Nuclear Translocation of SRPKs Is Associated with 5-FU and Cisplatin Sensitivity in HeLa and T24 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and the Expression of Recombinant Proteins
- 326 Thr→Ala (s): 5′-GAACCCACCTAATAAAATGGCCCAAGAAAAACTTGAAGAGTCAAG-3′;
- 326 Thr→Ala (a): 5′-CTTGACTCTTCAAGTTTTTCTTGGGCCATTTTATTAGGTGGGTTC-3′;
- 326 Thr→Asp (s): 5′-GAACCCACCTAATAAAATGGACCAAGAAAAACTTGAAGAGTCAAG-3′;
- 326 Thr→Asp (a): 5′-CTTGACTCTTCAAGTTTTTCTTGGTCCATTTTATTAGGTGGGTTC-3′;
- 408 Ser→Ala (s): 5′-ATGGAGACAGCAGCACAGCTCAAGAAACAGACTC-3′;
- 408 Ser→Ala (a): 5′-GAGTCTGTTTCTTGAGCTGTGCTGCTGTCTCCAT-3′;
- 408 Ser→Asp (s): 5′-ATGGAGACAGCAGCACAGATCAAGAAACAGACTC-3′;
- 408 Ser→Asp (a): 5′-GAGTCTGTTTCTTGATCTGTGCTGCTGTCTCCAT-3′.
2.2. Cell Culture and Transfection and Drug Treatments
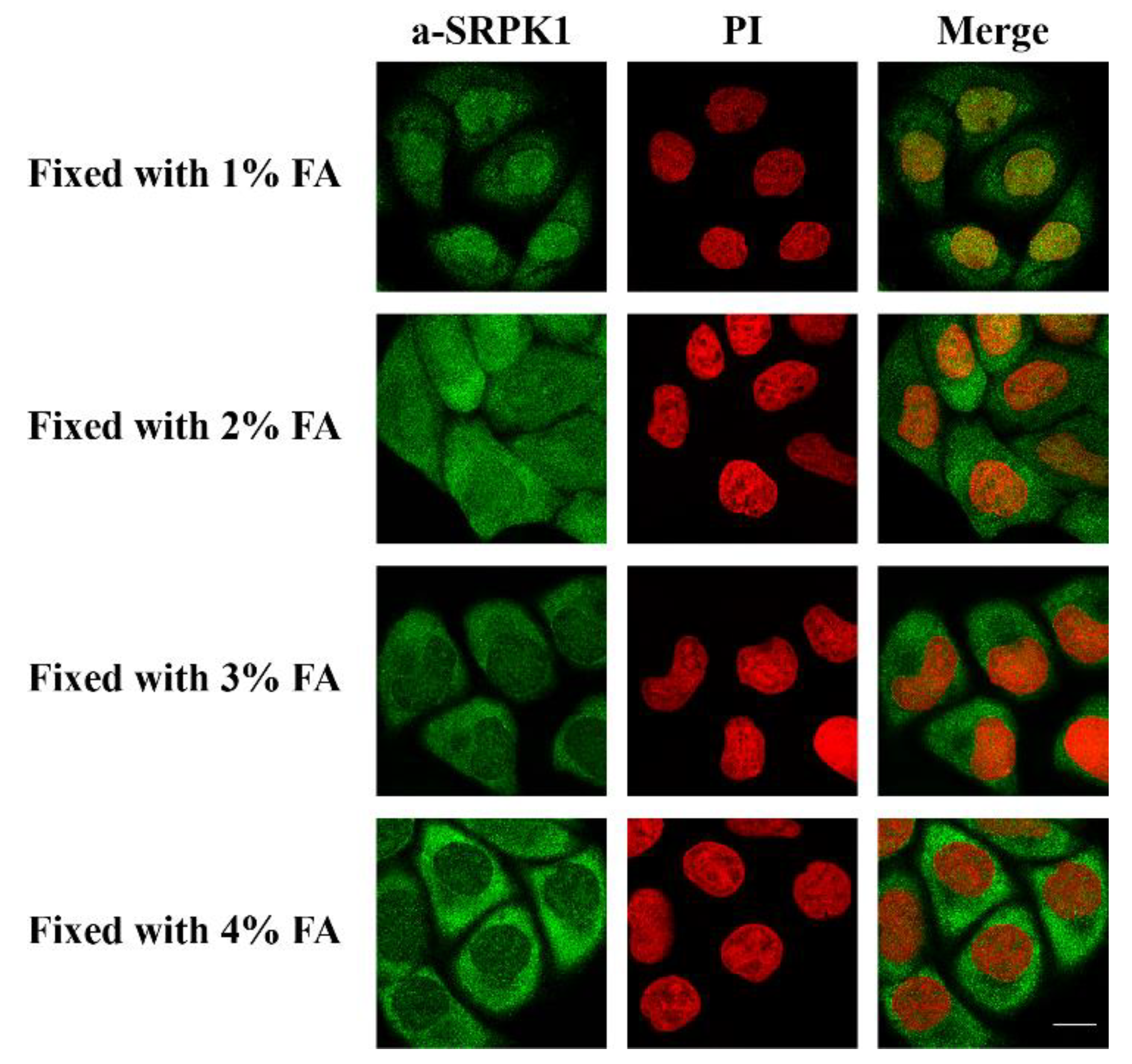
2.3. Immunofluorescence Microscopy
2.4. MTT Assays—Optical Microscopy
2.5. Cell Fractionation, SDS-PAGE, and Western Blotting
2.6. In Vitro Kinase Assays
3. Results
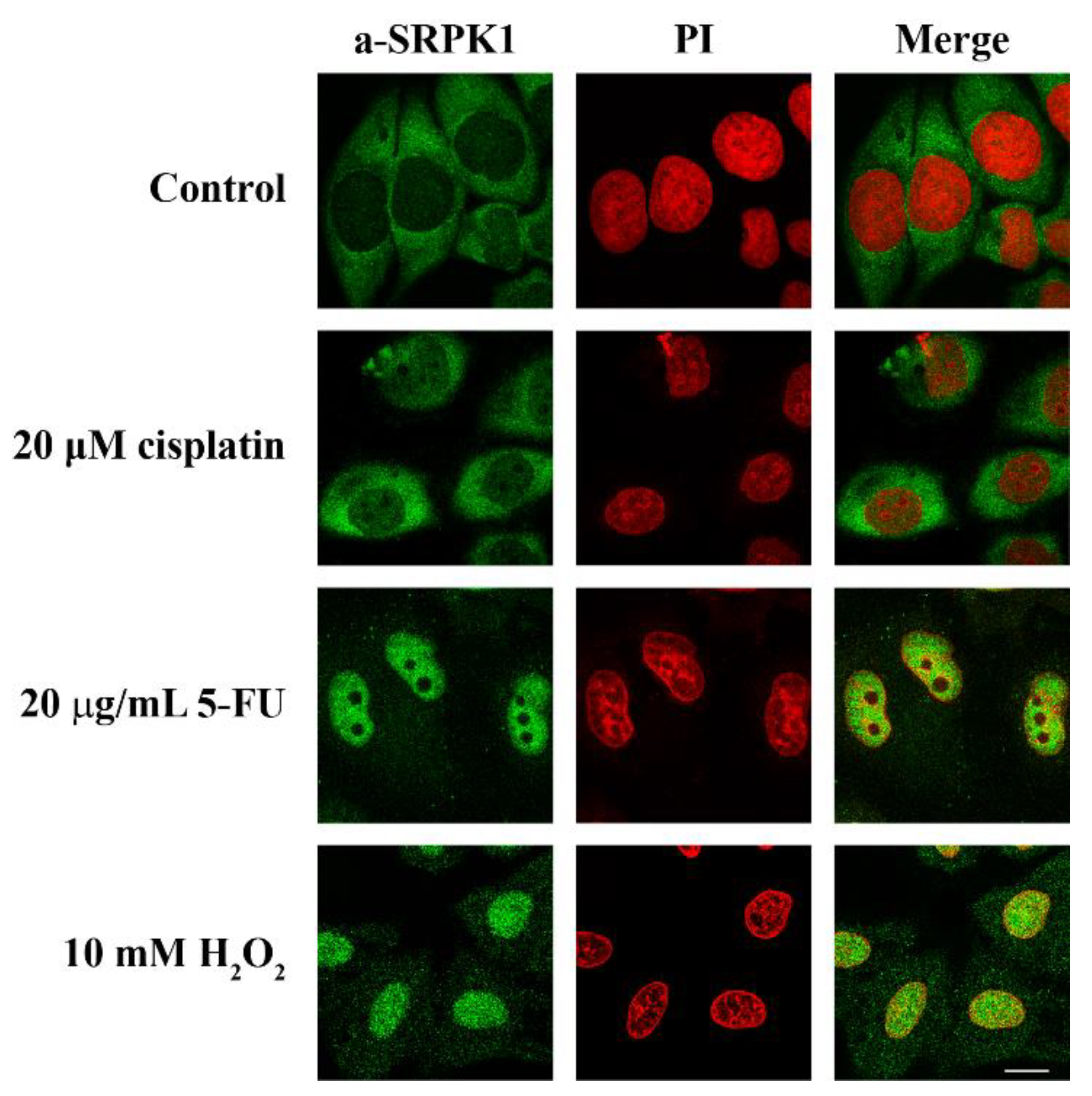
3.1. DNA Damage Inducers Trigger the Nuclear Translocation of SRPK1
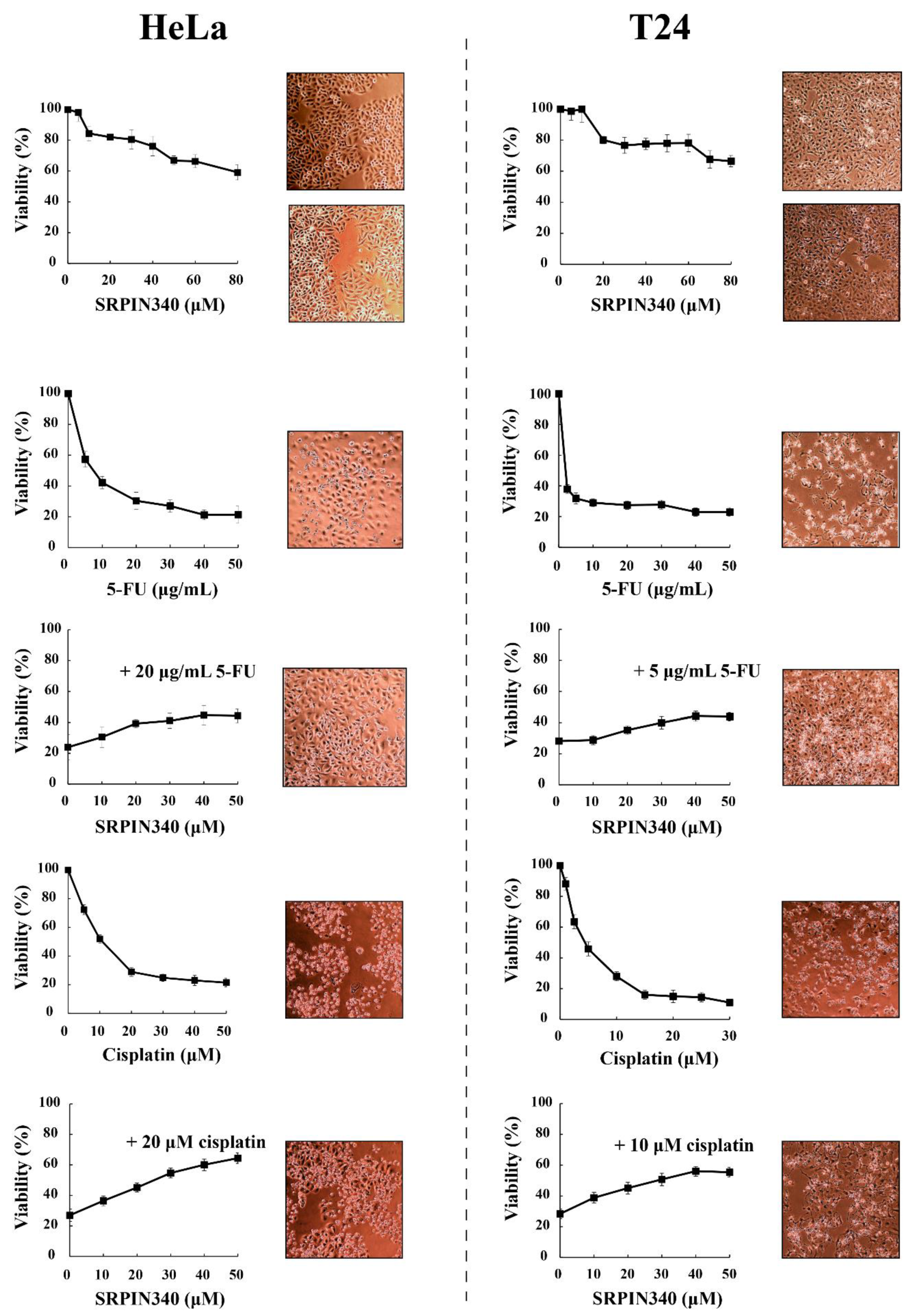
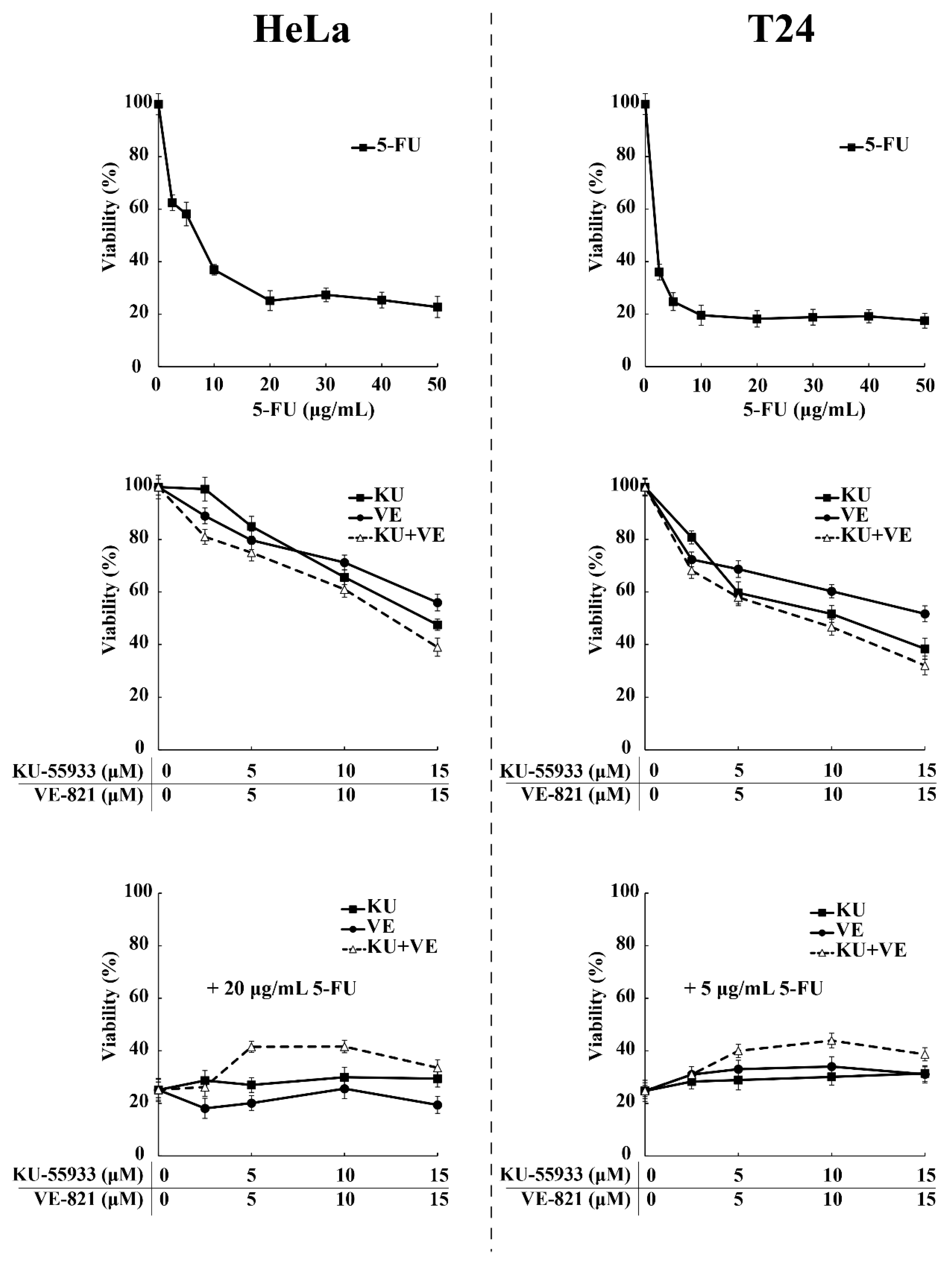
3.2. SRPIN340 Protected HeLa and T24 Cells from the Cytotoxic Effects of 5-FU and Cisplatin
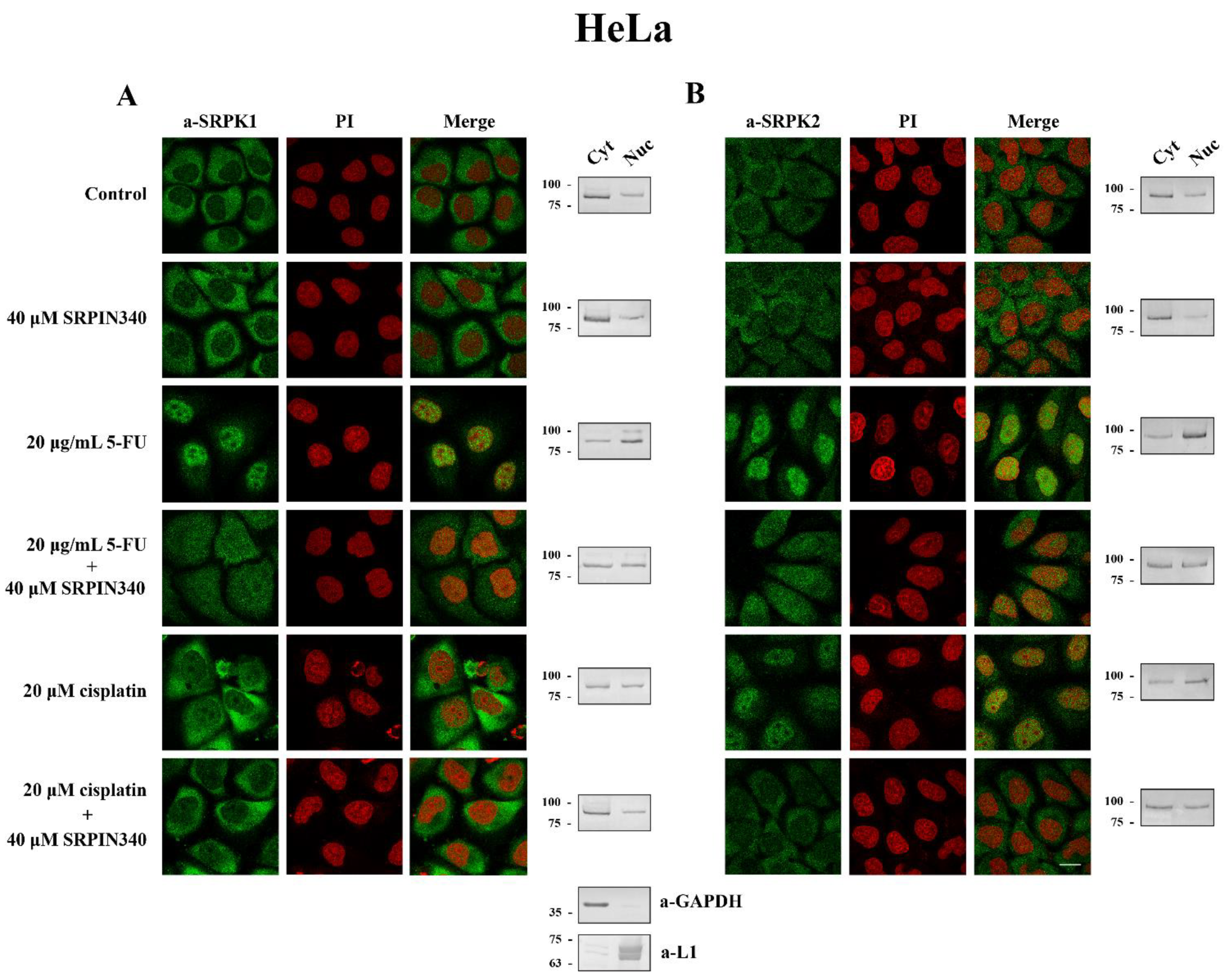
3.3. SRPIN340 Prevented the Nuclear Translocation of SRPK1 and SRPK2 in 5-FU- and Cisplatin-Treated Cells
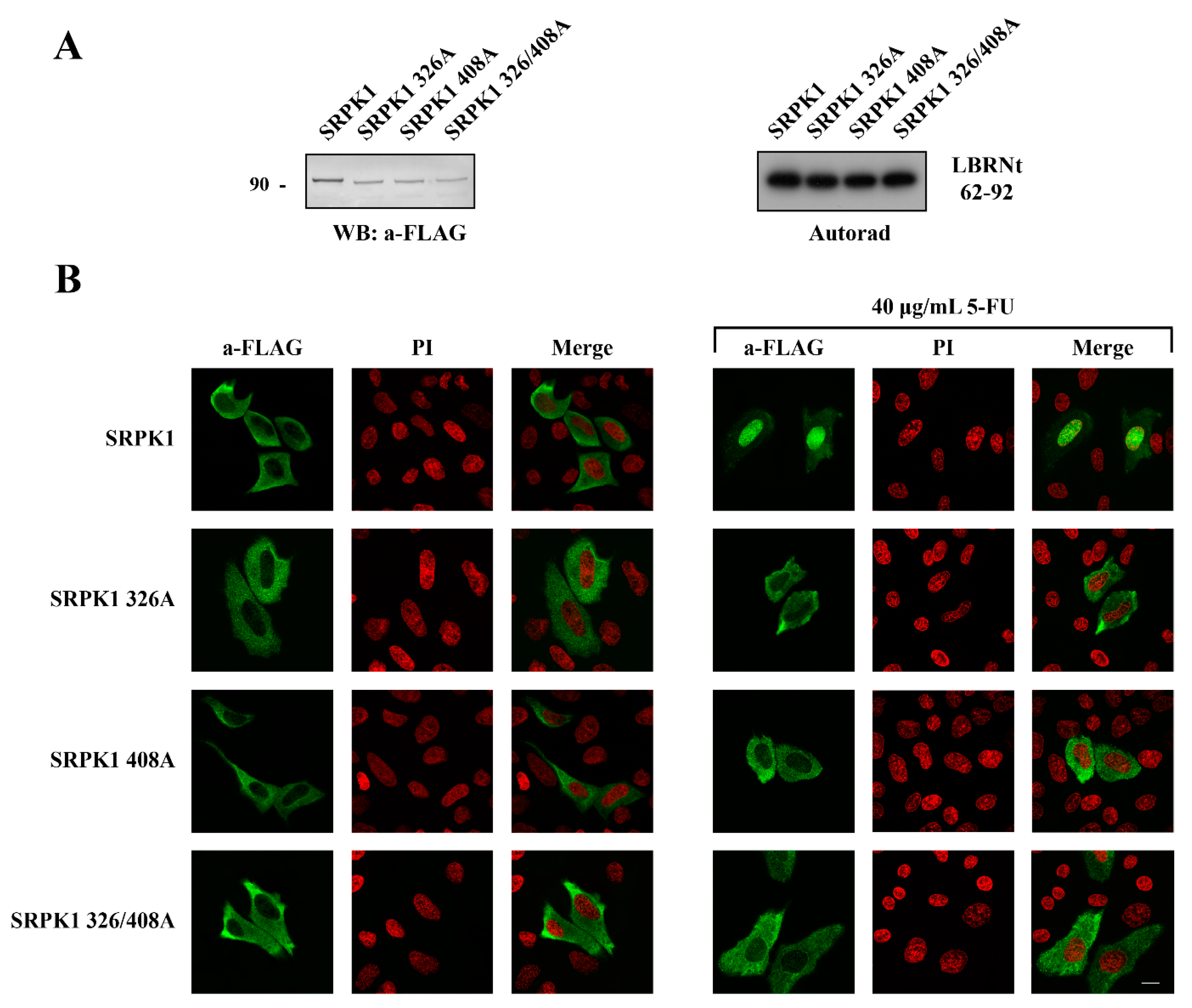
3.4. Phosphorylation of Thr326 and Ser408 Was Necessary but Not Sufficient for the Nuclear Translocation of SRPK1
3.5. ATR/ATM-Dependent Phosphorylation of Thr326 and Ser408
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Giannakouros, T.; Nikolakaki, E.; Mylonis, I.; Georgatsou, E. Serine-arginine protein kinases: A small protein kinase family with a large cellular presence. FEBS J. 2011, 278, 570–586. [Google Scholar] [CrossRef]
- Zhou, Z.; Fu, X.D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013, 122, 191–207. [Google Scholar] [CrossRef]
- Bullock, N.; Oltean, S. The many faces of SRPK1. J. Pathol. 2017, 241, 437–440. [Google Scholar] [CrossRef]
- Calarco, J.A.; Superina, S.; O’Hanlon, D.; Gabut, M.; Raj, B.; Pan, Q.; Skalska, U.; Clarke, L.; Gelinas, D.; van der Kooy, D.; et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 2009, 138, 898–910. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, G.; Adams, J.A. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011, 278, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Koutroumani, M.; Papadopoulos, G.E.; Vlassi, M.; Nikolakaki, E.; Giannakouros, T. Evidence for disulfide bonds in SR Protein Kinase 1 (SRPK1) that are required for activity and nuclear localization. PLoS ONE 2017, 12, e0171328. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.F.; Lane, W.S.; Fu, X.D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 1994, 369, 678–682. [Google Scholar] [CrossRef]
- Ding, J.H.; Zhong, X.Y.; Hagopian, J.C.; Cruz, M.M.; Ghosh, G.; Feramisco, J.; Adams, J.A.; Fu, X.D. Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol. Biol. Cell 2006, 17, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.Y.; Ding, J.H.; Adams, J.A.; Ghosh, G.; Fu, X.D. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009, 23, 482–495. [Google Scholar] [CrossRef]
- Siebel, C.W.; Feng, L.; Guthrie, C.; Fu, X.D. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5440–5445. [Google Scholar] [CrossRef]
- Zhou, Z.; Qiu, J.; Liu, W.; Zhou, Y.; Plocinik, R.M.; Li, H.; Hu, Q.; Ghosh, G.; Adams, J.A.; Rosenfeld, M.G.; et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell 2012, 47, 422–433. [Google Scholar] [CrossRef]
- Lee, G.; Zheng, Y.; Cho, S.; Jang, C.; England, C.; Dempsey, J.M.; Yu, Y.; Liu, X.; He, L.; Cavaliere, P.M.; et al. Post-transcriptional regulation of de novo lipogenesis by mTORC1-S6K1-SRPK2 signaling. Cell 2017, 171, 1545–1558. [Google Scholar] [CrossRef]
- Vivarelli, S.; Lenzken, S.C.; Ruepp, M.D.; Ranzini, F.; Maffioletti, A.; Alvarez, R.; Mühlemann, O.; Barabino, S.M. Paraquat modulates alternative pre-mRNA splicing by modifying the intracellular distribution of SRPK2. PLoS ONE 2013, 8, e61980. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Yanagida, M. A mitotic role for a novel fission yeast protein kinase dsk1 with cell cycle stage dependent phosphorylation and localization. Mol. Biol. Cell 1993, 4, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Hayes, G.M.; Carrigan, P.E.; Beck, A.M.; Miller, L.J. Targeting the RNA splicing machinery as a novel treatment strategy for pancreatic carcinoma. Cancer Res. 2006, 66, 3819–3827. [Google Scholar] [CrossRef]
- Hayes, G.M.; Carrigan, P.E.; Miller, L.J. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007, 67, 2072–2080. [Google Scholar] [CrossRef]
- Schenk, P.W.; Boersma, A.W.; Brandsma, J.A.; den Dulk, H.; Burger, H.; Stoter, G.; Brouwer, J.; Nooter, K. SKY1 is involved in cisplatin-induced cell kill in Saccharomyces cerevisiae and inactivation of its human homologue, SRPK1, induces cisplatin resistance in a human ovarian carcinoma cell line. Cancer Res. 2001, 61, 6982–6986. [Google Scholar]
- Schenk, P.W.; Stoop, H.; Bokemeyer, C.; Mayer, F.; Stoter, G.; Oosterhuis, J.W.; Wiemer, E.; Looijenga, L.H.; Nooter, K. Resistance to platinum-containing chemotherapy in testicular germ cell tumors is associated with downregulation of the protein kinase SRPK1. Neoplasia 2004, 6, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Plasencia, C.; Martínez-Balibrea, E.; Martinez-Cardús, A.; Quinn, D.I.; Abad, A.; Neamati, N. Expression analysis of genes involved in oxaliplatin response and development of oxaliplatin-resistant HT29 colon cancer cells. Int. J. Oncol. 2006, 29, 225–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krishnakumar, S.; Mohan, A.; Kandalam, M.; Ramkumar, H.L.; Venkatesan, N.; Das, R.R. SRPK1: A cisplatin sensitive protein expressed in retinoblastoma. Pediatr. Blood Cancer 2008, 50, 402–406. [Google Scholar] [CrossRef]
- Odunsi, K.; Mhawech-Fauceglia, P.; Andrews, C.; Beck, A.; Amuwo, O.; Lele, S.; Black, J.D.; Huang, R.Y. Elevated expression of the serine-arginine protein kinase 1 gene in ovarian cancer and its role in cisplatin cytotoxicity in vitro. PLoS ONE 2012, 7, e51030. [Google Scholar] [CrossRef]
- Edmond, V.; Moysan, E.; Khochbin, S.; Matthias, P.; Brambilla, C.; Brambilla, E.; Gazzeri, S.; Eymin, B. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 2011, 30, 510–523. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, Z.; Subhramanyam, C.S.; Cao, Q.; Heng, Z.S.L.; Liu, W.; Fu, X.; Hu, Q. SRPK1 acetylation modulates alternative splicing to regulate cisplatin resistance in breast cancer cells. Commun. Biol. 2020, 3, 1–13. [Google Scholar] [CrossRef]
- Boeing, S.; Williamson, L.; Encheva, V.; Gori, I.; Saunders, R.E.; Instrell, R.; Aygün, O.; Rodriguez-Martinez, M.; Weems, J.C.; Kelly, G.P.; et al. Multiomic analysis of the UV-induced DNA damage response. Cell Rep. 2016, 15, 1597–1610. [Google Scholar] [CrossRef]
- Nikolakaki, E.; Kohen, R.; Hartmann, A.M.; Stamm, S.; Georgatsou, E.; Giannakouros, T. Cloning and characterization of an alternatively spliced form of SR protein kinase 1 that interacts specifically with scaffold attachment factor-B. J. Biol. Chem. 2001, 276, 40175–40182. [Google Scholar] [CrossRef]
- Mylonis, I.; Giannakouros, T. Protein kinase CK2 phosphorylates and activates the SR protein-specific kinase 1. Biochem. Biophys. Res. Commun. 2003, 301, 650–656. [Google Scholar] [CrossRef]
- Voukkalis, N.; Koutroumani, M.; Zarkadas, C.; Nikolakaki, E.; Vlassi, M.; Giannakouros, T. SRPK1 and Akt protein kinases phosphorylate the RS domain of Lamin B Receptor with distinct specificity: A combined biochemical and in silico approach. PLoS ONE 2016, 11, e0154198. [Google Scholar] [CrossRef] [PubMed]
- Sigala, I.; Tsamis, K.I.; Gousia, A.; Alexiou, G.; Voulgaris, S.; Giannakouros, T.; Kyritsis, A.P.; Nikolakaki, E. Expression of SRPK1 in gliomas and its role in glioma cell lines viability. Tumour Biol. 2016, 37, 8699–8707. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Bose, P.; Leong-Quong, R.Y.; Fujita, D.J.; Riabowol, K. REAP: A two minute cell fractionation method. BMC Res. Notes 2010, 3, 1–6. [Google Scholar] [CrossRef]
- Törmänen Persson, H.; Aksaas, A.K.; Kvissel, A.K.; Punga, T.; Engström, Å.; Skålhegg, B.S.; Akusjärvi, G. Two cellular protein kinases, DNA-PK and PKA, phosphorylate the adenoviral L4-33K protein and have opposite effects on L1 alternative RNA splicing. PLoS ONE 2012, 7, e31871. [Google Scholar] [CrossRef]
- Sanidas, I.; Kotoula, V.; Ritou, E.; Daans, J.; Lenz, C.; Mairhofer, M.; Daniilidou, M.; Kolbus, A.; Kruft, V.; Ponsaerts, P.; et al. The ratio of SRPK1/SRPK1a regulates erythroid differentiation in K562 leukaemic cells. Biochim. Biophys. Acta 2010, 1803, 1319–13131. [Google Scholar] [CrossRef]
- Maison, C.; Horstmann, H.; Georgatos, S.D. Regulated docking of nuclear membrane vesicles to vimentin filaments during mitosis. J. Cell Biol. 1993, 123, 1491–1505. [Google Scholar] [CrossRef]
- Wang, H.Y.; Lin, W.; Dyck, J.A.; Yeakley, J.M.; Songyang, Z.; Cantley, L.C.; Fu, X.D. SRPK2: A differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol. 1998, 140, 737–750. [Google Scholar] [CrossRef]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef] [PubMed]
- Georgoulis, A.; Vorgias, C.E.; Chrousos, G.P.; Rogakou, E.P. Genome instability and γH2AX. Int. J. Mol. Sci. 2017, 18, 1979. [Google Scholar] [CrossRef]
- Fukuhara, T.; Hosoya, T.; Shimizu, S.; Sumi, K.; Oshiro, T.; Yoshinaka, Y.; Suzuki, M.; Yamamoto, N.; Herzenberg, L.A.; Herzenberg, L.A.; et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. USA 2006, 103, 11329–11333. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, M.B.; Leparc, G.G.; Lai, J.; Obata, T.; Volinia, S.; Cantley, L.C. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 2001, 19, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Jang, S.W.; Liu, X.; Fu, H.; Rees, H.; Yepes, M.; Levey, A.; Ye, K. Interaction of Akt-phosphorylated SRPK2 with 14-3-3 mediates cell cycle and cell death in neurons. J. Biol. Chem. 2009, 284, 24512–24525. [Google Scholar] [CrossRef] [PubMed]
- Moreira, G.A.; Lima, G.D.A.; Siqueira, R.P.; Barros, M.V.A.; Adjanohoun, A.L.M.; Santos, V.C.; Barbosa, É.A.A.; Loterio, R.K.; Paiva, J.C.; Gonçalves, V.H.S.; et al. Antimetastatic effect of the pharmacological inhibition of serine/arginine-rich protein kinases (SRPK) in murine melanoma. Toxicol. Appl. Pharmacol. 2018, 356, 214–223. [Google Scholar] [CrossRef]
- Huang, J.; Zhou, Y.; Xue, X.; Jiang, L.; Du, J.; Cui, Y.; Zhao, H. SRPIN340 protects heart muscle from oxidative damage via SRPK1/2 inhibition-mediated AKT activation. Biochem. Biophys. Res. Commun. 2019, 510, 97–103. [Google Scholar] [CrossRef]
- Lindenboim, L.; Zohar, H.; Worman, H.J.; Stein, R. The nuclear envelope: Target and mediator of the apoptotic process. Cell Death Discov. 2020, 6, 1–11. [Google Scholar] [CrossRef]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef]
- Nikolakaki, E.; Simos, G.; Georgatos, S.D.; Giannakouros, T. A nuclear envelope-associated kinase phosphorylates arginine-serine motifs and modulates interactions between the lamin B receptor and other nuclear proteins. J. Biol. Chem. 1996, 271, 8365–8372. [Google Scholar] [CrossRef] [PubMed]
- Sellis, D.; Drosou, V.; Vlachakis, D.; Voukkalis, N.; Giannakouros, T.; Vlassi, M. Phosphorylation of the arginine/serine repeats of lamin B receptor by SRPK1-insights from molecular dynamics simulations. Biochim. Biophys. Acta 2012, 1820, 44–55. [Google Scholar] [CrossRef]
- Makatsori, D.; Kourmouli, N.; Polioudaki, H.; Shultz, L.D.; McLean, K.; Theodoropoulos, P.A.; Singh, P.B.; Georgatos, S.D. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J. Biol. Chem. 2004, 279, 25567–25573. [Google Scholar] [CrossRef]
- Nikolakaki, E.; Drosou, V.; Sanidas, I.; Peidis, P.; Papamarcaki, T.; Iakoucheva, L.M.; Giannakouros, T. RNA association or phosphorylation of the RS domain prevents aggregation of RS domain-containing proteins. Biochim. Biophys. Acta 2008, 1780, 214–225. [Google Scholar] [CrossRef]
- Clever, M.; Funakoshi, T.; Mimura, Y.; Takagi, M.; Imamoto, N. The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells. Nucleus 2012, 3, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Mimura, Y.; Takagi, M.; Clever, M.; Imamoto, N. ELYS regulates the localization of LBR by modulating its phosphorylation state. J. Cell Sci. 2016, 129, 4200–4212. [Google Scholar] [CrossRef]
- Karnitz, L.M.; Zou, L. Molecular Pathways: Targeting ATR in Cancer Therapy. Clin. Cancer Res. 2015, 21, 4780–4785. [Google Scholar] [CrossRef]
- Sarkaria, J.N.; Busby, E.C.; Tibbetts, R.S.; Roos, P.; Taya, Y.; Karnitz, L.M.; Abraham, R.T. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999, 59, 4375–4382. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sigala, I.; Koutroumani, M.; Koukiali, A.; Giannakouros, T.; Nikolakaki, E. Nuclear Translocation of SRPKs Is Associated with 5-FU and Cisplatin Sensitivity in HeLa and T24 Cells. Cells 2021, 10, 759. https://doi.org/10.3390/cells10040759
Sigala I, Koutroumani M, Koukiali A, Giannakouros T, Nikolakaki E. Nuclear Translocation of SRPKs Is Associated with 5-FU and Cisplatin Sensitivity in HeLa and T24 Cells. Cells. 2021; 10(4):759. https://doi.org/10.3390/cells10040759
Chicago/Turabian StyleSigala, Ioanna, Maria Koutroumani, Anastasia Koukiali, Thomas Giannakouros, and Eleni Nikolakaki. 2021. "Nuclear Translocation of SRPKs Is Associated with 5-FU and Cisplatin Sensitivity in HeLa and T24 Cells" Cells 10, no. 4: 759. https://doi.org/10.3390/cells10040759
APA StyleSigala, I., Koutroumani, M., Koukiali, A., Giannakouros, T., & Nikolakaki, E. (2021). Nuclear Translocation of SRPKs Is Associated with 5-FU and Cisplatin Sensitivity in HeLa and T24 Cells. Cells, 10(4), 759. https://doi.org/10.3390/cells10040759





