Cadmium (II)-Induced Oxidative Stress Results in Replication Stress and Epigenetic Modifications in Root Meristem Cell Nuclei of Vicia faba
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
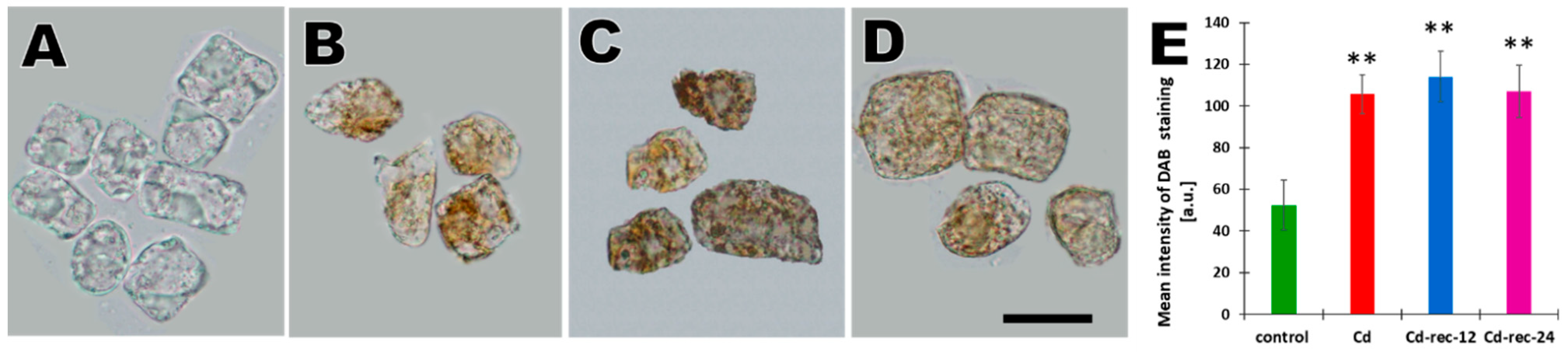
2.2. Detection of H2O2 Using DAB
2.3. Feulgen DNA Staining
2.4. EdU Labeling
2.5. Immunocytochemical Detection of Phosphorylated H3 (Ser10) Histones
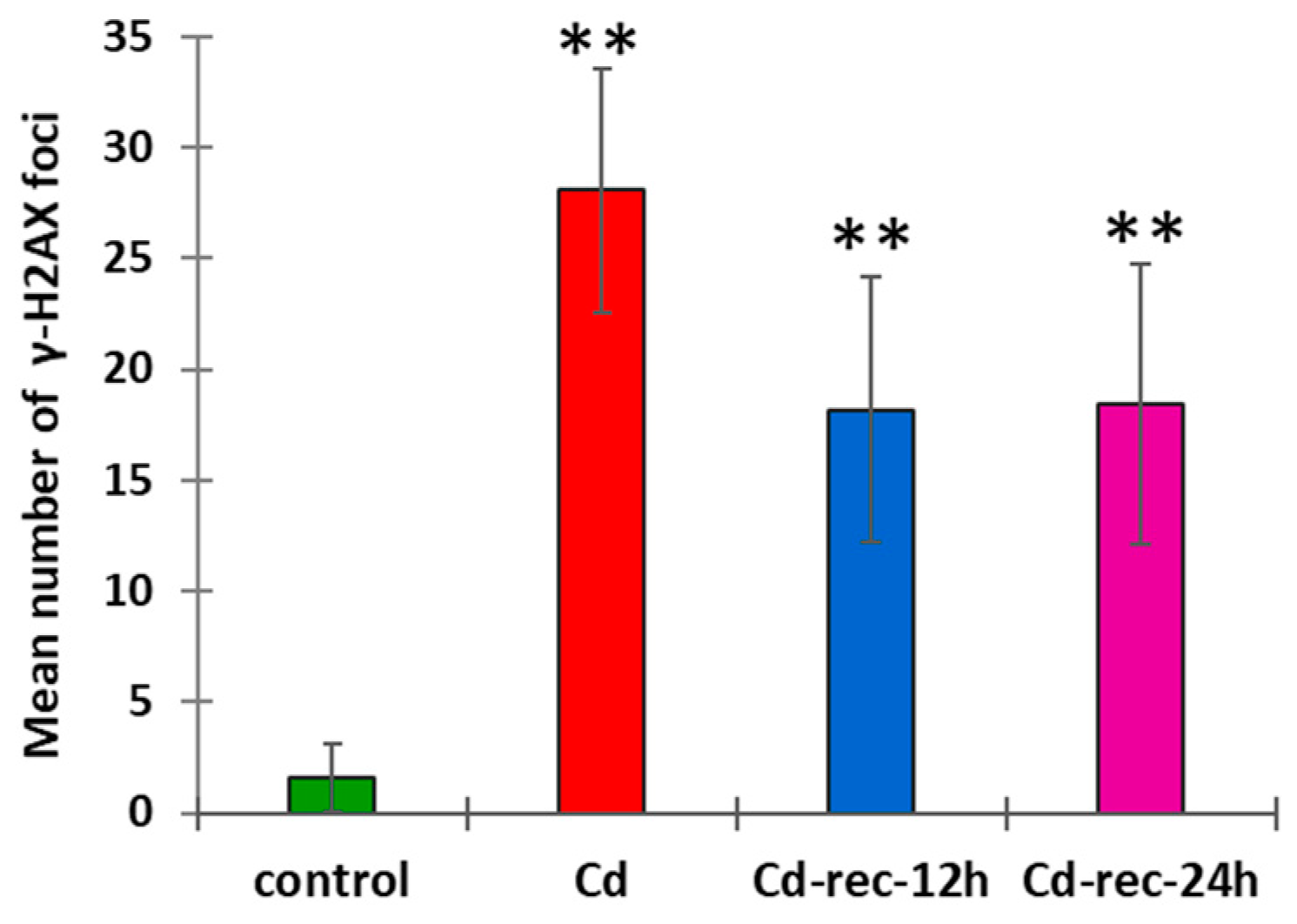
2.6. Immunocytochemical Detection of γ-Phosphorylated H2AX Histones
2.7. Immunocytochemical Staining of H4K5 Acetylated Histones
2.8. AgNOR Staining
2.9. Observations and Analyses
3. Results
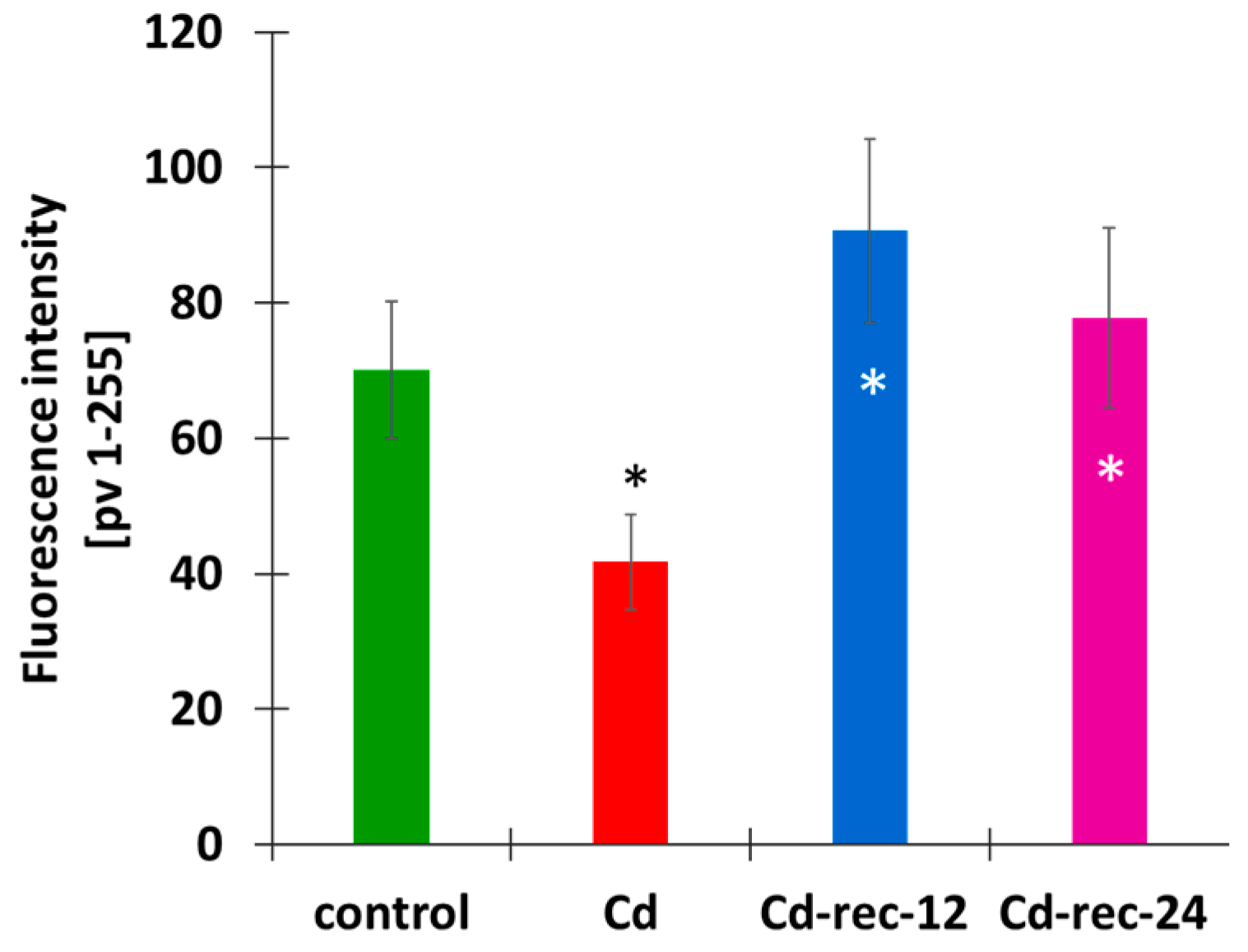
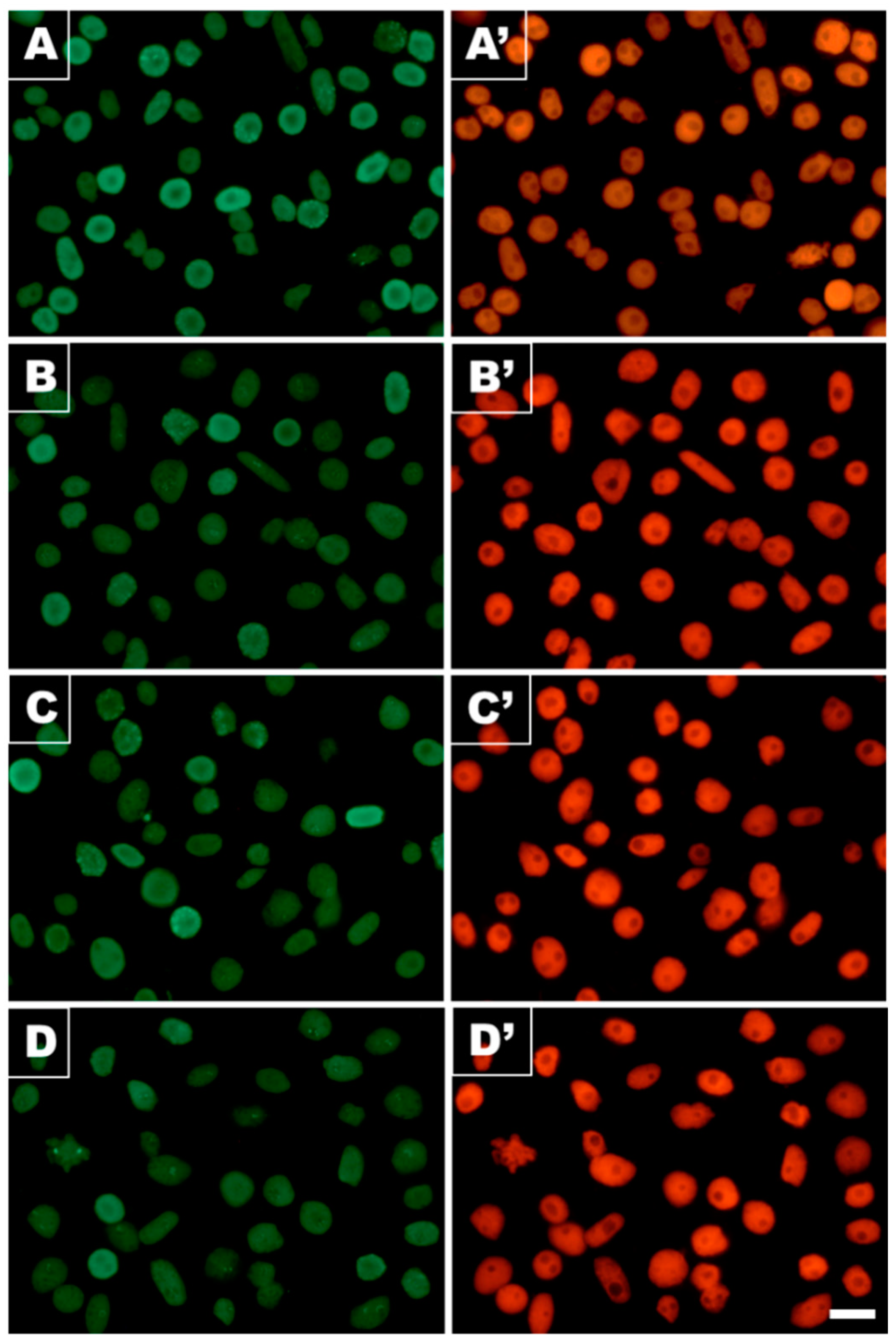
3.1. Cd(II) Ions Generate Hydrogen Peroxide and γ-Phosphorylation of H2AX Histones
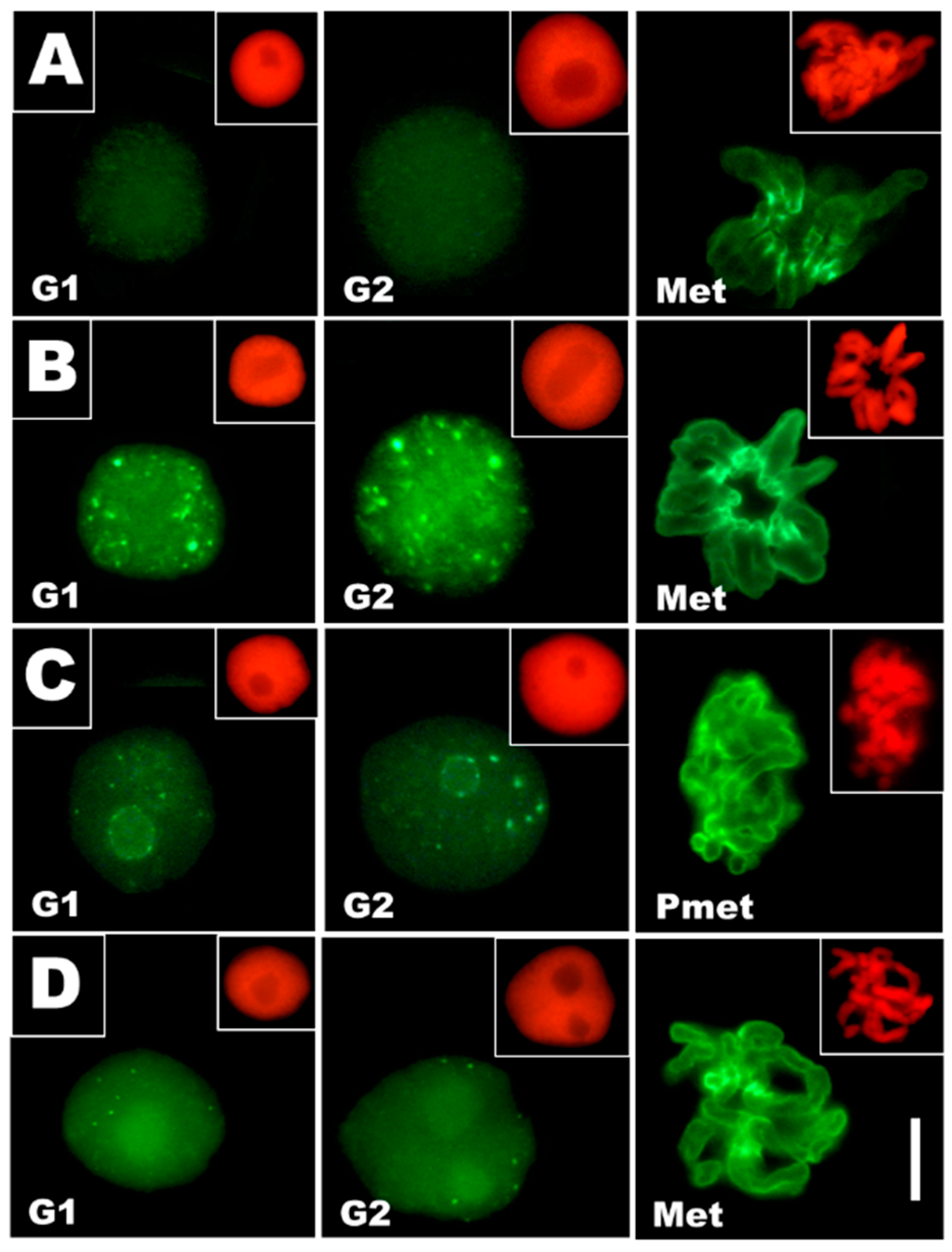
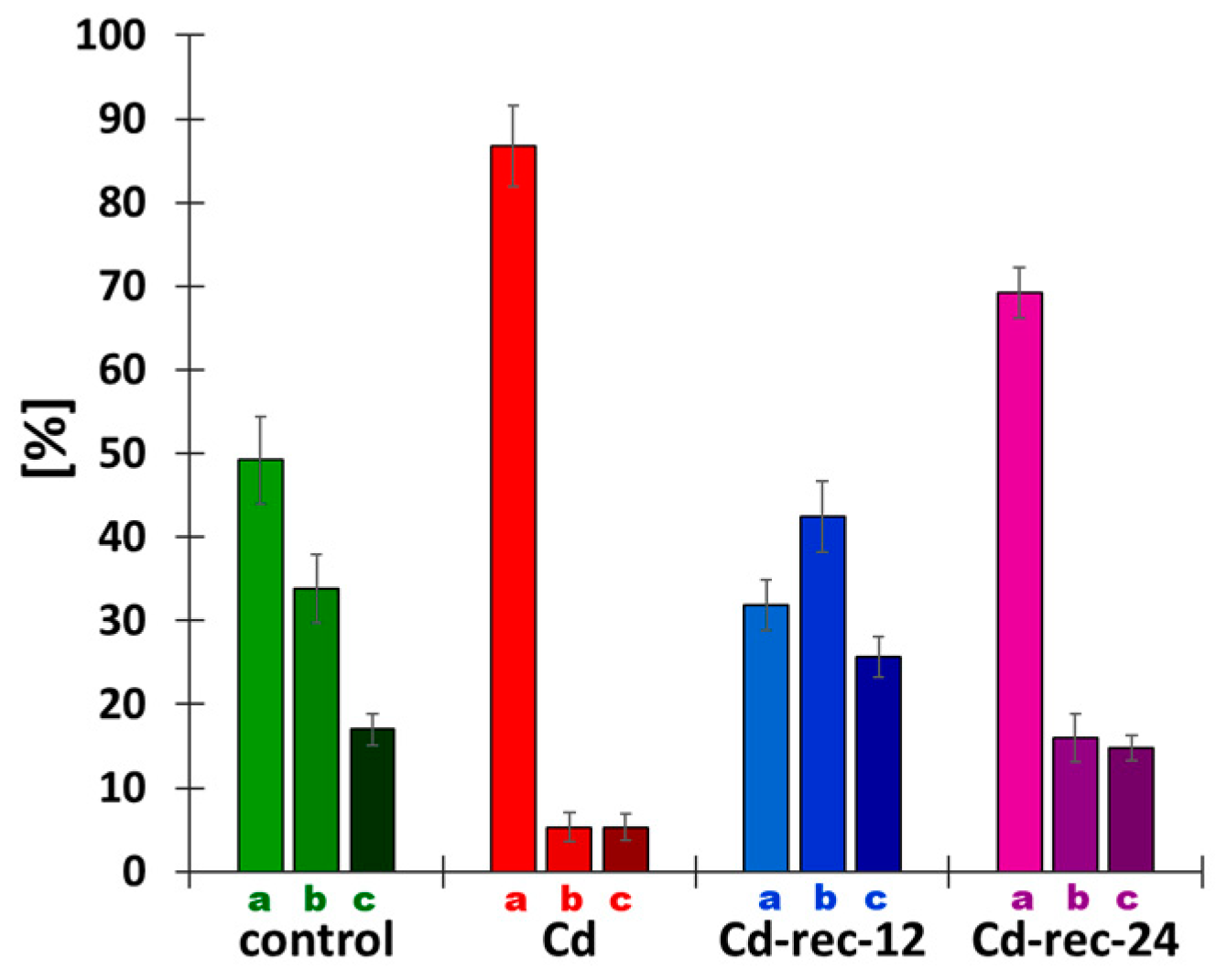
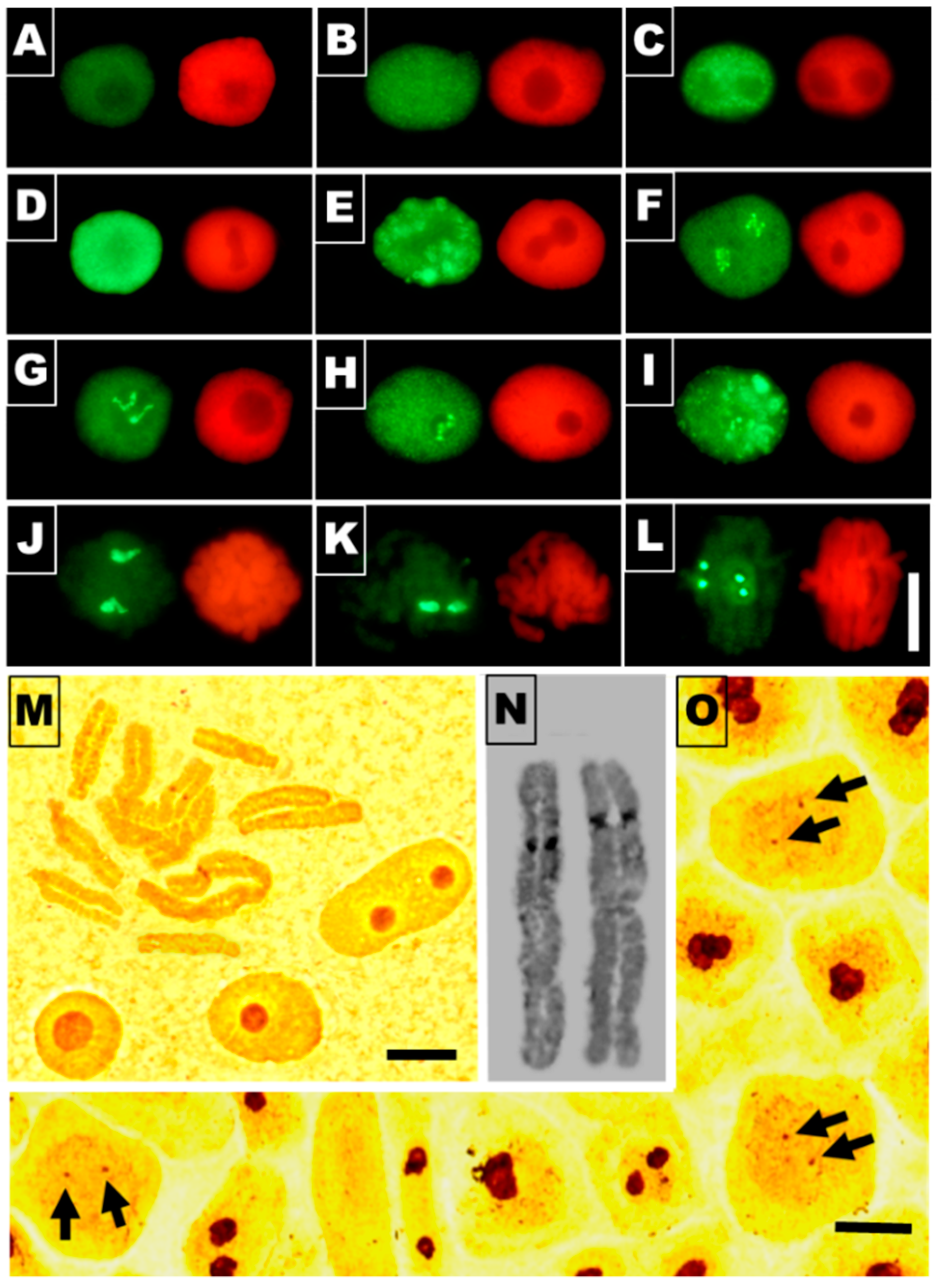
3.2. Cadmium-Stress Increased Condensation of Chromatin Is Correlated with Ectopic Histone H3S10 Phosphorylation
3.3. Cadmium-Induced Changes in DNA Replication and S-Phase Progression
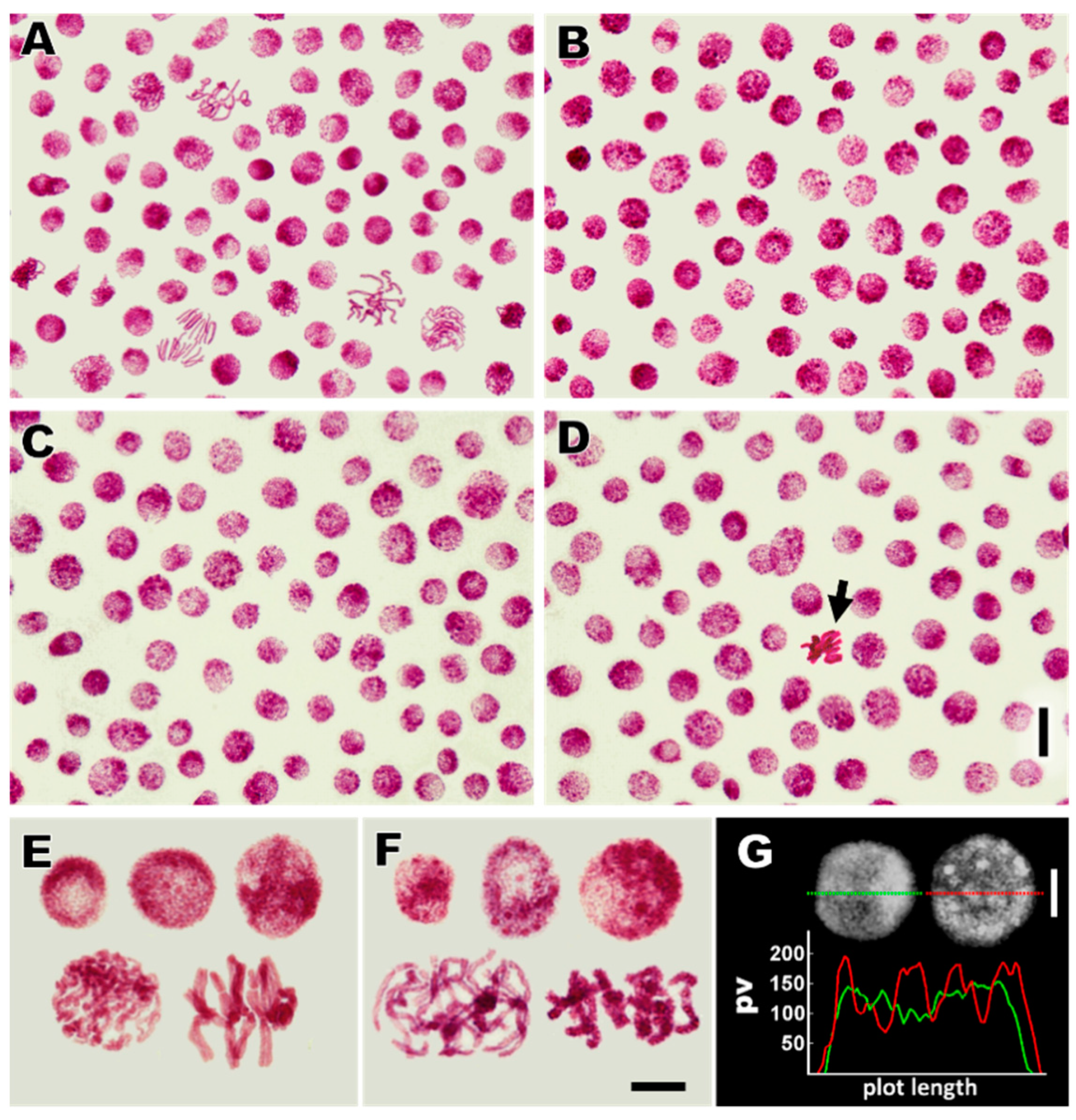
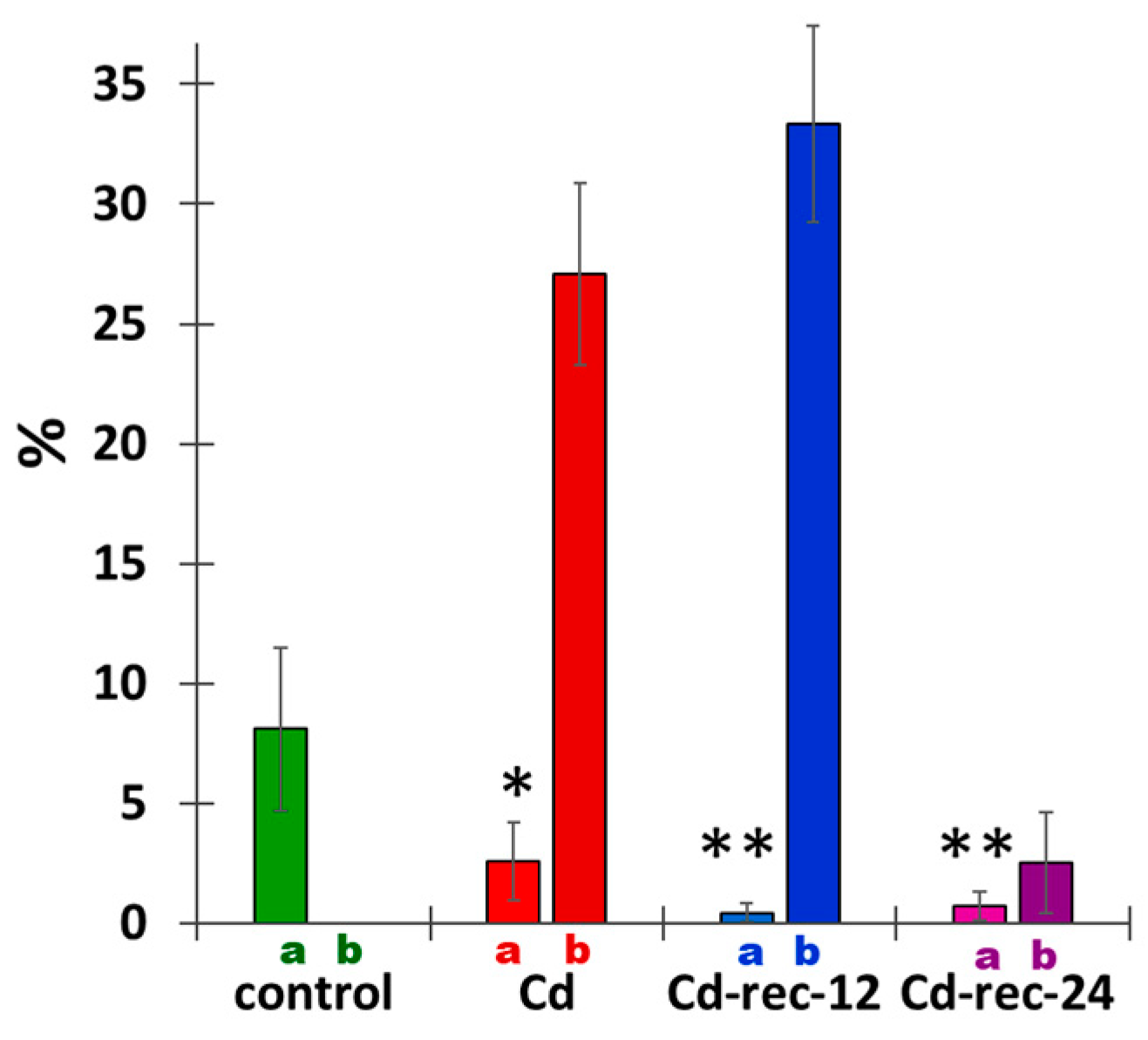
3.4. Cadmium Stress Reduces Mitotic Activity of RAM Cells and Generates Chromosomal Defects
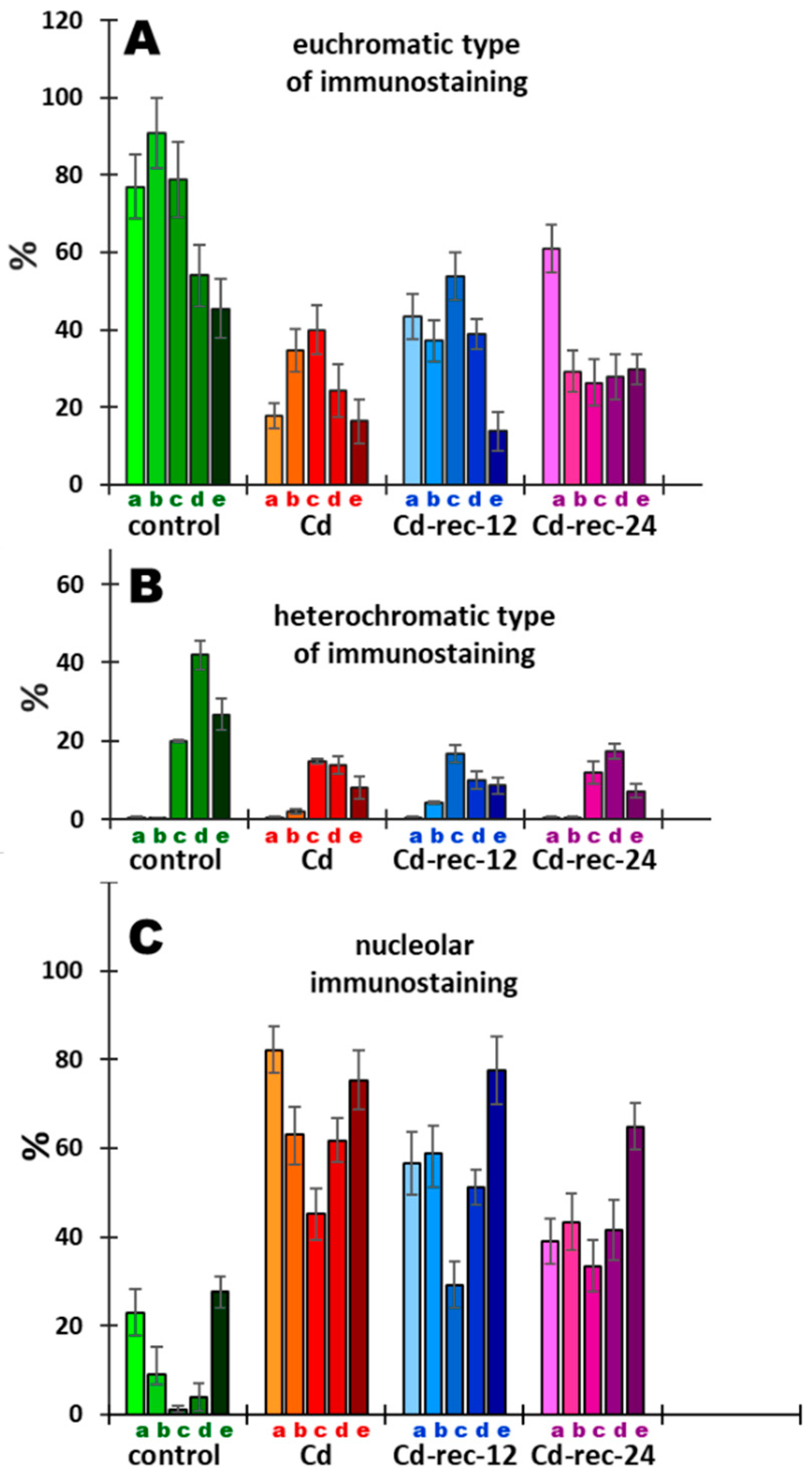
3.5. Acetylation of H4K5
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bhat, J.A.; Shivaraj, S.M.; Singh, P.; Navadagi, D.B.; Tripathi, D.K.; Dash, P.K.; Solanke, A.U.; Sonah, H.; Deshmukh, R. Role of silicon in mitigation of heavy metal stresses in crop plants. Plants 2019, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Duan, S.; Wu, Q.; Yu, M.; Shabala, S. Reducing cadmium accumulation in plants: Structure-function relations and tissue-specific operation of transporters in the spotlight. Plants 2020, 9, 223–240. [Google Scholar] [CrossRef]
- Nadgórska-Socha, A.; Kafel, A.; Kandziora-Ciupa, M.; Gospodarek, J.; Zawisza-Raszka, A. Accumulation of heavy metals and antioxidant responses in Vicia faba plants grown on monometallic contaminated soil. Environ. Sci. Pollut. Res. 2013, 20, 1124–1134. [Google Scholar] [CrossRef]
- Singh, S.; Parihar, P.; Singh, R.; Singh, V.P.; Prasad, S.M. Heavy metal tolerance in plants: Role of transcriptomics, proteomics, metabolomics, and ionomics. Front. Plant Sci. 2015, 6, 1143. [Google Scholar] [CrossRef] [PubMed]
- Bücker-Net, L.; Paiva, A.L.S.; Machado, R.D.; Arenhart, R.A.; Margis-Pinheiro, M. Interactions between plant hormones and heavy metals responses. Genet. Mol. Biol. 2017, 40, 373–386. [Google Scholar] [CrossRef]
- DalCorso, G.; Farinati, S.; Furini, A. Regulatory networks of cadmium stress in plants. Plant Signal. Behav. 2010, 5, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, T.; Ragu, S.; Magdalou, I.; Machon, C.; Dardillac, E.; Techer, H.; Guitton, J.; Debatisse, M.; Lopez, B.S. Slow replication fork velocity of homologous recombination-defective cells results from endogenous oxidative stress. PLoS Genet. 2016, 12, e1006007. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metals toxicity and the environment. EXS 2012, 101, 133–164. [Google Scholar]
- Song, Y.; Jin, L.; Wang, X. Cadmium absorption and transportation pathways in plants. Int. J. Phytoremediat. 2017, 19, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Xu, H.; Lu, J.; Chen, Q.; Li, W.; Wu, L.; Tang, L.; Ma, L. The immobilization of soil cadmium by the combined amendment of bacteria and hydroxyapatite. Sci. Rep. 2020, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Yang, Z.; Xie, Y.; Nie, L.; Cui, J.; Shen, W.B. Arabidopsis HY1 confers cadmium tolerance by decreasing nitric oxide production and improving iron homeostasis. Mol. Plant 2014, 7, 388–403. [Google Scholar] [CrossRef]
- Fan, J.L.; Wei, X.Z.; Wan, L.C.; Zhang, H.Y.; Zhao, X.Q.; Liu, W.Z.; Zhang, H.Y. Disarrangement of actin filaments and Ca2+ gradient by CdCl2 alters cell wall construction in Arabidopsis thaliana root hairs by inhibiting vesicular trafficking. J. Plant Physiol. 2011, 168, 1157–1167. [Google Scholar] [CrossRef]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 2012, 63, 695–709. [Google Scholar] [CrossRef]
- Bruno, L.; Pacenza, M.; Forgione, I.; Lamerton, L.R.; Greco, M.; Chiappetta, A.; Bitonti, M.B. In Arabidopsis thaliana cadmium impact on the growth of primary root by altering SCR expression and auxin-cytokinin cross-talk. Front. Plant Sci. 2017, 8, 1323. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Sarkar, A.; Singh, S.; Singh, P.; de Araujo, A.S.F.; Singh, R.P. Agroecological responses of heavy metal pollution with special emphasis on soil health and plant performances. Front. Environ. Sci. 2017, 5, 64. [Google Scholar] [CrossRef]
- Dutta, S.; Mitra, M.; Agarwal, P.; Mahapatra, K.; De, S.; Sett, U.; Roy, S. Oxidative and genotoxic damages in plants in response to heavy metal stress and maintenance of genome stability. Plant Signal. Behav. 2018, 13, e1460048. [Google Scholar] [CrossRef] [PubMed]
- Huybrechts, M.; Cuypers, A.; Deckers, J.; Iven, V.; Vandionant, S.; Jozefczak, M.; Hendrix, S. Cadmium and plant development: An agony from seed to seed. Int. J. Mol. Sci. 2019, 20, 3971. [Google Scholar] [CrossRef]
- Sobkowiak, R.; Deckert, J. Cadmium-induced changes in growth and cell cycle gene expression in suspension-culture cells of soybean. Plant Physiol. Biochem. 2003, 41, 767–772. [Google Scholar] [CrossRef]
- Pena, L.B.; Azpilicueta, C.E.; Benavides, M.P.; Gallego, S.M. Protein oxidative modifications. In Metal Toxicity in Plants: Perception, Signaling and Remediation; Gupta, D.K., Sandalio, L.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 207–225. [Google Scholar]
- Ünyayar, S.; Çelik, A.; Çekiç, F.O.; Gözel, A. Cadmium-induced genotoxicity, cytotoxicity and lipid peroxidation in Allium sativum and Vicia faba. Mutagenesis 2006, 21, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Souguir, D.; Ferjani, E.; Ledoigt, G.; Pascale, G. Sequential effects of cadmium on genotoxicity and lipoperoxidation in Vicia faba roots. Ecotoxicology 2011, 20, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Thordal-Christensen, H.; Zhang, Z.; Wei, Y.; Collinge, D.B. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997, 11, 1187–1194. [Google Scholar] [CrossRef]
- Żabka, A.; Polit, J.T.; Maszewski, J. DNA replication stress induces deregulation of the cell cycle events in root meristems of Allium cepa. Ann. Bot. 2012, 110, 1581–1591. [Google Scholar] [CrossRef] [PubMed]
- Żabka, A.; Polit, J.T.; Maszewski, J. Inter- and intrachromosomal asynchrony of cell division cycle events in root meristem cells of Allium cepa: Possible connection with gradient of cyclin B-like proteins. Plant Cell Rep. 2010, 29, 845–856. [Google Scholar] [CrossRef]
- Żabka, A.; Winnicki, K.; Polit, J.T.; Maszewski, J. The effects of anti-DNA topoisomerase II drugs, etoposide and ellipticine, are modified in root merystem cells of Allium cepa by MG132, an inhibitor of 26S proteasomes. Plant Physiol. Biochem. 2015, 96, 72–82. [Google Scholar] [CrossRef]
- Giménez-Abián, J.F.; Clarke, D.J.; Mullinger, A.M.; Downes, C.S.; Johnson, R.T. A postprophase topoisomerase II–dependent chromatid core separation step in the formation of metaphase chromosomes. J. Cell Biol. 1995, 131, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Howell, W.M.; Black, A.D. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 1980, 36, 1014–1015. [Google Scholar] [CrossRef]
- Abraham, R.T. Checkpoint signalling: Focusing on 53BP1. Nat. Cell Biol. 2002, 4, E277–E279. [Google Scholar] [CrossRef]
- Rybaczek, D.; Bodys, A.; Maszewski, J. H2AX foci in late S/G2- and M-phase cells after hydroxyurea- and aphidicolin-induced DNA replication stress in Vicia. Histochem. Cell Biol. 2007, 128, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Block, W.D.; Yu, Y.; Merkle, D.; Gifford, J.L.; Ding, Q.; Meek, K.; Lees-Miller, S.P. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004, 32, 4351–4357. [Google Scholar] [CrossRef] [PubMed]
- Paull, T.T.; Rogakou, E.P.; Yamazaki, V.; Kirchgessner, C.U.; Gellert, M.; Bonner, W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000, 10, 886–895. [Google Scholar] [CrossRef]
- Jasencakova, Z.; Meister, A.; Walter, J.; Turner, B.M. Schubert I Histone acetylation of euchromatin and heterochromatin is correlated with replication rather than with transcription. Plant Cell 2000, 12, 2087–2100. [Google Scholar] [CrossRef]
- Jasencakova, Z.; Meister, A.; Schubert, I. Chromatin organization and its relation to replication and histone acetylation along the cell cycle in barley. Chromosoma 2001, 110, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine, 4th ed.; Clarendon Press: Oxford, UK, 2006. [Google Scholar]
- Foyer, C.H.; Ruban, A.V.; Noctor, G. Viewing oxidative stress through the lens of oxidative signalling rather than damage. Biochem. J. 2017, 474, 877–883. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- De Michele, R.; Vurro, E.; Rigo, C.; Costa, A.; Elviri, L.; Di Valentin, M.; Careri, M.; Zottini, M.; Sanità di Toppi, L.; Lo Schiavo, F. Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol. 2009, 150, 217–228. [Google Scholar] [CrossRef]
- Arasimowicz-Jelonek, M.; Floryszak-Wieczorek, J.; Deckert, J.; Rucinska-Sobkowiak, R.; Gzyl, J.; Pawlak-Sprada, S.; Abramowski, D.; Jelonek, T.; Gwozdz, E.A. Nitric oxide implication in cadmium-induced programmed cell death in roots and signaling response of yellow lupine plants. Plant Physiol. Biochem. 2012, 58, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Gallego, S.M.; Benavides, M.P. Cadmium-induced oxidative and nitrosative stress in plants. In Cadmium Toxicity and Tolerance in Plants; Academic Press: Cambridge, MA, USA, 2019; pp. 233–274. [Google Scholar]
- Mullineaux, P.; Creissen, G.P. Glutathione reductase: Regulation and role in oxidative stress. In Oxidative Stress and the Molecular Biology of Antioxidant Defenses; Scandalios, J.G., Ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; pp. 667–713. [Google Scholar]
- Schützendübel, A.; Schwanz, P.; Teichmann, T.; Gross, K.; Langenfeld-Heyser, R.; Godbold, D.L.; Polle, A. Cadmium-induced changes in antioxidative systems, H2O2 content and differentiation in pine (Pinus sylvestris) roots. Plant Physiol. 2001, 127, 887–892. [Google Scholar] [CrossRef]
- Ortega-Villasante, C.; Hernández, L.E.; Rellán-Álvarez, R.; del Campo, F.F.; Carpena-Ruiz, R.O. Rapid alteration of cellular redox homeostasis upon exposure to cadmium and mercury in alfalfa seedlings. New Phytol. 2007, 176, 96–107. [Google Scholar] [CrossRef]
- Deng, X.; Xia, Y.; Hu, W.; Zhang, H.; Shen, Z. Cadmium-induced oxidative damage and protective effects of N-acetyl-l-cysteine against cadmium toxicity in Solanum nigrum L. J. Hazard. Mater. 2010, 180, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Chaca, M.V.; Rodriguez-Serrano, M.; Molina, A.S.; Pedranzani, H.E.; Zirulnik, F.; Sandalio, L.M.; Romero-Puertas, M.C. Cadmium induces two waves of reactive oxygen species in Glycine max (L.) roots. Plant Cell Environ. 2014, 37, 1672–1687. [Google Scholar] [CrossRef] [PubMed]
- Romero-Puertas, M.C.; Rodríguez-Serrano, M.; Corpas, F.J.; Gómez, M.; del Río, L.A.; Sandalio, L.M. Cd-induced subcellular accumulation of O2.− and H2O2 in pea leaves. Plant Cell Environ. 2004, 27, 1122–1134. [Google Scholar] [CrossRef]
- Rodriguez-Serrano, M.; Romero-Puertas, M.C.; Zabalza, A.; Corpas, F.J.; Gomez, M.; Del Rio, L.A.; Sandalio, L.M. Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant Cell Environ. 2006, 29, 1532–1544. [Google Scholar] [CrossRef] [PubMed]
- Olmos, E.; Martinez-Solano, J.R.; Piqueras, A.; Hellin, E. Early steps in the oxidative burst induced by cadmium in cultured tobacco cells (BY-2 line). J. Exp. Bot. 2003, 54, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Chao, Y.; Hong, C.; Kao, C.H. The role of hydrogen peroxide in cadmium-inhibited root growth of rice seedlings. Plant Growth Regul. 2012, 66, 27–35. [Google Scholar] [CrossRef]
- Lin, Z.P.; Belcourt, M.F.; Carbone, R.; Eaton, J.S.; Penketh, P.G.; Shadel, G.S.; Cory, J.G.; Sartorelli, A.C. Excess ribonucleotide reductase R2 subunits coordinate the S phase checkpoint to facilitate DNA damage repair and recovery from replication stress. Biochem. Pharmacol. 2007, 73, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hunt, C.P.; Wadhwa, M.; Sanders, L.H. DNA damage by oxidative stress: Measurement strategies for two genomes. Curr. Opin. Toxicol. 2018, 7, 87–94. [Google Scholar] [CrossRef]
- Luo, T.; Yu, Q.; Zou, H.; Zhao, H.; Gu, J.; Yuan, Y.; Zhu, J.; Bian, J.; Liu, Z. Role of poly (ADP-ribose) polymerase-1 in cadmium-induced cellular DNA damage and cell cycle arrest in rat renal tubular epithelial cell line NRK-52E. Environ. Pollut. 2020, 261, 114149. [Google Scholar] [CrossRef] [PubMed]
- Friesner, J.D.; Liu, B.; Culligan, K.; Britt, A.B. Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell 2005, 16, 2566–2576. [Google Scholar] [CrossRef]
- Rybaczek, D.; Maszewski, J. Phosphorylation of H2AX histones in response to double-strand breaks and induction of premature chromatin condensation in hydroxyurea-treated root merystem cells of Raphanus sativus, Vicia faba, and Allium porum. Protoplasma 2007, 230, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Rybaczek, D.; Maszewski, J. Induction of foci of phosphorylated H2AX histones and premature chromosome condensation after DNA damage in Vicia faba root meristem. Biol. Plant. 2007, 51, 443–450. [Google Scholar] [CrossRef]
- Mah, L.J.; El-Osta, A.; Karagiannis, T.C. gammaH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef]
- Żabka, A.; Trzaskoma, P.; Maszewski, J. Dissimilar effects of β-lapachone- and hydroxyurea-induced DNA replication stress in root meristem cells of Allium cepa. Plant Physiol. Biochem. 2013, 73, 282–293. [Google Scholar] [CrossRef]
- Ward, I.M.; Chen, J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001, 276, 47759–47762. [Google Scholar] [CrossRef] [PubMed]
- Elledge, S.J. Cell cycle checkpoints: Preventing an identity crisis. Science 1996, 274, 1664–1672. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.B.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef]
- Esnault, M.A.; Legue, F.; Chenal, C. Ionizing radiation: Advances in plant response. Environ. Exp. Bot. 2010, 68, 231–237. [Google Scholar] [CrossRef]
- Bártová, E.; Lochmanová, G.; Legartová, S.; Suchánková, J.; Fedr, R.; Krejčí, J.; Zdráhal, Z. Irradiation by γ-rays reduces the level of H3S10 phosphorylation and weakens the G2 phase-dependent interaction between H3S10 phosphorylation and γH2AX. Biochimie 2018, 154, 86–98. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Mukherjee, A. Investigating the underlying mechanism of cadmium-induced plant adaptive response to genotoxic stress. Ecotoxicol. Environ. Saf. 2021, 209, 111817. [Google Scholar] [CrossRef]
- Somyajit, K.; Gupta, R.; Sedlackova, H.; Neelsen, K.J.; Ochs, F.; Rask, M.B.; Choudhary, C.; Lukas, J. Redox-sensitive alteration of replisome architecture safeguards genome integrity. Science 2017, 358, 797–802. [Google Scholar] [CrossRef]
- Klattenhoff, A.W.; Thakur, M.; Chu, C.S.; Ray, D.; Habib, S.L.; Kidane, D. Loss of NEIL3 DNA glycosylase markedly increases replication associated double strand breaks and enhances sensitivity to ATR inhibitor in glioblastoma cells. Oncotarget 2017, 8, 112942–112958. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. Biological consequences of free radical-damaged, DNA bases. Free Radic. Biol. Med. 2002, 33, 1–14. [Google Scholar] [CrossRef]
- Sanders, C.M.; Sizov, D.; Seavers, P.R.; Ortiz-Lombardia, M.; Antson, A.A. Transcription activator structure reveals redox control of a replication initiation reaction. Nucleic Acids Res. 2007, 35, 3504–3515. [Google Scholar] [CrossRef]
- Houben, A.; Schroeder-Reiter, E.; Nagaki, K.; Nasuda, S.; Wanner, G.; Murata, M.; Endo, T.R. CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 2007, 116, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Nowak, S.J.; Corces, V.G. Phosphorylation of histone H3: A balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004, 20, 214–220. [Google Scholar] [CrossRef]
- Chen, C.C.L.; Goyal, P.; Karimi, M.M.; Abildgaard, M.H.; Kimura, H.; Lorincz, M.C. H3S10ph broadly marks early-replicating domains in interphase ESCs and shows reciprocal antagonism with H3K9me2. Genome Res. 2018, 28, 37–51. [Google Scholar] [CrossRef]
- Zippo, A.; Serafini, R.; Rocchigiani, M.; Pennacchini, S.; Krepelova, A.; Oliviero, S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 2009, 138, 1122–1136. [Google Scholar] [CrossRef]
- Schubert, I.; Rieger, R. Asymmetric banding of Vicia faba chromosomes after BrdU incorporation. Chromosoma 1979, 70, 385–391. [Google Scholar] [CrossRef]
- Fuchs, J.; Strehl, S.; Brandes, A.; Schweizer, D.; Schubert, I. Molecular-cytogenetic characterization of the Vicia faba genome—Heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res. 1998, 6, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Idei, S.; Kondo, K.; Turner, B.M.; Fukui, K. Tomographic distribution of acetylated histone H4 in plant chromosomes, nuclei and nucleoli. Chromosoma 1996, 105, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, P. Histone acetylation: A possible mechanism for the inheritance of cell memory at mitosis. Bioessays 1997, 19, 67–74. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Żabka, A.; Winnicki, K.; Polit, J.T.; Wróblewski, M.; Maszewski, J. Cadmium (II)-Induced Oxidative Stress Results in Replication Stress and Epigenetic Modifications in Root Meristem Cell Nuclei of Vicia faba. Cells 2021, 10, 640. https://doi.org/10.3390/cells10030640
Żabka A, Winnicki K, Polit JT, Wróblewski M, Maszewski J. Cadmium (II)-Induced Oxidative Stress Results in Replication Stress and Epigenetic Modifications in Root Meristem Cell Nuclei of Vicia faba. Cells. 2021; 10(3):640. https://doi.org/10.3390/cells10030640
Chicago/Turabian StyleŻabka, Aneta, Konrad Winnicki, Justyna Teresa Polit, Mateusz Wróblewski, and Janusz Maszewski. 2021. "Cadmium (II)-Induced Oxidative Stress Results in Replication Stress and Epigenetic Modifications in Root Meristem Cell Nuclei of Vicia faba" Cells 10, no. 3: 640. https://doi.org/10.3390/cells10030640
APA StyleŻabka, A., Winnicki, K., Polit, J. T., Wróblewski, M., & Maszewski, J. (2021). Cadmium (II)-Induced Oxidative Stress Results in Replication Stress and Epigenetic Modifications in Root Meristem Cell Nuclei of Vicia faba. Cells, 10(3), 640. https://doi.org/10.3390/cells10030640






