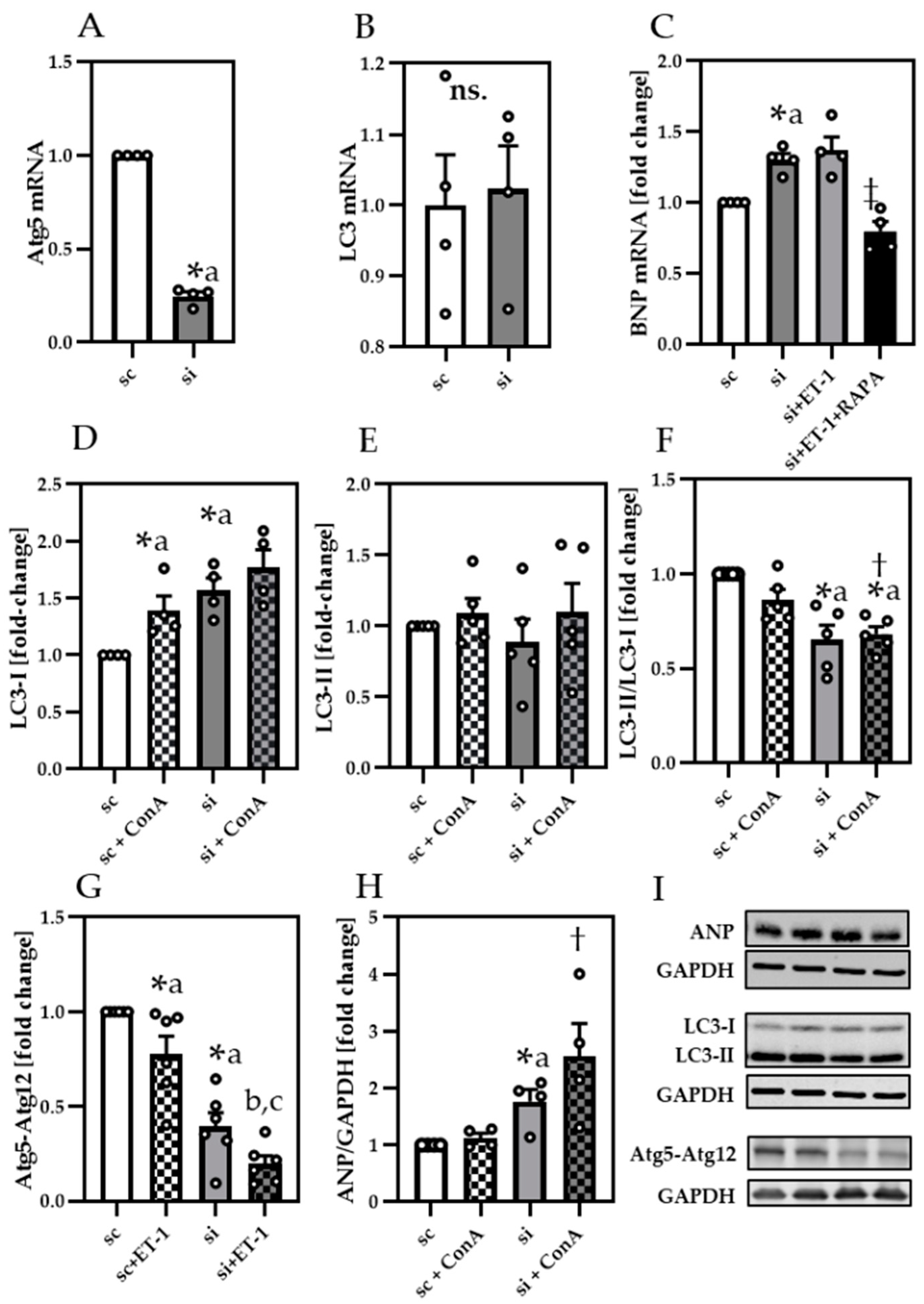
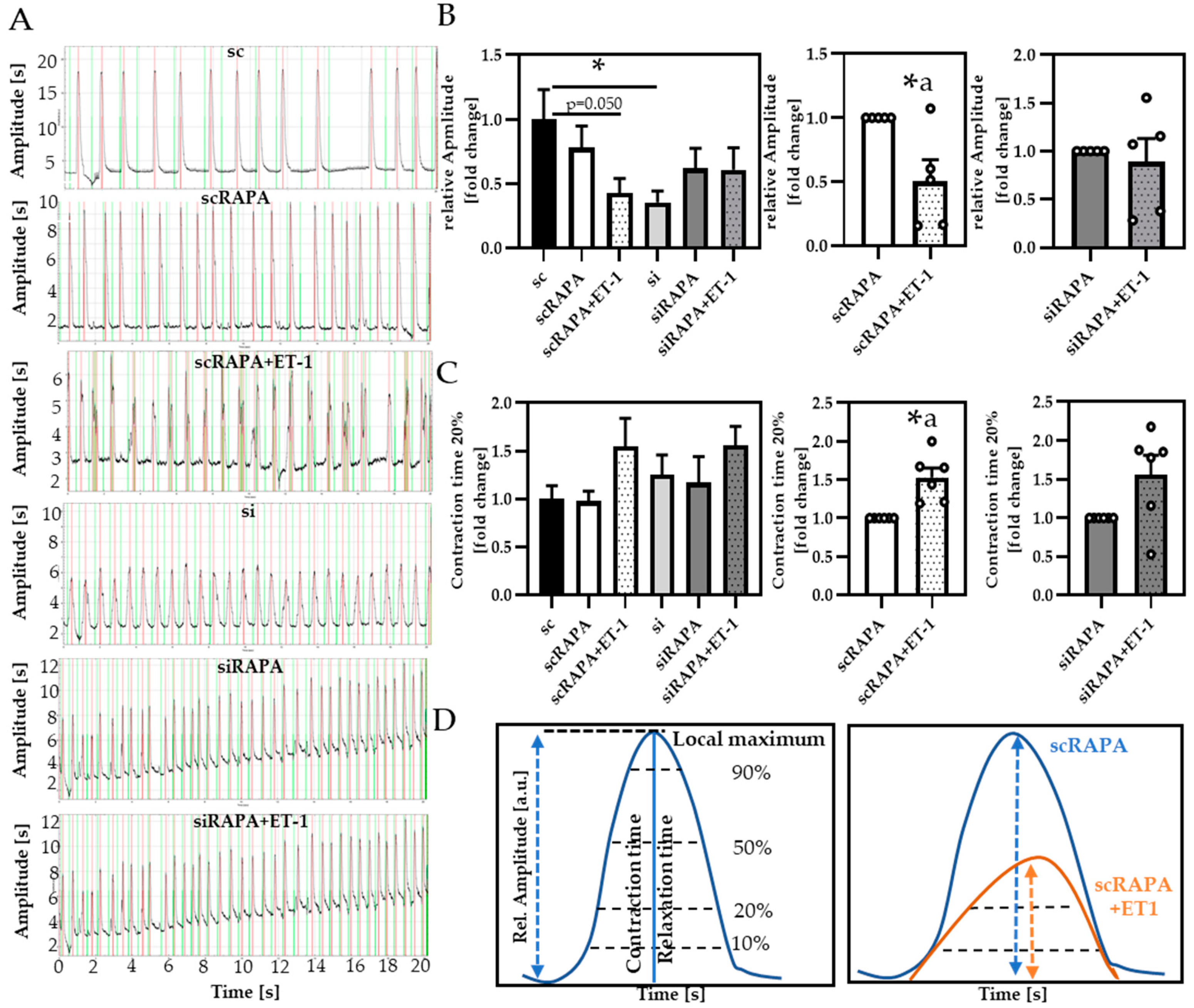
3.4. Impaired Autophagy Led to Decreased Cardiomyocyte Contractility
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Virani, S.S.; Alonso, A.; Aparicio, H.J.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation 2017, 135, e146–e603. [Google Scholar]
- Rame, J.E.; Dries, D.L. Heart failure and cardiac hypertrophy. Curr. Treat. Options Cardiovasc. Med. 2007, 9, 289–301. [Google Scholar] [CrossRef]
- Taegtmeyer, H.; Sen, S.; Vela, D. Return to the fetal gene program: A suggested metabolic link to gene expression in the heart. Ann. N. Y. Acad. Sci. 2010, 1188, 191–198. [Google Scholar] [CrossRef] [Green Version]
- Dai, D.F.; Hsieh, E.J.; Liu, Y.; Chen, T.; Beyer, R.P.; Chin, M.T.; MacCoss, M.J.; Rabinovitch, P.S. Mitochondrial proteome remodelling in pressure overload-induced heart failure: The role of mitochondrial oxidative stress. Cardiovasc. Res. 2012, 93, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Nakai, A.; Yamaguchi, O.; Takeda, T.; Higuchi, Y.; Hikoso, S.; Taniike, M.; Omiya, S.; Mizote, I.; Matsumura, Y.; Asahi, M.; et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007, 13, 619–624. [Google Scholar] [CrossRef]
- Lampert, M.A.; Gustafsson, Å.B. Balancing autophagy for a healthy heart. Curr. Opin. Physiol. 2018, 1, 21–26. [Google Scholar] [CrossRef]
- Gottlieb, R.A.; Mentzer, R.M. Autophagy: An affair of the heart. Heart Fail. Rev. 2013, 18, 575–584. [Google Scholar] [CrossRef] [Green Version]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef] [PubMed]
- Höhn, A.; Weber, D.; Jung, T.; Ott, C.; Hugo, M.; Kochlik, B.; Kehm, R.; König, J.; Grune, T.; Castro, J.P. Happily (n)ever after: Aging in the context of oxidative stress, proteostasis loss and cellular senescence. Redox Biol. 2017, 11, 482–501. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Arozena, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Lawrence, J.C.; Sturgill, T.W.; Harris, T.E. Mammalian Target of Rapamycin Complex 1 (mTORC1) Activity Is Associated with Phosphorylation of Raptor by mTOR. J. Biol. Chem. 2009, 284, 14693–14697. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Huang, S. The complexes of mammalian target of rapamycin. Curr. Protein Pept. Sci. 2010, 11, 409–424. [Google Scholar] [CrossRef] [Green Version]
- Lamming, D.W. Inhibition of the Mechanistic Target of Rapamycin (mTOR)-Rapamycin and Beyond. Cold Spring Harb. Perspect. Med. 2016, 6, a025924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef]
- Parzych, K.R.; Klionsky, D.J. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otomo, C.; Metlagel, Z.; Takaesu, G.; Otomo, T. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013, 20, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef] [Green Version]
- Ichimura, Y.; Kumanomidou, T.; Sou, Y.-S.; Mizushima, T.; Ezaki, J.; Ueno, T.; Kominami, E.; Yamane, T.; Tanaka, K.; Komatsu, M. Structural Basis for Sorting Mechanism of p62 in Selective Autophagy. J. Biol. Chem. 2008, 283, 22847–22857. [Google Scholar] [CrossRef] [Green Version]
- DeAlmeida, A.C.; Van Oort, R.J.; Wehrens, X.H.T. Transverse Aortic Constriction in Mice. J. Vis. Exp. 2010, 10, e1729. [Google Scholar] [CrossRef]
- Zhu, H.; Tannous, P.; Johnstone, J.L.; Kong, Y.; Shelton, J.M.; Richardson, J.A.; Le, V.; Levine, B.; Rothermel, B.A.; Hill, J.A. Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. Investig. 2007, 117, 1782–1793. [Google Scholar] [CrossRef]
- Matsui, Y.; Takagi, H.; Qu, X.; Abdellatif, M.; Sakoda, H.; Asano, T.; Levine, B.; Sadoshima, J. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ. Res. 2007, 100, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, X.; Hu, X.; Fassett, J.; Zhu, G.; Tao, Y.; Li, J.; Huang, Y.; Zhang, P.; Zhao, B.; et al. PGC-1α Regulates Expression of Myocardial Mitochondrial Antioxidants and Myocardial Oxidative Stress After Chronic Systolic Overload. Antioxid. Redox Signal. 2010, 13, 1011–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Xu, X.; Hu, X.; Lee, S.; Traverse, J.H.; Zhu, G.; Fassett, J.; Tao, Y.; Zhang, P.; dos Remedios, C.; et al. Oxidative Stress Regulates Left Ventricular PDE5 Expression in the Failing Heart. Circulation 2010, 121, 1474–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grune, J.; Blumrich, A.; Brix, S.; Jeuthe, S.; Drescher, C.; Grune, T.; Foryst-Ludwig, A.; Messroghli, D.; Kuebler, W.M.; Ott, C.; et al. Evaluation of a commercial multi-dimensional echocardiography technique for ventricular volumetry in small animals. Cardiovasc. Ultrasound 2018, 16, 1–13. [Google Scholar] [CrossRef]
- Garcia-Menendez, L.; Karamanlidis, G.; Kolwicz, S.; Tian, R. Substrain specific response to cardiac pressure overload in C57BL/6 mice. Am. J. Physiol. Circ. Physiol. 2013, 305, H397–H402. [Google Scholar] [CrossRef] [Green Version]
- Widdop, R.E.; Li, X.C. A Simple Versatile Method for Measuring Tail Cuff Systolic Blood Pressure in Conscious Rats. Clin. Sci. 1997, 93, 191–194. [Google Scholar] [CrossRef] [Green Version]
- Beyhoff, N.; Brix, S.; Betz, I.R.; Klopfleisch, R.; Foryst-Ludwig, A.; Krannich, A.; Stawowy, P.; Knebel, F.; Grune, J.; Kintscher, U. Application of Speckle-Tracking Echocardiography in an Experimental Model of Isolated Subendocardial Damage. J. Am. Soc. Echocardiogr. 2017, 30, 1239–1250.e2. [Google Scholar] [CrossRef]
- Kehm, R.; König, J.; Nowotny, K.; Jung, T.; Deubel, S.; Gohlke, S.; Schulz, T.J.; Höhn, A. Age-related oxidative changes in pancreatic islets are predominantly located in the vascular system. Redox Biol. 2018, 15, 387–393. [Google Scholar] [CrossRef]
- Grune, T.; Ott, C.; Haseli, S.; Hohn, A.; Jung, T. The “MYOCYTER”—Convert cellular and cardiac contractions into numbers with Image. Sci. Rep. 2019, 9, 15112. [Google Scholar] [CrossRef] [Green Version]
- Takagi, H.; Matsui, Y.; Hirotani, S.; Sakoda, H.; Asano, T.; Sadoshima, J. AMPK mediates autophagy during myocardial ischemia in vivo. Autophagy 2007, 3, 405–407. [Google Scholar] [CrossRef] [Green Version]
- Lavandero, S.; Troncoso, R.; Rothermel, B.A.; Martinet, W.; Sadoshima, J.; Hill, J.A. Cardiovascular autophagy: Concepts, controversies, and perspectives. Autophagy 2013, 9, 1455–1466. [Google Scholar] [CrossRef] [Green Version]
- De Meyer, G.R.Y.; De Keulenaer, G.W.; Martinet, W. Role of autophagy in heart failure associated with aging. Heart Fail. Rev. 2010, 15, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Häseli, S.; Deubel, S.; Jung, T.; Grune, T.; Ott, C. Cardiomyocyte Contractility and Autophagy in a Premature Senescence Model of Cardiac Aging. Oxidative Med. Cell. Longev. 2020, 2020, 1–14. [Google Scholar] [CrossRef]
- Meyer, K.; Hodwin, B.; Ramanujam, D.; Engelhardt, S.; Sarikas, A. Essential Role for Premature Senescence of Myofibroblasts in Myocardial Fibrosis. J. Am. Coll. Cardiol. 2016, 67, 2018–2028. [Google Scholar] [CrossRef]
- Bishu, K.; Ogut, O.; Kushwaha, S.; Mohammed, S.F.; Ohtani, T.; Xu, X.; Brozovich, F.V.; Redfield, M.M. Anti-Remodeling Effects of Rapamycin in Experimental Heart Failure: Dose Response and Interaction with Angiotensin Receptor Blockade. PLoS ONE 2013, 8, e81325. [Google Scholar] [CrossRef] [PubMed]
- Sala-Mercado, J.A.; Wider, J.; Undyala, V.V.R.; Jahania, S.; Yoo, W.; Mentzer, J.R.M.; Gottlieb, R.A.; Przyklenk, K. Profound Cardioprotection With Chloramphenicol Succinate in the Swine Model of Myocardial Ischemia-Reperfusion Injury. Circulation 2010, 122, S179–S184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirakabe, A.; Zhai, P.; Ikeda, Y.; Saito, T.; Maejima, Y.; Hsu, C.-P.; Nomura, M.; Egashira, K.; Levine, B.; Sadoshima, J. Drp1-Dependent Mitochondrial Autophagy Plays a Protective Role Against Pressure Overload–Induced Mitochondrial Dysfunction and Heart Failure. Circulation 2016, 133, 1249–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, B.C.; Tyagi, N.; Ovechkin, A.; Kartha, G.K.; Moshal, K.S.; Tyagi, S.C. Oxidative remodeling in pressure overload induced chronic heart failure. Eur. J. Heart Fail. 2007, 9, 450–457. [Google Scholar] [CrossRef]
- Tan, J.M.; Wong, E.S.; Kirkpatrick, D.S.; Pletnikova, O.; Ko, H.S.; Tay, S.-P.; Ho, M.W.; Troncoso, J.; Gygi, S.P.; Lee, M.K.; et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 2008, 17, 431–439. [Google Scholar] [CrossRef]
- Shaid, S.; Brandts, C.H.; Serve, H.; Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 2013, 20, 21–30. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kominami, E.; Tanaka, K.; Komatsu, M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy 2008, 4, 1063–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flynn, J.M.; O’Leary, M.N.; Zambataro, C.A.; Academia, E.C.; Presley, M.P.; Garrett, B.J.; Zykovich, A.; Mooney, S.D.; Strong, R.; Rosen, C.J.; et al. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell 2013, 12, 851–862. [Google Scholar] [CrossRef]
- Urfer, S.R.; Kaeberlein, T.L.; Mailheau, S.; Bergman, P.J.; Creevy, K.E.; Promislow, D.E.L.; Kaeberlein, M. A randomized controlled trial to establish effects of short-term rapamycin treatment in 24 middle-aged companion dogs. GeroScience 2017, 39, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Lagerqvist, E.; Finnin, B.; Pouton, C.; Haynes, J. Endothelin-1 and angiotensin II modulate rate and contraction amplitude in a subpopulation of mouse embryonic stem cell-derived cardiomyocyte-containing bodies. Stem Cell Res. 2011, 6, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Ceylan-Isik, A.F.; Dong, M.; Zhang, Y.; Dong, F.; Turdi, S.; Nair, S.; Yanagisawa, M.; Ren, J. Cardiomyocyte-specific deletion of endothelin receptor A rescues aging-associated cardiac hypertrophy and contractile dysfunction: Role of autophagy. Basic Res. Cardiol. 2013, 108, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, R.; Wang, Y.; Li, J.; Wu, J.; Wang, W.; Li, B.; Sun, C.; Wang, Z.; Shi, C.; Zhou, Y.; et al. Rapamycin Induced Autophagy Inhibits Inflammation-Mediated Endplate Degeneration by Enhancing Nrf2/Keap1 Signaling of Cartilage Endplate Stem Cells. Stem Cells 2019, 37, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Giordano, A.; Avellino, R.; Ferraro, P.; Romano, S.; Corcione, N.; Romano, M.F. Rapamycin antagonizes NF-kappaB nuclear translocation activated by TNF-alpha in primary vascular smooth muscle cells and enhances apoptosis. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H2459–H2465. [Google Scholar] [CrossRef]
- Das, A.; Salloum, F.N.; Durrant, D.; Ockaili, R.; Kukreja, R.C. Rapamycin protects against myocardial ischemia–reperfusion injury through JAK2–STAT3 signaling pathway. J. Mol. Cell. Cardiol. 2012, 53, 858–869. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Qi, H.; Zhou, J.; Xu, S.; Gao, Y. P27 Protects Cardiomyocytes from Sepsis via Activation of Autophagy and Inhibition of Apoptosis. Med Sci. Monit. 2018, 24, 8565–8576. [Google Scholar] [CrossRef]
- Engedal, N.; Torgersen, M.L.; Guldvik, I.J.; Barfeld, S.J.; Bakula, D.; Sætre, F.; Hagen, L.K.; Patterson, J.B.; Proikas-Cezanne, T.; Seglen, P.O.; et al. Modulation of intracellular calcium homeostasis blocks autophagosome formation. Autophagy 2013, 9, 1475–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tykocki, N.R.; Watts, S.W. The interdependence of endothelin-1 and calcium: A review. Clin. Sci. 2010, 119, 361–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| VEH | RAPA | |||
|---|---|---|---|---|
| Cardiac Function and Dimension | SHAM | TAC | SHAM | TAC |
| No. of animals | 7 | 9 | 6 | 8 |
| HR (bpm) | 489 ± 22.2 | 512.5 ± 16.5 | 536.2 ± 13.3 | 529.6 ± 19.5 |
| LVM (mg) | 66.9 ± 7.9 | 105.2 ± 7.4 * | 52.5 ± 6.1 † | 64.3 ± 3.7 † |
| LVID-d (mm) | 4± ± 0.1 | 4.3 ± 0.1 | 3.9 ± 0.1b | 3.9 ± 0.1 † |
| LVID-s (mm) | 2.9 ± 0.2 | 3.6 ± 0.2 * | 3.0 ± 0.1 | 3.1 ± 0.1 |
| LVAW-d (mm) | 0.6 ± 0.04 | 0.8 ± 0.03 * | 0.5 ± 0.02 † | 0.6 ± 0.03 † |
| LVAW-s (mm) | 0.7 ± 0.1 | 1 ± 0.04 * | 0.7 ± 0.02 † | 0.8 ± 0.02 † |
| LVPW-d (mm) | 0.6 ± 0.02 | 0.7 ± 0.2 * | 0.5 ± 0.02*, † | 0.6 ± 0.1 † |
| LVPW-s (mm) | 0.8 ± 0.03 | 0.9 ± 0.03 | 0.7 ± 0.03 † | 0.8 ± 0.03 |
| EDV (µL) | 70 ± 4.4 | 85.5 ± 5.5 * | 66.8 ± 2.8 † | 67.2 ± 2.1 † |
| ESV (µL) | 33.2 ± 4.1 | 55.7 ± 7.6 * | 34.5 ± 2.9 | 36.9 ± 2.3 |
| SV (µL) | 36.9 ± 1.2 | 29.8 ± 3.1 | 32.3 ± 1.5 | 30.3 ± 1.9 |
| CO (mL/min) | 18.1 ± 1.1 | 15.1 ± 1.5 | 17.4 ± 0.9 | 15.9 ± 0.9 |
| EF (%) | 53.8 ± 3.1 | 36.9 ± 4.8 * | 48.7 ± 2.9 | 45.3 ± 2.7 |
| FS (%) | 27.7 ± 1.9 | 18 ± 2.5 * | 24.2 ± 0.8 | 22.3 ± 1.6 |
| Aortic vel. desc (mm/s) | 1199 ± 72 | 1268 ± 139 | 1143 ± 100 | 1097 ± 105 |
| Aortic vel. asc (mm/s) | −946 ± 74 | −3107 ± 142 * | −887 ± 62 † | −2543 ± 257 *, ‡ |
| Aortic Peak Pressure (mmHg) | 3.7 ± 0.6 | 39.3 ± 3.4 * | 3.2 ± 0.5 † | 28 ± 5.3 *, ‡ |
| Pressure Gradient P (mmHg) | −2.2 ± 1.1 | 32.2 ± 3.2 * | −2.2 ± 1 † | 22.8 ± 5.8 *, ‡ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ott, C.; Jung, T.; Brix, S.; John, C.; Betz, I.R.; Foryst-Ludwig, A.; Deubel, S.; Kuebler, W.M.; Grune, T.; Kintscher, U.; et al. Hypertrophy-Reduced Autophagy Causes Cardiac Dysfunction by Directly Impacting Cardiomyocyte Contractility. Cells 2021, 10, 805. https://doi.org/10.3390/cells10040805
Ott C, Jung T, Brix S, John C, Betz IR, Foryst-Ludwig A, Deubel S, Kuebler WM, Grune T, Kintscher U, et al. Hypertrophy-Reduced Autophagy Causes Cardiac Dysfunction by Directly Impacting Cardiomyocyte Contractility. Cells. 2021; 10(4):805. https://doi.org/10.3390/cells10040805
Chicago/Turabian StyleOtt, Christiane, Tobias Jung, Sarah Brix, Cathleen John, Iris R. Betz, Anna Foryst-Ludwig, Stefanie Deubel, Wolfgang M. Kuebler, Tilman Grune, Ulrich Kintscher, and et al. 2021. "Hypertrophy-Reduced Autophagy Causes Cardiac Dysfunction by Directly Impacting Cardiomyocyte Contractility" Cells 10, no. 4: 805. https://doi.org/10.3390/cells10040805
APA StyleOtt, C., Jung, T., Brix, S., John, C., Betz, I. R., Foryst-Ludwig, A., Deubel, S., Kuebler, W. M., Grune, T., Kintscher, U., & Grune, J. (2021). Hypertrophy-Reduced Autophagy Causes Cardiac Dysfunction by Directly Impacting Cardiomyocyte Contractility. Cells, 10(4), 805. https://doi.org/10.3390/cells10040805







