MUSE Stem Cells Can Be Isolated from Stromal Compartment of Mouse Bone Marrow, Adipose Tissue, and Ear Connective Tissue: A Comparative Study of Their In Vitro Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Mouse MSCs and Fibroblasts’ Isolation
2.3. Harvest of Mouse Tissues and Organs
2.4. Culture of MUSE Cells
2.5. Flow Cytometry Analysis
2.6. Cell Cycle Analysis
2.7. Cell Proliferation Assay
2.8. Apoptosis Detection by Annexin V Assay
2.9. Senescence Detection by Acid Beta-Galactosidase Assay
2.10. Spontaneous Commitment to Differentiation of MUSE Cells
2.11. Immunocytochemistry (ICC) and Immunohistochemistry (IHC)
2.12. RT-qPCR
2.13. Statistical Analysis
3. Results
3.1. MUSE Cells Were Successfully Isolated from Mouse Stromal Tissues
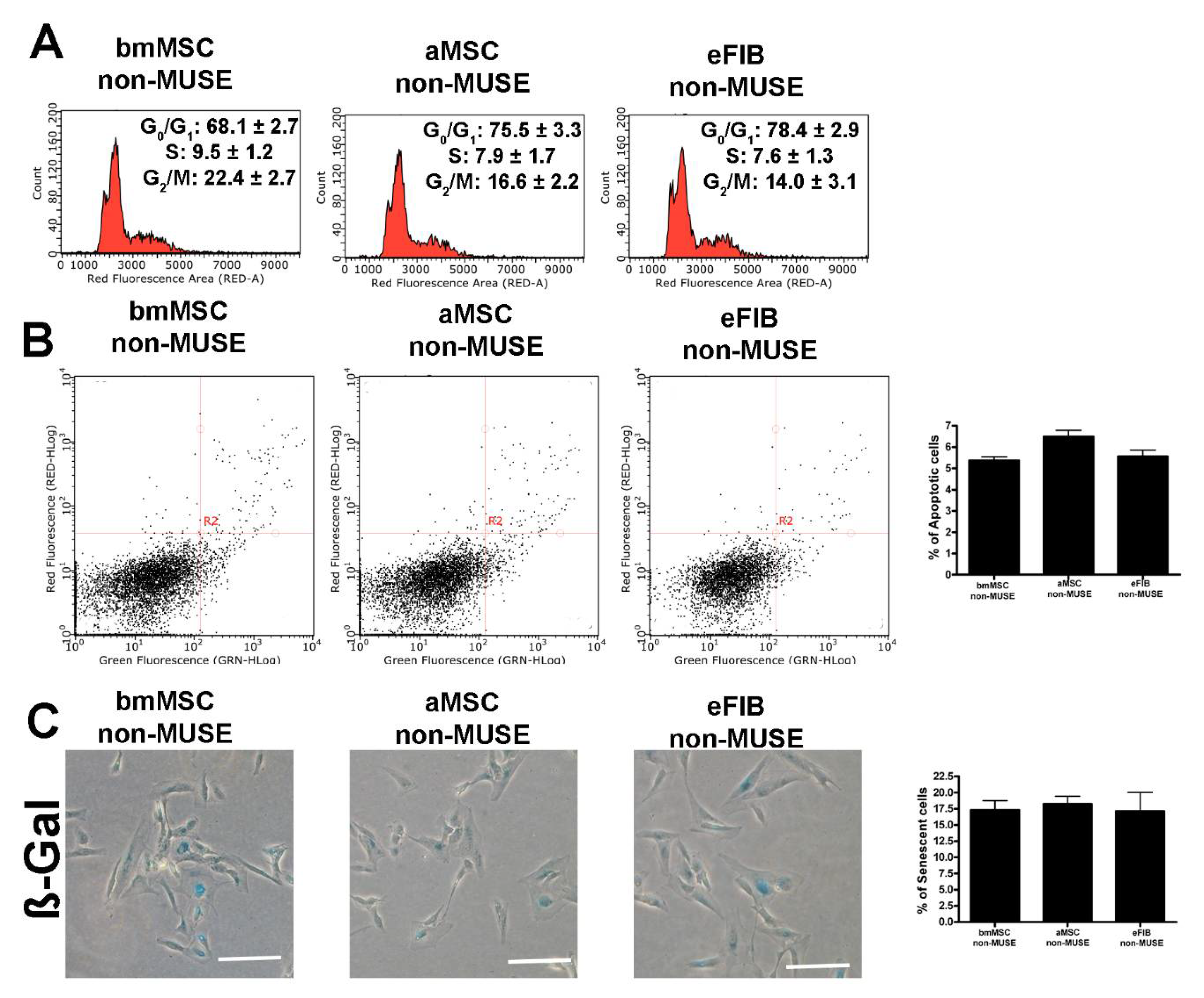
3.2. The MUSE Cells from Different Tissues Showed Similar In Vitro Biological Properties
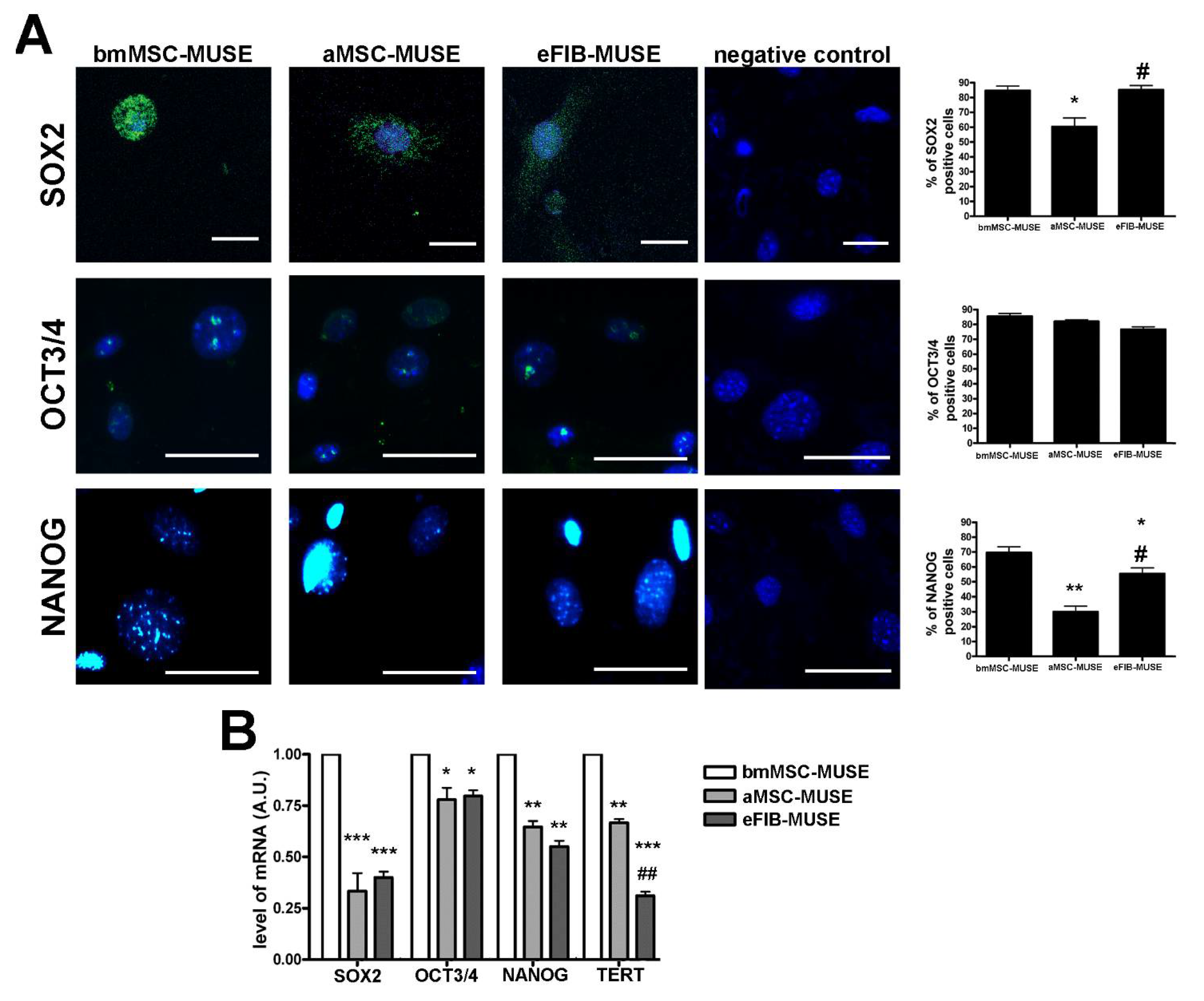
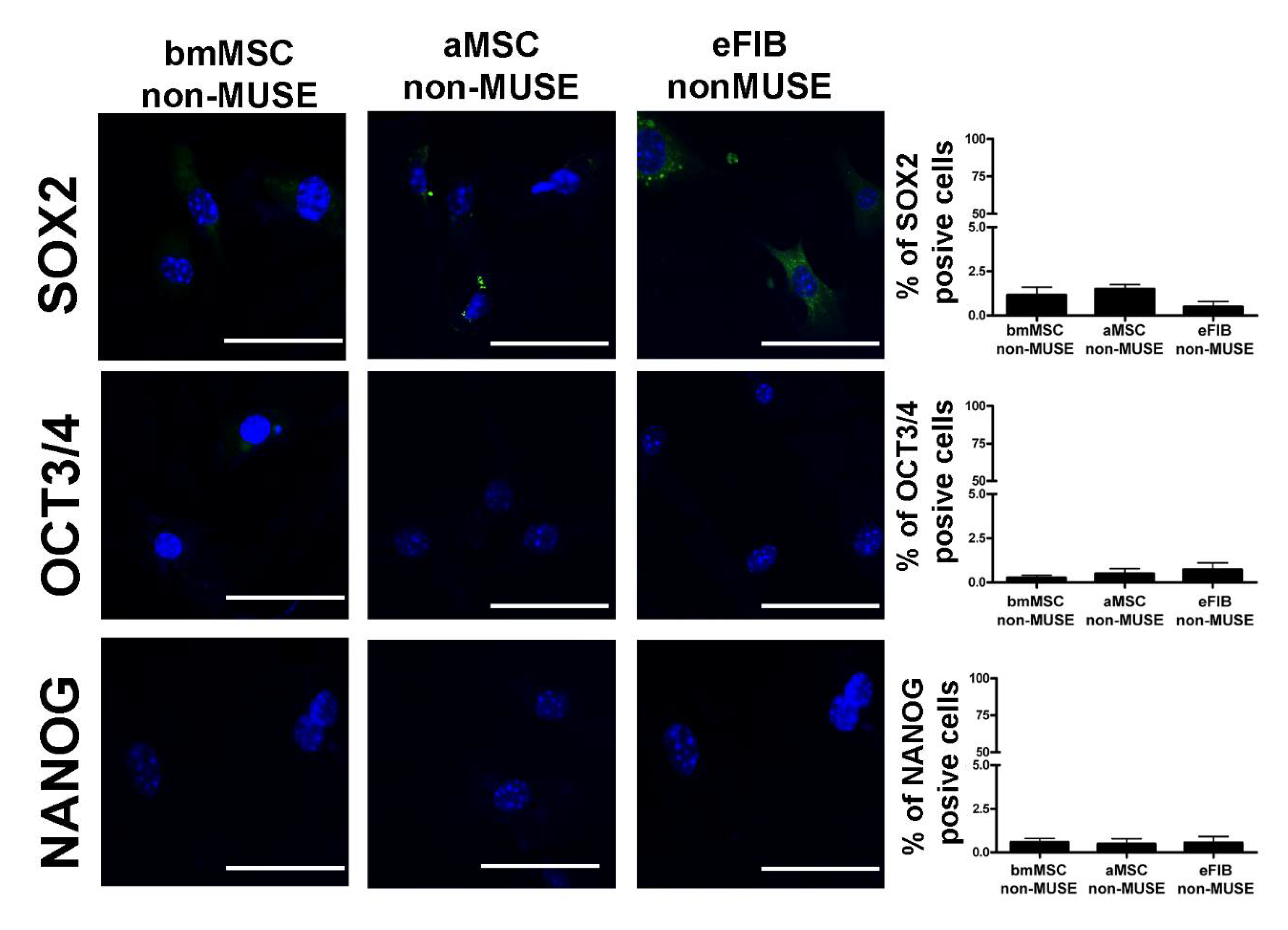
3.3. MUSE Cells Expressed Stem Cell Markers
3.4. MUSE Cells Showed Spontaneous Commitment to Differentiation in Meso/Ecto/Endodermal Derivatives
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bernardo, M.E.; Locatelli, F.; Fibbe, W.E. Mesenchymal stromal cells. Ann. N. Y. Acad. Sci. 2009, 1176, 101–117. [Google Scholar] [CrossRef]
- Kuroda, Y.; Kitada, M.; Wakao, S.; Nishikawa, K.; Tanimura, Y.; Makinoshima, H.; Goda, M.; Akashi, H.; Inutsuka, A.; Niwa, A.; et al. Unique multipotent cells in adult human mesenchymal cell populations. Proc. Natl. Acad. Sci. USA 2010, 107, 8639–8643. [Google Scholar] [CrossRef] [PubMed]
- Alessio, N.; Squillaro, T.; Ozcan, S.; Di Bernardo, G.; Venditti, M.; Melone, M.; Peluso, G.; Galderisi, U. Stress and stem cells: Adult Muse cells tolerate extensive genotoxic stimuli better than mesenchymal stromal cells. Oncotarget 2018, 9, 19328–19341. [Google Scholar] [CrossRef] [PubMed]
- Fisch, S.C.; Gimeno, M.L.; Phan, J.D.; Simerman, A.A.; Dumesic, D.A.; Perone, M.J.; Chazenbalk, G.D. Pluripotent nontumorigenic multilineage differentiating stress enduring cells (Muse cells): A seven-year retrospective. Stem Cell Res. Ther. 2017, 8, 227. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Yamasaki, K.; Tsuchiyama, K.; Koike, S.; Aiba, S. A quantitative analysis of multilineage-differentiating stress-enduring (Muse) cells in human adipose tissue and efficacy of melanocytes induction. J. Dermatol. Sci. 2017, 86, 198–205. [Google Scholar] [CrossRef]
- Dezawa, M. Muse Cells Provide the Pluripotency of Mesenchymal Stem Cells: Direct Contribution of Muse Cells to Tissue Regeneration. Cell Transplant. 2016, 25, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Simerman, A.A.; Phan, J.D.; Dumesic, D.A.; Chazenbalk, G.D. Muse Cells: Nontumorigenic Pluripotent Stem Cells Present in Adult Tissues-A Paradigm Shift in Tissue Regeneration and Evolution. Stem Cells Int. 2016, 2016, 1463258. [Google Scholar] [CrossRef] [PubMed]
- Uchida, H.; Niizuma, K.; Kushida, Y.; Wakao, S.; Tominaga, T.; Borlongan, C.V.; Dezawa, M. Human Muse Cells Reconstruct Neuronal Circuitry in Subacute Lacunar Stroke Model. Stroke 2017, 48, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Wakao, S.; Kushida, Y.; Minatoguchi, S.; Mikami, A.; Higashi, K.; Baba, S.; Shigemoto, T.; Kuroda, Y.; Kanamori, H.; et al. S1P-S1PR2 Axis Mediates Homing of Muse Cells Into Damaged Heart for Long-Lasting Tissue Repair and Functional Recovery After Acute Myocardial Infarction. Circ. Res. 2018, 122, 1069–1083. [Google Scholar] [CrossRef]
- D’Ippolito, G.; Diabira, S.; Howard, G.A.; Menei, P.; Roos, B.A.; Schiller, P.C. Marrow-isolated adult multilineage inducible (MIAMI) cells, a unique population of postnatal young and old human cells with extensive expansion and differentiation potential. J. Cell Sci. 2004, 117, 2971–2981. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Zuba-Surma, E.K.; Machalinski, B.; Ratajczak, J.; Kucia, M. Very small embryonic-like (VSEL) stem cells: Purification from adult organs, characterization, and biological significance. Stem Cell Rev. 2008, 4, 89–99. [Google Scholar] [CrossRef]
- Nitobe, Y.; Nagaoki, T.; Kumagai, G.; Sasaki, A.; Liu, X.; Fujita, T.; Fukutoku, T.; Wada, K.; Tanaka, T.; Kudo, H.; et al. Neurotrophic Factor Secretion and Neural Differentiation Potential of Multilineage-differentiating Stress-enduring (Muse) Cells Derived from Mouse Adipose Tissue. Cell Transplant. 2019, 28, 1132–1139. [Google Scholar] [CrossRef]
- Alessio, N.; Ozcan, S.; Tatsumi, K.; Murat, A.; Peluso, G.; Dezawa, M.; Galderisi, U. The secretome of MUSE cells contains factors that may play a role in regulation of stemness, apoptosis and immunomodulation. Cell Cycle 2017, 16, 33–44. [Google Scholar] [CrossRef]
- Kuroda, Y.; Wakao, S.; Kitada, M.; Murakami, T.; Nojima, M.; Dezawa, M. Isolation, culture and evaluation of multilineage-differentiating stress-enduring (Muse) cells. Nat. Protoc. 2013, 8, 1391–1415. [Google Scholar] [CrossRef] [PubMed]
- Zanichelli, F.; Capasso, S.; Di Bernardo, G.; Cipollaro, M.; Pagnotta, E.; Carteni, M.; Casale, F.; Iori, R.; Giordano, A.; Galderisi, U. Low concentrations of isothiocyanates protect mesenchymal stem cells from oxidative injuries, while high concentrations exacerbate DNA damage. Apoptosis Int. J. Program. Cell Death 2012, 17, 964–974. [Google Scholar] [CrossRef]
- Ogura, F.; Wakao, S.; Kuroda, Y.; Tsuchiyama, K.; Bagheri, M.; Heneidi, S.; Chazenbalk, G.; Aiba, S.; Dezawa, M. Human adipose tissue possesses a unique population of pluripotent stem cells with nontumorigenic and low telomerase activities: Potential implications in regenerative medicine. Stem Cells Dev. 2014, 23, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Turinetto, V.; Vitale, E.; Giachino, C. Senescence in Human Mesenchymal Stem Cells: Functional Changes and Implications in Stem Cell-Based Therapy. Int. J. Mol. Sci. 2016, 17, 1164. [Google Scholar] [CrossRef]
- Furusawa, T.; Ikeda, M.; Inoue, F.; Ohkoshi, K.; Hamano, T.; Tokunaga, T. Gene expression profiling of mouse embryonic stem cell subpopulations. Biol. Reprod. 2006, 75, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Wakao, S.; Kushida, Y.; Dezawa, M. Basic Characteristics of Muse Cells. Adv. Exp. Med. Biol. 2018, 1103, 13–41. [Google Scholar] [PubMed]
- Hiyama, E.; Hiyama, K. Telomere and telomerase in stem cells. Br. J. Cancer 2007, 96, 1020–1024. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Lee, J.S.; Chu, I.S.; Takahama, Y.; Thorgeirsson, S.S. Spontaneous differentiation of mouse embryonic stem cells in vitro: Characterization by global gene expression profiles. Biochem. Biophys. Res. Commun. 2005, 332, 1061–1069. [Google Scholar] [CrossRef]
- Shafa, M.; Yang, F.; Fellner, T.; Rao, M.S.; Baghbaderani, B.A. Human-Induced Pluripotent Stem Cells Manufactured Using a Current Good Manufacturing Practice-Compliant Process Differentiate Into Clinically Relevant Cells From Three Germ Layers. Front. Med. 2018, 5, 69. [Google Scholar] [CrossRef]
- Thorens, B. GLUT2, glucose sensing and glucose homeostasis. Diabetologia 2015, 58, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Yang, L.; Cao, H.; Shen, Z.Y.; Song, H.L. Study of the protective effect on damaged intestinal epithelial cells of rat multilineage-differentiating stress-enduring (Muse) cells. Cell Biol. Int. 2020, 44, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Galderisi, U.; Giordano, A. The gap between the physiological and therapeutic roles of mesenchymal stem cells. Med. Res. Rev. 2014, 34, 1100–1126. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Carrillo-Galvez, A.B.; Garcia-Perez, A.; Cobo, M.; Martin, F. CD105 (endoglin)-negative murine mesenchymal stromal cells define a new multipotent subpopulation with distinct differentiation and immunomodulatory capacities. PLoS ONE 2013, 8, e76979. [Google Scholar] [CrossRef] [PubMed]
- Boward, B.; Wu, T.; Dalton, S. Concise Review: Control of Cell Fate Through Cell Cycle and Pluripotency Networks. Stem Cells 2016, 34, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Wolff, S.C.; Kedziora, K.M.; Dumitru, R.; Dungee, C.D.; Zikry, T.M.; Beltran, A.S.; Haggerty, R.A.; Cheng, J.; Redick, M.A.; Purvis, J.E. Inheritance of OCT4 predetermines fate choice in human embryonic stem cells. Mol. Syst. Biol. 2018, 14, e8140. [Google Scholar] [CrossRef]
- Kolodziejczyk, A.A.; Kim, J.K.; Tsang, J.C.; Ilicic, T.; Henriksson, J.; Natarajan, K.N.; Tuck, A.C.; Gao, X.; Buhler, M.; Liu, P.; et al. Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation. Cell Stem Cell 2015, 17, 471–485. [Google Scholar] [CrossRef]
- Iseki, M.; Kushida, Y.; Wakao, S.; Akimoto, T.; Mizuma, M.; Motoi, F.; Asada, R.; Shimizu, S.; Unno, M.; Chazenbalk, G.; et al. Muse Cells, Nontumorigenic Pluripotent-Like Stem Cells, Have Liver Regeneration Capacity Through Specific Homing and Cell Replacement in a Mouse Model of Liver Fibrosis. Cell Transplant. 2017, 26, 821–840. [Google Scholar] [CrossRef] [PubMed]
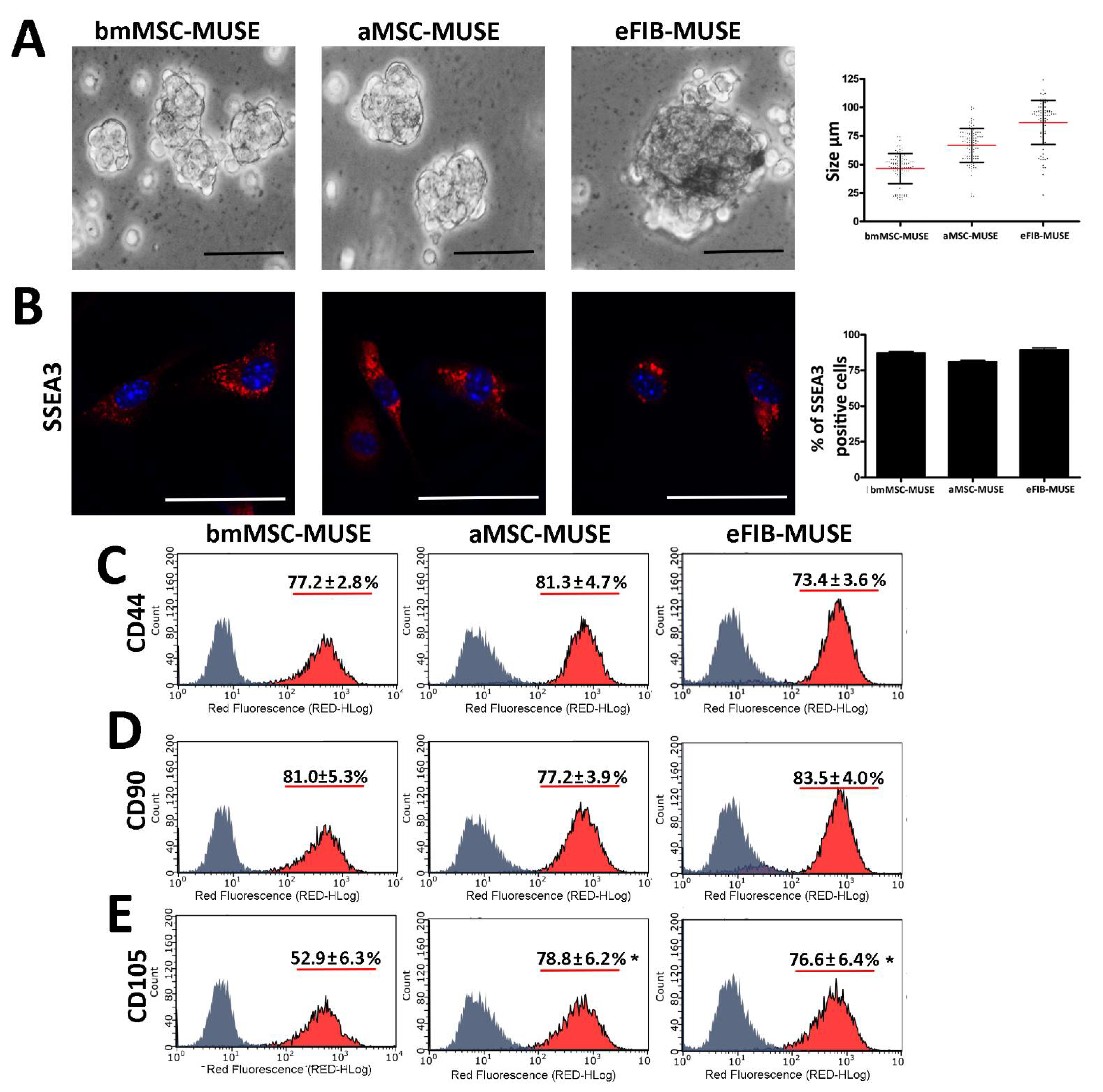

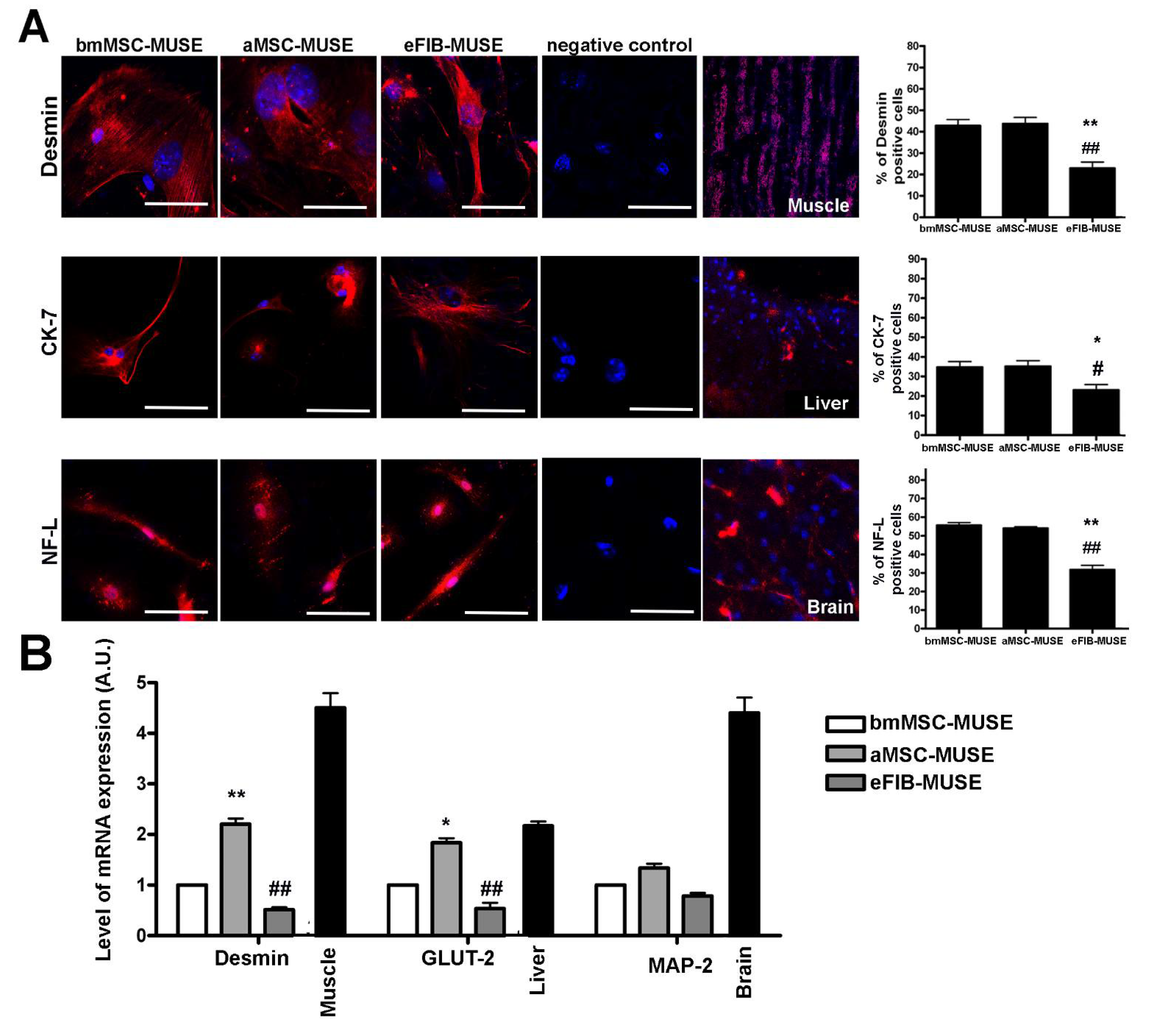
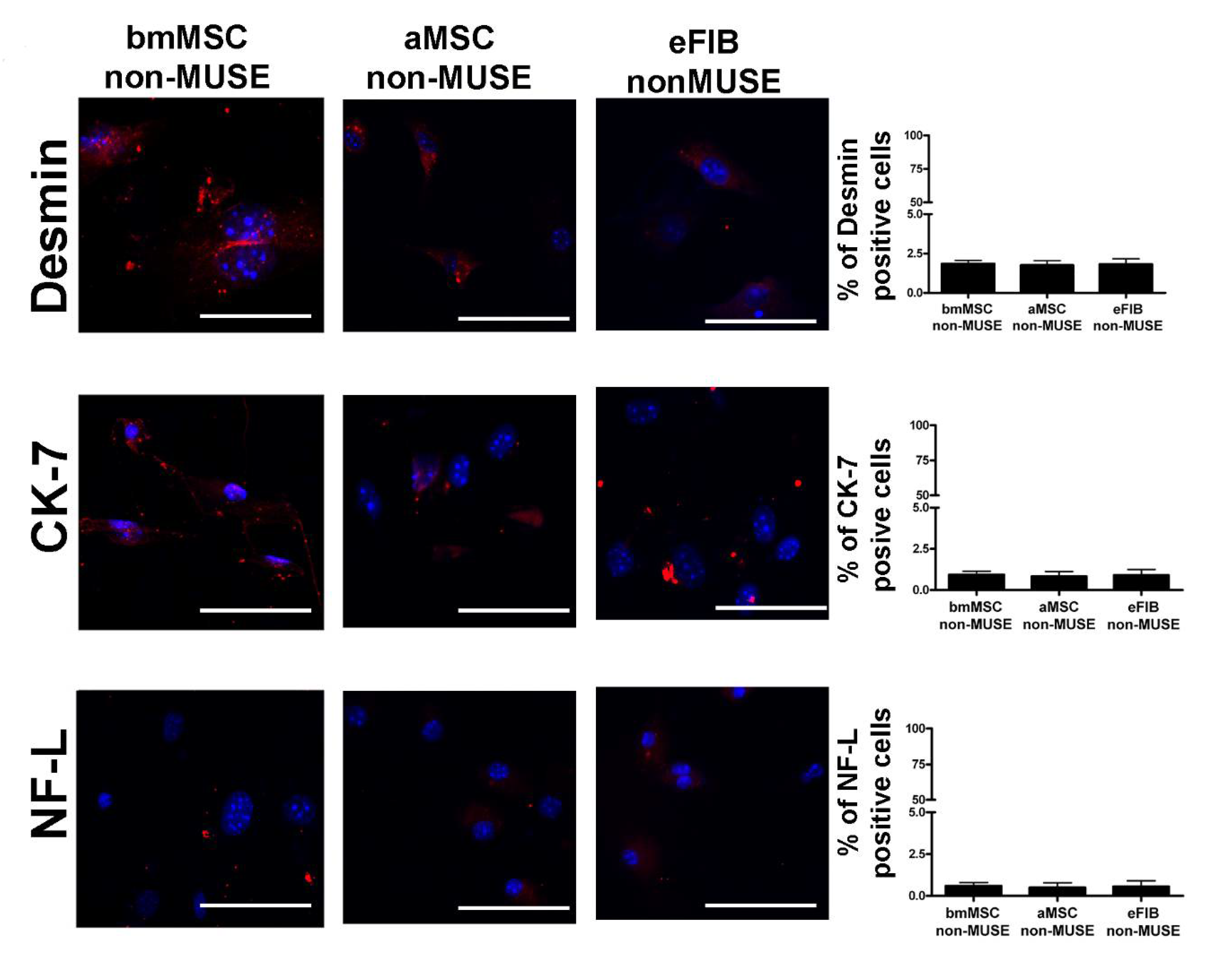
| bmMSC-MUSE | aMSC-MUSE | eFIB-MUSE | |
|---|---|---|---|
| Yield of SSEA3(+) cells after MACS sorting | 3.0% | 2.6% | 2.1% |
| Number of cells at 10 DIV/plate | 5 × 105 ± 4.1 × 104 | 5.2 × 105 ± 3.8 × 104 | 5.4 × 105 ± 4.5 × 104 |
| Number of cluster at 10 DIV/plate | 32 ± 4.5 | 27 ± 4.1 | 19 ± 3.7 |
| Cluster size at 10 DIV | 48 ± 17 μm | 67 ± 12 μm | 98 ± 15 μm |
| Cells per cluster at 10 DIV | 15.6 ± 5.1 | 19.4 ± 5.9 | 28.3 ± 9.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aprile, D.; Alessio, N.; Demirsoy, I.H.; Squillaro, T.; Peluso, G.; Di Bernardo, G.; Galderisi, U. MUSE Stem Cells Can Be Isolated from Stromal Compartment of Mouse Bone Marrow, Adipose Tissue, and Ear Connective Tissue: A Comparative Study of Their In Vitro Properties. Cells 2021, 10, 761. https://doi.org/10.3390/cells10040761
Aprile D, Alessio N, Demirsoy IH, Squillaro T, Peluso G, Di Bernardo G, Galderisi U. MUSE Stem Cells Can Be Isolated from Stromal Compartment of Mouse Bone Marrow, Adipose Tissue, and Ear Connective Tissue: A Comparative Study of Their In Vitro Properties. Cells. 2021; 10(4):761. https://doi.org/10.3390/cells10040761
Chicago/Turabian StyleAprile, Domenico, Nicola Alessio, Ibrahim H. Demirsoy, Tiziana Squillaro, Gianfranco Peluso, Giovanni Di Bernardo, and Umberto Galderisi. 2021. "MUSE Stem Cells Can Be Isolated from Stromal Compartment of Mouse Bone Marrow, Adipose Tissue, and Ear Connective Tissue: A Comparative Study of Their In Vitro Properties" Cells 10, no. 4: 761. https://doi.org/10.3390/cells10040761
APA StyleAprile, D., Alessio, N., Demirsoy, I. H., Squillaro, T., Peluso, G., Di Bernardo, G., & Galderisi, U. (2021). MUSE Stem Cells Can Be Isolated from Stromal Compartment of Mouse Bone Marrow, Adipose Tissue, and Ear Connective Tissue: A Comparative Study of Their In Vitro Properties. Cells, 10(4), 761. https://doi.org/10.3390/cells10040761










