Overexpression of an Agave Phosphoenolpyruvate Carboxylase Improves Plant Growth and Stress Tolerance
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Analysis of the PEPC Gene Family
2.2. Phylogenetic Analysis
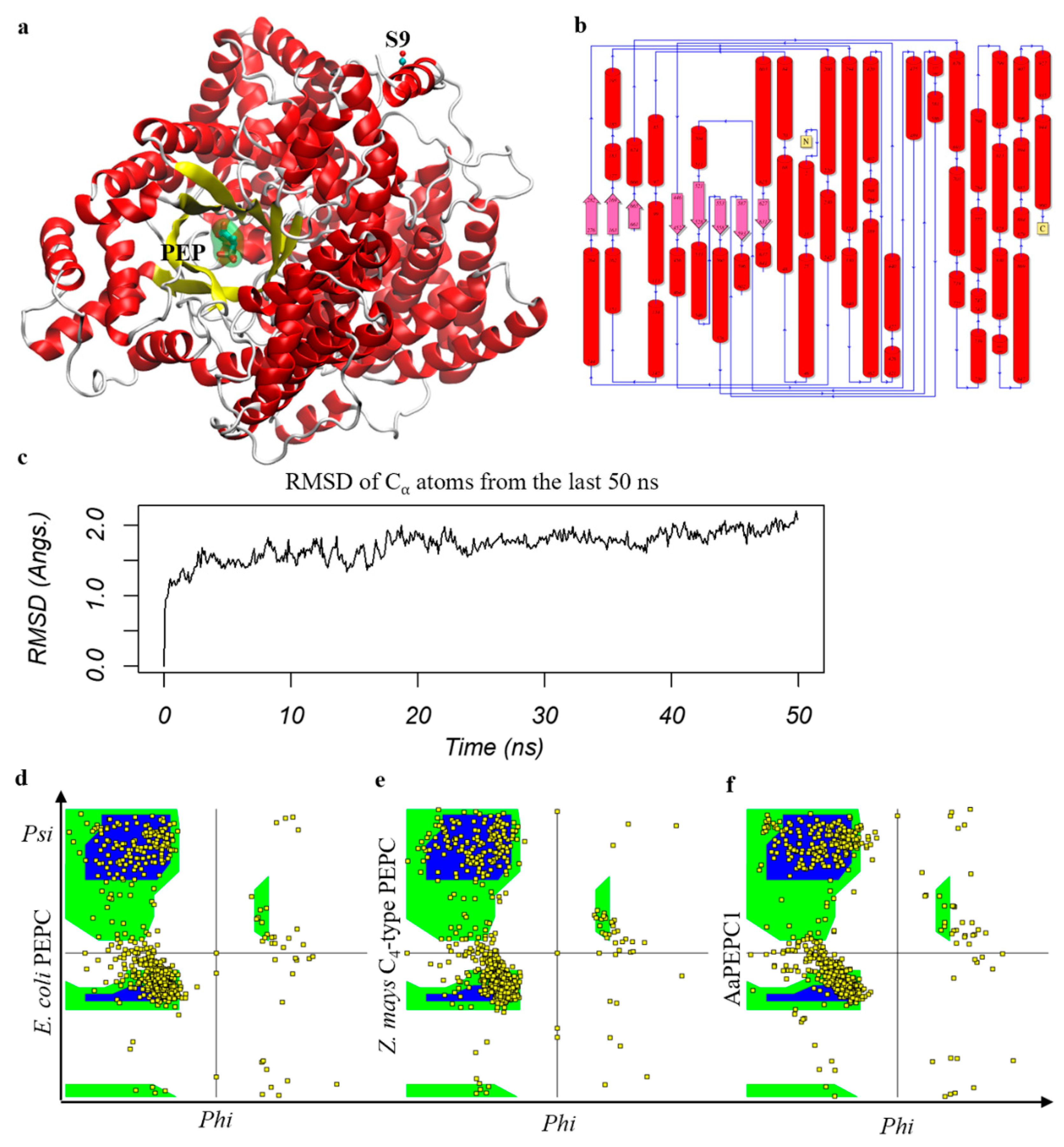
2.3. Structural Modeling and Molecular Dynamics Simulation
2.4. Plasmid Construction
2.5. Plant Transformation
2.6. Measurement of Photosynthesis
2.7. Analysis of Malate, Glucose and Proline Content
2.8. Carbon Isotope Ratio Analysis
2.9. Salt and Drought Stress Treatment
2.10. Expression Levels Analysis of the Related Genes
2.11. Statistical Analysis
3. Results
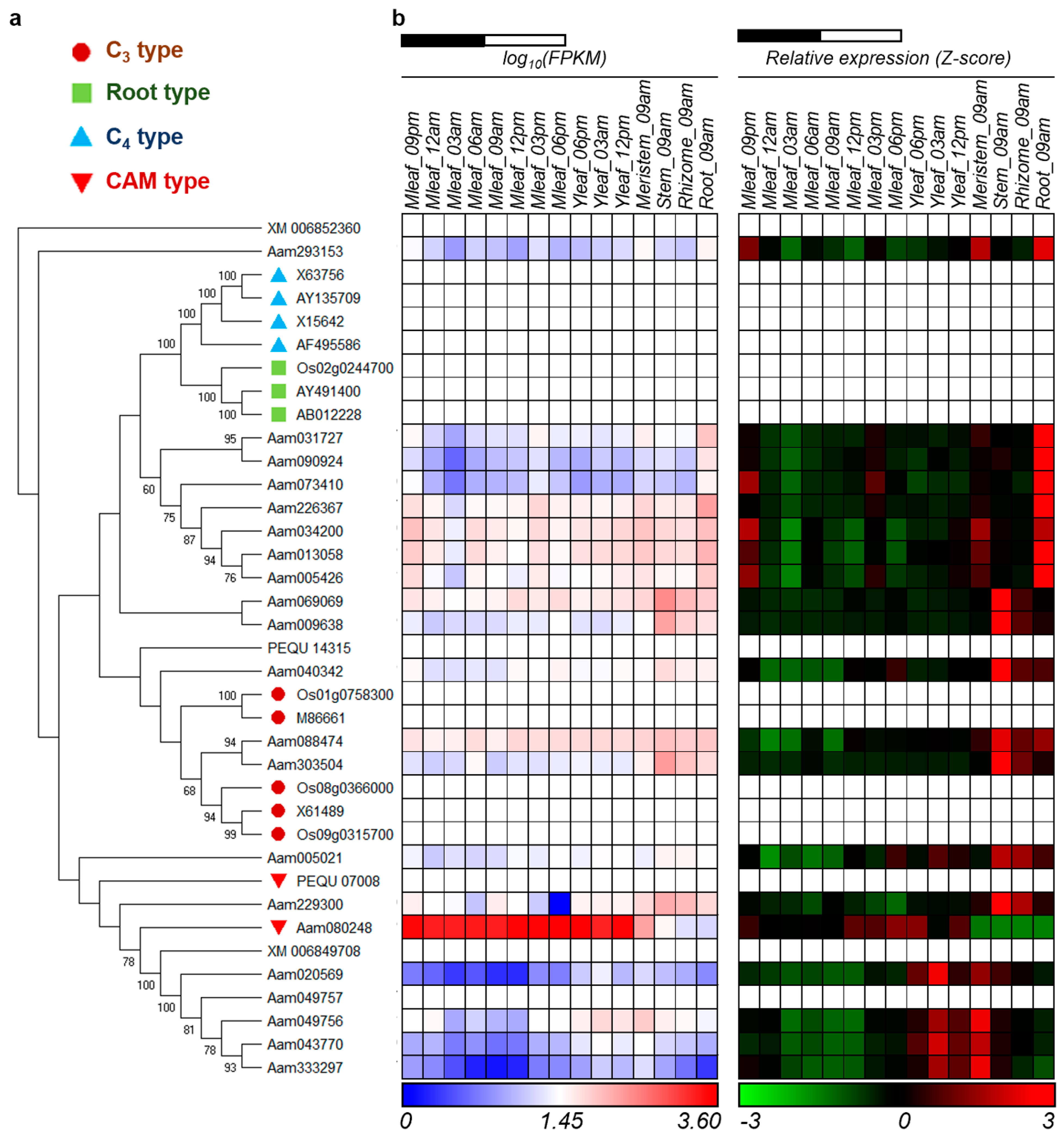
3.1. CAM-Specific PEPC in Agave americana
3.2. AaPEPC1 Binds to Phosphoenolpyruvate
3.3. Development of Transgenic Tobacco Lines Overexpressing AaPEPC1
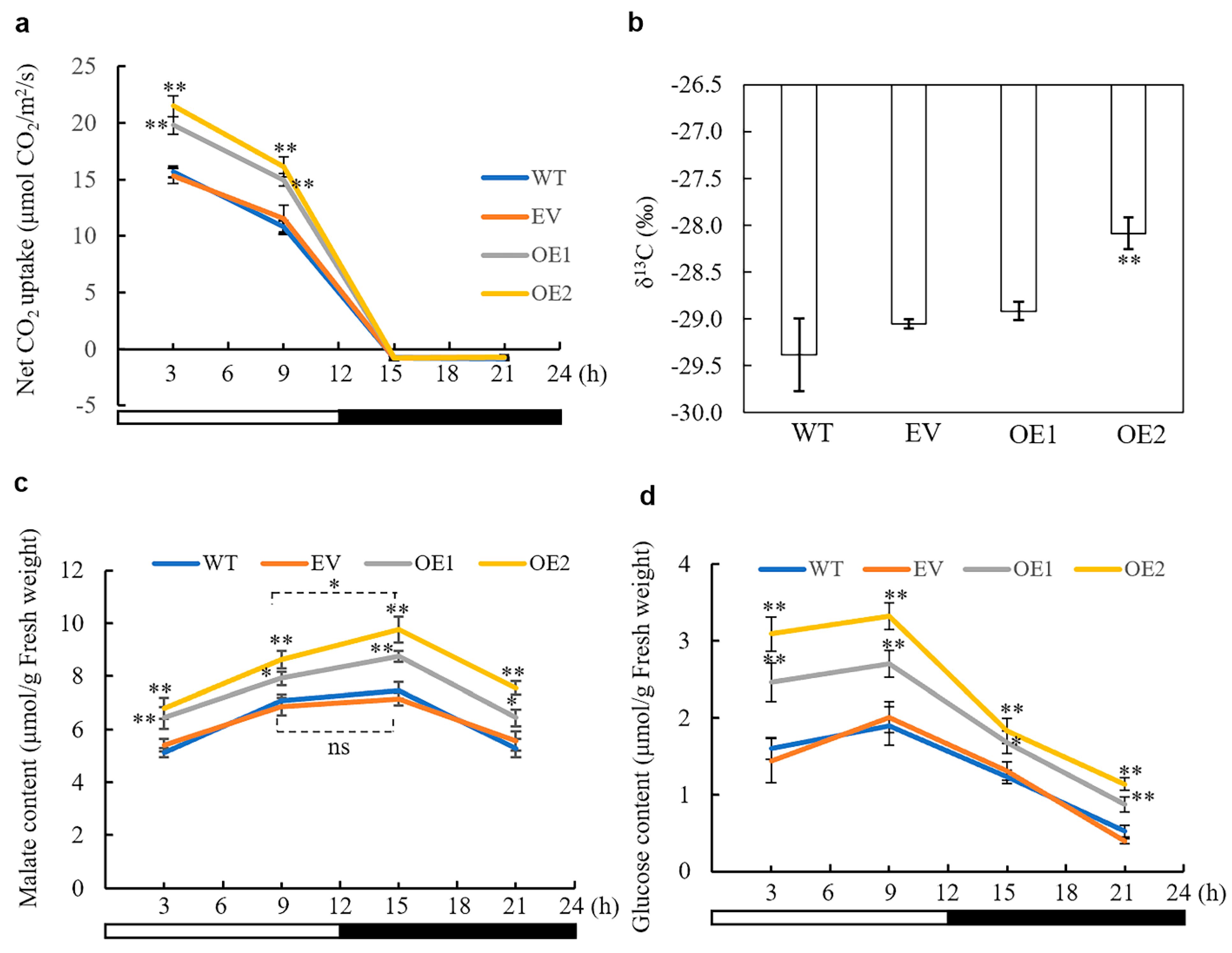
3.4. Overexpression of AaPEPC1 Increases Photosynthetic Rate and Changes Stable Carbon Isotope Ratio
3.5. The Impact of AaPEPC1 Overexpression on the Accumulation of Malate and Glucose
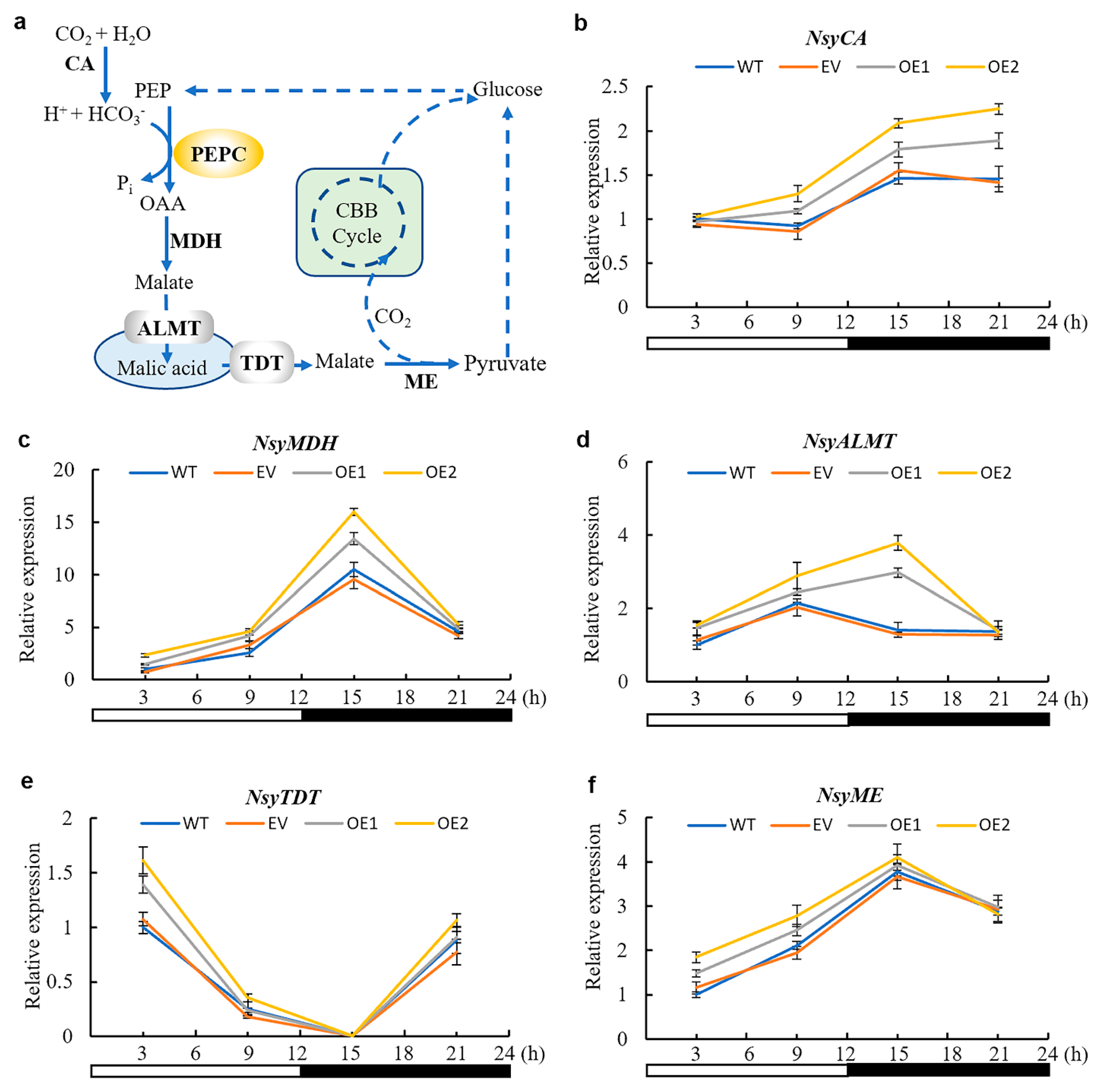
3.6. CAM-Related Genes Were Up-Regulated by AaPEPC1 Overexpression
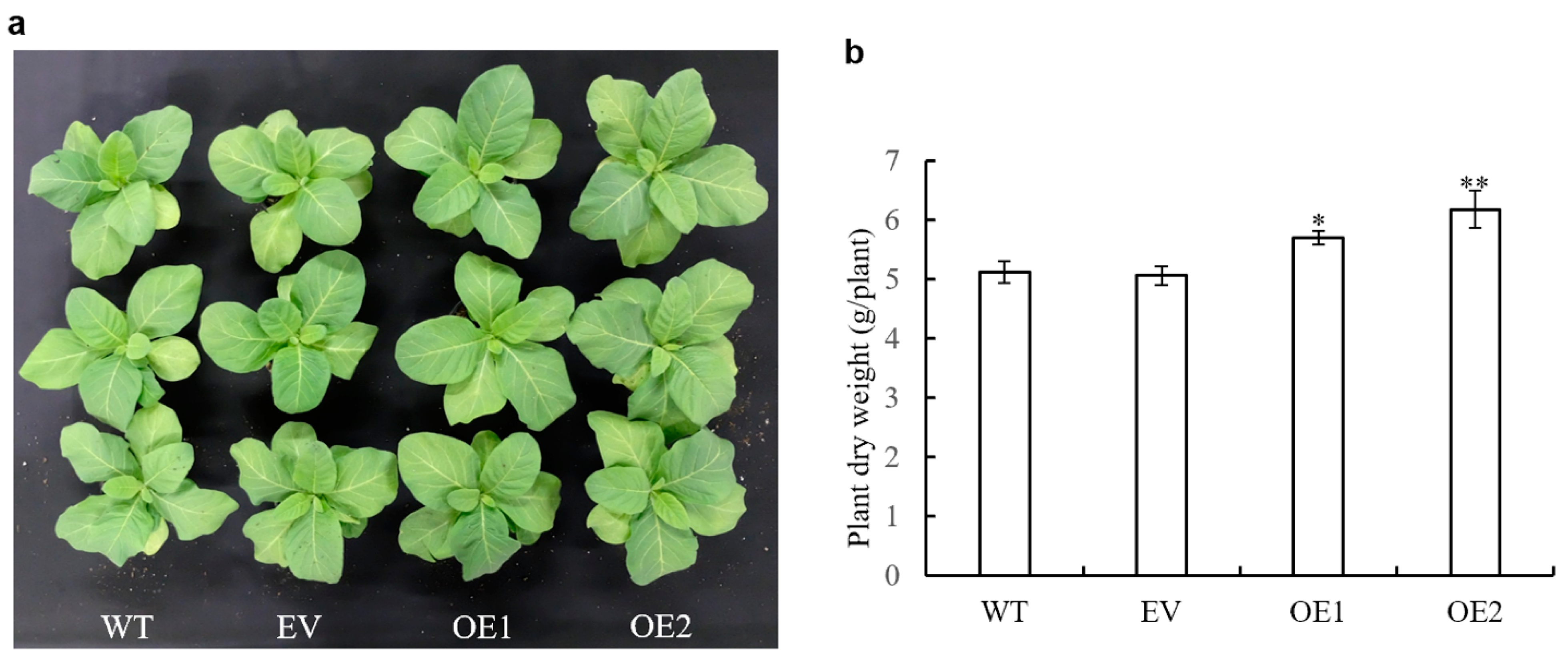
3.7. Impact of AaPEPC1 Overexpression on Biomass Production
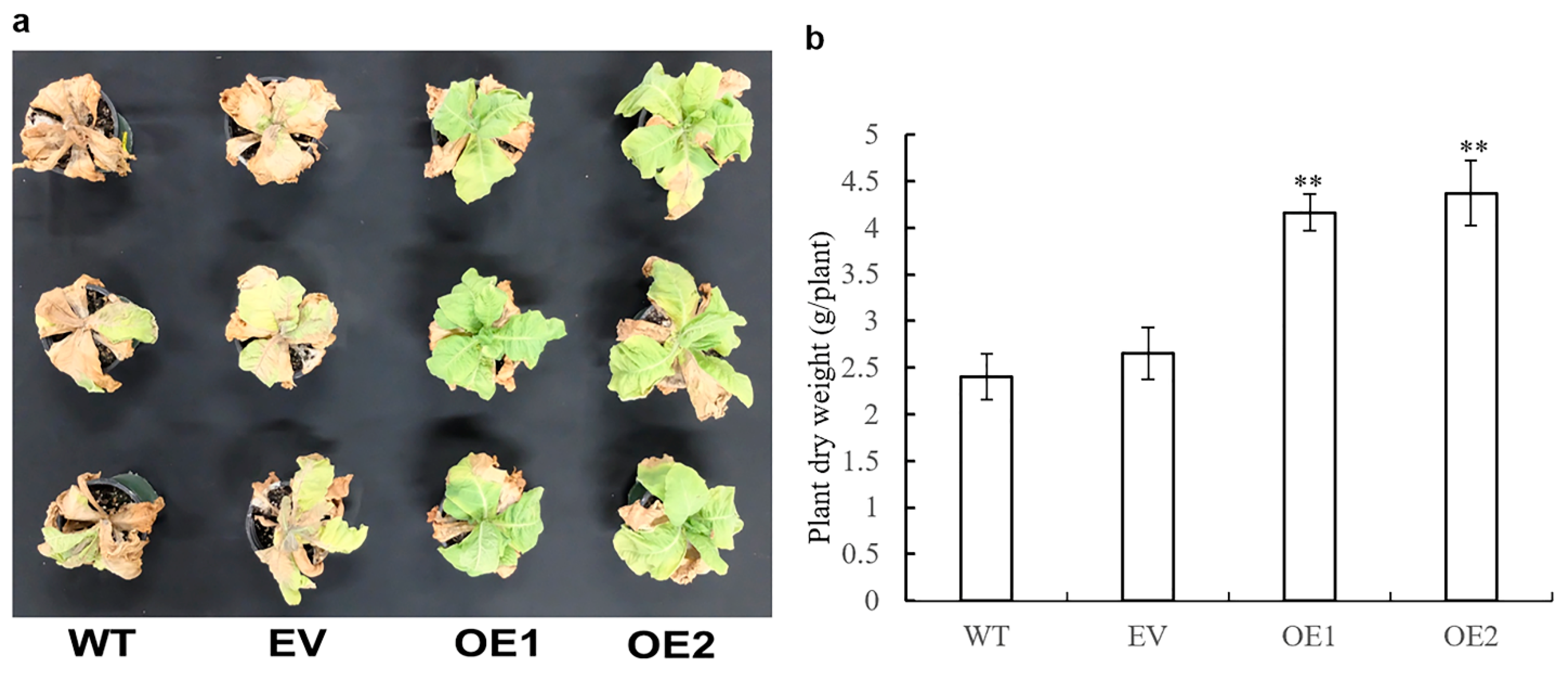
3.8. Impact of AaPEPC1 Overexpression on Salt and Drought Tolerance
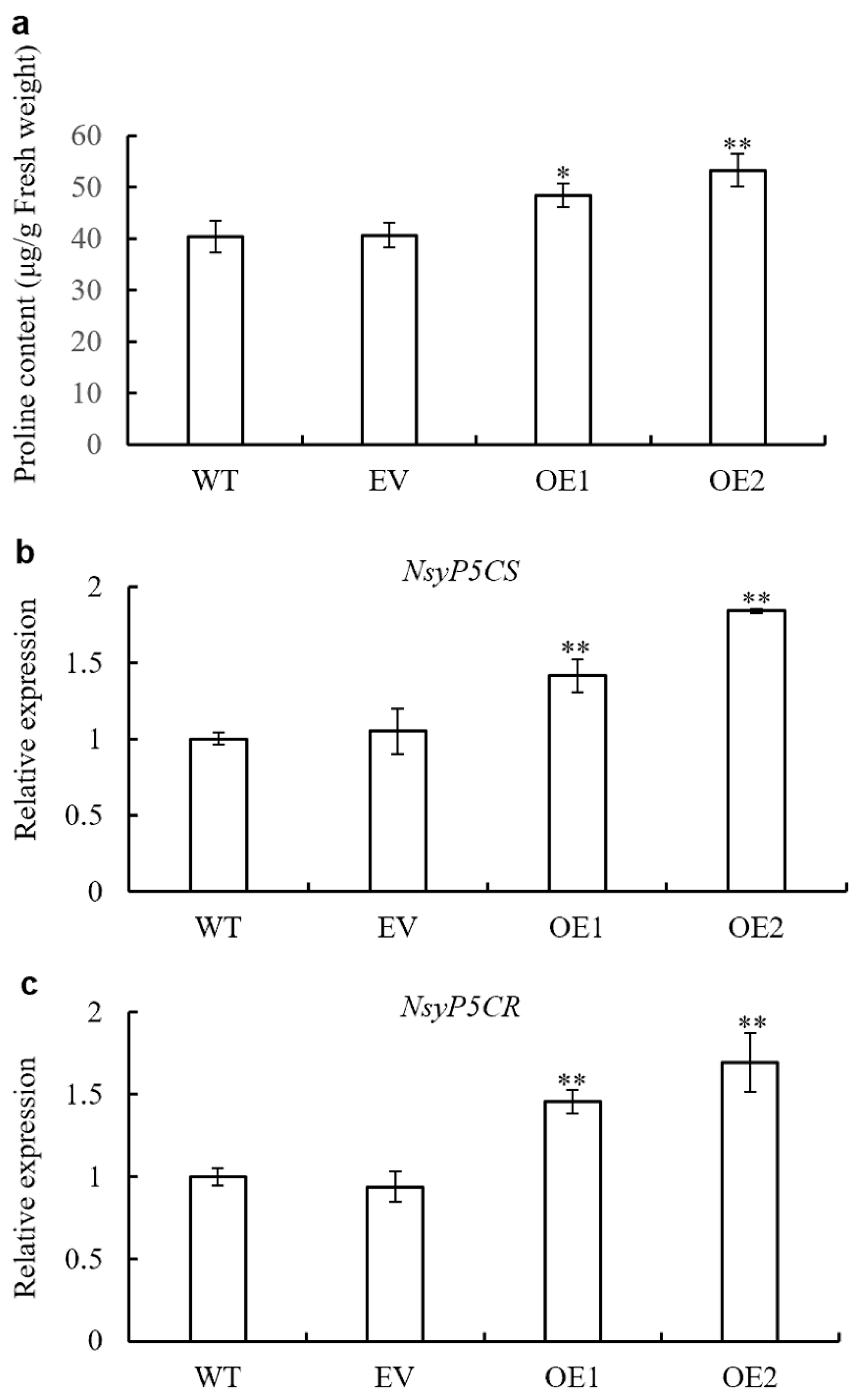
3.9. Proline Biosynthesis Is Enhanced by AaPEPC1 Overexpression
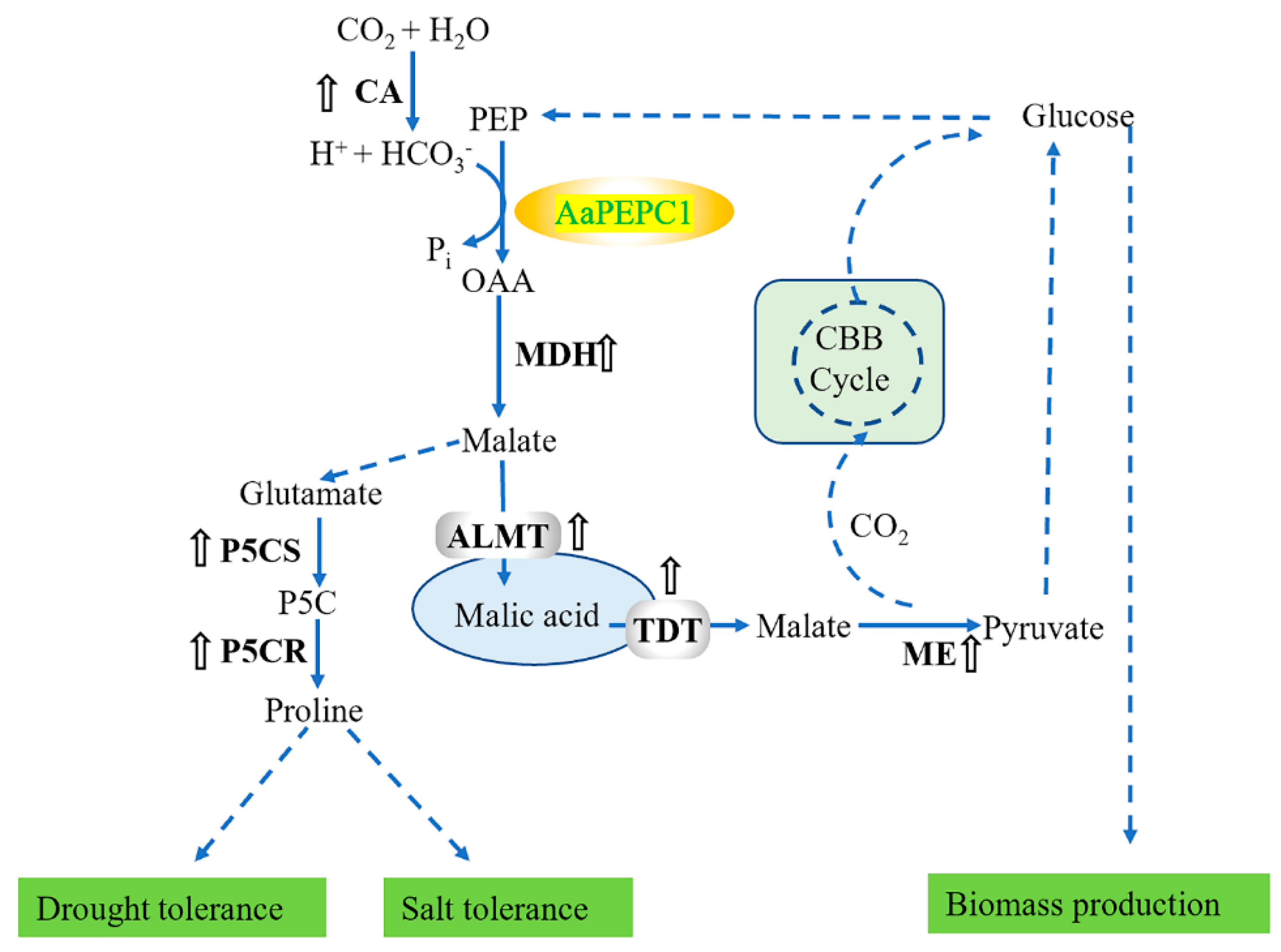
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Additional Information
References
- Yang, X.; Cushman, J.C.; Borland, A.M.; Edwards, E.J.; Wullschleger, S.D.; Tuskan, G.A.; Owen, N.A.; Griffiths, H.; Smith, J.A.C.; De Paoli, H.C. A roadmap for research on crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world. New Phytol. 2015, 207, 491–504. [Google Scholar] [CrossRef]
- Liu, D.; Palla, K.J.; Hu, R.; Moseley, R.C.; Mendoza, C.; Chen, M.; Abraham, P.E.; Labbé, J.L.; Kalluri, U.C.; Tschaplinski, T.J. Perspectives on the basic and applied aspects of crassulacean acid metabolism (CAM) research. Plant Sci. 2018, 274, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Hanjra, M.A.; Qureshi, M.E. Global water crisis and future food security in an era of climate change. Food Policy 2010, 35, 365–377. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef]
- South, P.F.; Cavanagh, A.P.; Liu, H.W.; Ort, D.R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 2019, 363, eaat9077. [Google Scholar] [CrossRef] [PubMed]
- Salesse-Smith, C.E.; Sharwood, R.E.; Busch, F.A.; Kromdijk, J.; Bardal, V.; Stern, D.B. Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat. Plants 2018, 4, 802. [Google Scholar] [CrossRef]
- Hibberd, J.M.; Sheehy, J.E.; Langdale, J.A. Using C4 photosynthesis to increase the yield of rice—Rationale and feasibility. Curr. Opin. Plant Biol. 2008, 11, 228–231. [Google Scholar] [CrossRef]
- Shameer, S.; Baghalian, K.; Cheung, C.M.; Ratcliffe, R.G.; Sweetlove, L.J. Computational analysis of the productivity potential of CAM. Nat. Plants 2018, 4, 165. [Google Scholar] [CrossRef] [PubMed]
- Borland, A.M.; Hartwell, J.; Weston, D.J.; Schlauch, K.A.; Tschaplinski, T.J.; Tuskan, G.A.; Yang, X.; Cushman, J.C. Engineering crassulacean acid metabolism to improve water-use efficiency. Trends Plant Sci. 2014, 19, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Khalid, S.; Abdullah, M.; Ahmed, Z.; Shah, M.K.N.; Ghafoor, A.; Du, X. Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 2019, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; He, S.; Zhai, H.; Wang, L.; Zhao, Y.; Wang, B.; Li, R.; Liu, Q. Overexpression of IbP5CR enhances salt tolerance in transgenic sweetpotato. Plant Cell. Tissue Organ Cult. 2013, 117, 1–16. [Google Scholar] [CrossRef]
- Zhai, H.; Wang, F.; Si, Z.; Huo, J.; Xing, L.; An, Y.; He, S.; Liu, Q. A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol. J. 2016, 14, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.M.; Nadira, U.A.; Qiu, C.-W.; Cao, F.; Chen, Z.-H.; Vincze, E.; Wu, F. The barley S-adenosylmethionine synthetase 3 gene HvSAMS3 positively regulates the tolerance to combined drought and salinity stress in Tibetan wild barley. Cells 2020, 9, 1530. [Google Scholar] [CrossRef]
- Movafeghi, A.; Khataee, A.; Abedi, M.; Tarrahi, R.; Dadpour, M.; Vafaei, F. Effects of TiO2 nanoparticles on the aquatic plant Spirodela polyrrhiza: Evaluation of growth parameters, pigment contents and antioxidant enzyme activities. J. Environ. Sci. 2018, 64, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Tarrahi, R.; Khataee, A.; Movafeghi, A.; Rezanejad, F.; Gohari, G. Toxicological implications of selenium nanoparticles with different coatings along with Se4+ on Lemna minor. Chemosphere 2017, 181, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Turbat, A.; Rakk, D.; Vigneshwari, A.; Kocsubé, S.; Thu, H.; Szepesi, Á.; Bakacsy, L.; D Škrbić, B.; Jigjiddorj, E.-A.; Vágvölgyi, C. Characterization of the plant growth-promoting activities of endophytic fungi isolated from Sophora flavescens. Microorganisms 2020, 8, 683. [Google Scholar] [CrossRef]
- Ehleringer, J.R.; Monson, R.K. Evolutionary and ecological aspects of photosynthetic pathway variation. Annu. Rev. Ecol. Syst. 1993, 24, 411–439. [Google Scholar] [CrossRef]
- Lüttge, U. CO2-concentrating: consequences in crassulacean acid metabolism. J. Exp. Bot. 2002, 53, 2131–2142. [Google Scholar] [CrossRef]
- Owen, N.A.; Griffiths, H. A system dynamics model integrating physiology and biochemical regulation predicts extent of crassulacean acid metabolism (CAM) phases. New Phytol. 2013, 200, 1116–1131. [Google Scholar] [CrossRef]
- Boxall, S.F.; Dever, L.V.; Knerova, J.; Gould, P.D.; Hartwell, J. Phosphorylation of phosphoenolpyruvate carboxylase is essential for maximal and sustained dark CO2 fixation and core circadian clock operation in the obligate crassulacean acid metabolism species Kalanchoë fedtschenkoi. Plant Cell 2017, 29, 2519–2536. [Google Scholar] [CrossRef]
- Eisenhut, M.; Weber, A.P. Improving crop yield. Science 2019, 363, 32–33. [Google Scholar] [CrossRef]
- Miyamoto, S. Salt Tolerance of Landscape Plants Common to the Southwest; Texas Water Resources Institute: El Paso, TX, USA, 2008. [Google Scholar]
- Davis, S.C.; Kuzmick, E.R.; Niechayev, N.; Hunsaker, D.J. Productivity and water use efficiency of Agave americana in the first field trial as bioenergy feedstock on arid lands. GCB Bioenergy 2016, 9, 314–325. [Google Scholar] [CrossRef]
- Yang, X.; Hu, R.; Yin, H.; Jenkins, J.; Shu, S.; Tang, H.; Liu, D.; Weighill, D.A.; Yim, W.C.; Ha, J.; et al. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. 2017, 8, 1899. [Google Scholar] [CrossRef]
- Black, C.C.; Osmond, C.B. Crassulacean acid metabolism photosynthesis: 'Working the night shift'. Photosynth. Res. 2003, 76, 329–341. [Google Scholar] [CrossRef]
- Klavsen, S.K.; Madsen, T.V.; Maberly, S.C. Crassulacean acid metabolism in the context of other carbon-concentrating mechanisms in freshwater plants: A review. Photosynth. Res. 2011, 109, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Silvera, K.; Neubig, K.M.; Whitten, W.M.; Williams, N.H.; Winter, K.; Cushman, J.C. Evolution along the crassulacean acid metabolism continuum. Funct. Plant Biol. 2010, 37, 995–1010. [Google Scholar] [CrossRef]
- Abraham, P.E.; Yin, H.; Borland, A.M.; Weighill, D.; Lim, S.D.; De Paoli, H.C.; Engle, N.; Jones, P.C.; Agh, R.; Weston, D.J.; et al. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat. Plants 2016, 2, 16178. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Masumoto, C.; Miyazawa, S.-I.; Ohkawa, H.; Fukuda, T.; Taniguchi, Y.; Murayama, S.; Kusano, M.; Saito, K.; Fukayama, H.; Miyao, M. Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc. Natl. Acad. Sci. USA 2010, 107, 5226–5231. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Mirjalili, V.; Noyes, K.; Feig, M. Physics-based protein structure refinement through multiple molecular dynamics trajectories and structure averaging. Proteins 2014, 82, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and Side-Chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jablonska, J.; Pravda, L.; Varekova, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Terpe, K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biot. 2003, 60, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Höfgen, R.; Willmitzer, L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988, 16, 9877. [Google Scholar] [CrossRef]
- Zhang, L.; Jia, J.; Xu, Y.; Wang, Y.; Hao, J.; Li, T. Production of transgenic Nicotiana sylvestris plants expressing melatonin synthetase genes and their effect on UV-B-induced DNA damage. Vitr. Cell. Dev. Biol. Plant 2011, 48, 275–282. [Google Scholar] [CrossRef]
- He, S.; Han, Y.; Wang, Y.; Zhai, H.; Liu, Q. In vitro selection and identification of sweetpotato (Ipomoea batatas (L.) Lam.) plants tolerant to NaCl. Plant Cell. Tissue Organ Cult. 2008, 96, 69–74. [Google Scholar] [CrossRef]
- Xia, Z.; Wei, Y.; Sun, K.; Wu, J.; Wang, Y.; Wu, K. The maize AAA-type protein SKD1 confers enhanced salt and drought stress tolerance in transgenic tobacco by interacting with Lyst-interacting protein 5. PLoS ONE 2013, 8, e69787. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- O’Leary, B.; Park, J.; Plaxton, W.C. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): Recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem. J. 2011, 436, 15–34. [Google Scholar] [CrossRef]
- Shi, J.; Yi, K.; Liu, Y.; Xie, L.; Zhou, Z.; Chen, Y.; Hu, Z.; Zheng, T.; Liu, R.; Chen, Y.; et al. Phosphoenolpyruvate carboxylase in Arabidopsis leaves plays a crucial role in carbon and nitrogen metabolism. Plant Physiol. 2015, 167, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Kai, Y.; Matsumura, H.; Izui, K. Phosphoenolpyruvate carboxylase: Three-dimensional structure and molecular mechanisms. Arch. Biochem. Biophys. 2003, 414, 170–179. [Google Scholar] [CrossRef]
- González-Segura, L.; Mújica-Jiménez, C.; Juárez-Díaz, J.A.; Güemez-Toro, R.; Martínez-Castilla, L.P.; Muñoz-Clares, R.A. Identification of the allosteric site for neutral amino acids in the maize C4-isozyme of phosphoenolpyruvate carboxylase: The critical role of Ser-100. J. Biol. Chem. 2018, 293, 9945–9957. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, H.; Xie, Y.; Shirakata, S.; Inoue, T.; Yoshinaga, T.; Ueno, Y.; Izui, K.; Kai, Y. Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases. Structure 2002, 10, 1721–1730. [Google Scholar] [CrossRef]
- Kai, Y.; Matsumura, H.; Inoue, T.; Terada, K.; Nagara, Y.; Yoshinaga, T.; Kihara, A.; Tsumura, K.; Izui, K. Three-dimensional structure of phosphoenolpyruvate carboxylase: A proposed mechanism for allosteric inhibition. 1999, 96, 823–828. Proc. Natl. Acad. Sci. USA 1999, 96, 823–828. [Google Scholar] [CrossRef]
- Winter, K.; Holtum, J.A. How closely do the δ13C values of crassulacean acid metabolism plants reflect the proportion of CO2 fixed during fay and night? Plant Physiol. 2002, 129, 1843–1851. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zhang, G.Q.; Zhang, Y.Q.; Niu, S.; Xiong, J.-S.; Lin, Z.; Cheng, Z.M.; Liu, Z.J. Origin and mechanism of crassulacean acid metabolism in orchids as implied by comparative transcriptomics and genomics of the carbon fixation pathway. Plant J. 2016, 86, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Holtum, J.A.; Aranda, J.; Virgo, A.; Gehrig, H.H.; Winter, K. δ13C values and crassulacean acid metabolism in Clusia species from Panama. Trees 2004, 18, 658–668. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Araújo, W.L.; Fernie, A.R. Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol. 2010, 155, 101–107. [Google Scholar] [CrossRef]
- Bräutigam, A.; Schlüter, U.; Eisenhut, M.; Gowik, U. On the evolutionary origin of CAM photosynthesis. Plant Physiol. 2017, 174, 473–477. [Google Scholar] [CrossRef]
- Munns, R.; Termaat, A. Whole-plant responses to salinity. Funct. Plant Biol. 1986, 13, 143–160. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Patel, M.K.; Kumar, M.; Li, W.; Luo, Y.; Burritt, D.J.; Alkan, N.; Tran, L.-S.P. Enhancing salt tolerance of plants: From metabolic reprogramming to exogenous chemical treatments and molecular approaches. Cells 2020, 9, 2492. [Google Scholar] [CrossRef]
- Boxall, S.F.; Kadu, N.; Dever, L.V.; Knerova, J.; Waller, J.L.; Gould, P.D.; Hartwell, J. Kalanchoë PPC1 is essential for crassulacean acid metabolism and the regulation of core circadian clock and guard cell signaling genes. Plant Cell 2020, 32, 1136–1160. [Google Scholar] [CrossRef] [PubMed]
- Daloso, D.M.; Antunes, W.C.; Pinheiro, D.P.; Waquim, J.P.; AraÚJo, W.L.; Loureiro, M.E.; Fernie, A.R.; Williams, T. Tobacco guard cells fix CO 2 by both Rubisco and PEP case while sucrose acts as a substrate during light-induced stomatal opening. Plant Cell Environ. 2015, 38, 2353–2371. [Google Scholar] [CrossRef]
- Li, L.; Sheen, J.J. Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 2016, 33, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-P.; Lei, Q.-Y. Metabolite sensing and signaling in cell metabolism. Signal Transduct. Target. Ther. 2018, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; McCormack, M.P.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nat. Cell Biol. 2013, 496, 181–186. [Google Scholar] [CrossRef]
- Sheen, J. Master regulators in plant glucose signaling networks. J. Plant Biol. 2014, 57, 67–79. [Google Scholar] [CrossRef]
- Kandoi, D.; Mohanty, S.; Tripathy, B.C. Towards efficient photosynthesis: Overexpression of Zea mays phosphoenolpyruvate carboxylase in Arabidopsis thaliana. Photosynth. Res. 2016, 130, 47–72. [Google Scholar] [CrossRef]
- Zhang, C.; Li, X.; He, Y.; Zhang, J.; Yan, T.; Liu, X. Physiological investigation of C4-phosphoenolpyruvate-carboxylase-introduced rice line shows that sucrose metabolism is involved in the improved drought tolerance. Plant Physiol. Biochem. 2017, 115, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Li, X.; Liu, X.; Chen, P.; Ren, C.; Dai, C. Enhanced drought tolerance in transgenic rice over-expressing of maize C4 phosphoenolpyruvate carboxylase gene via NO and Ca2+. J. Plant Physiol. 2015, 175, 9–20. [Google Scholar] [CrossRef]
- Ku, M.S.; Agarie, S.; Nomura, M.; Fukayama, H.; Tsuchida, H.; Ono, K.; Hirose, S.; Toki, S.; Miyao, M.; Matsuoka, M. High-level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nat. Biotechnol. 1999, 17, 76–80. [Google Scholar] [CrossRef]
- Ding, Z.-S.; Huang, S.-H.; Zhou, B.-Y.; Sun, X.-F.; Zhao, M. Over-expression of phosphoenolpyruvate carboxylase cDNA from C4 millet (Seteria italica) increase rice photosynthesis and yield under upland condition but not in wetland fields. Plant Biotechnol. Rep. 2013, 7, 155–163. [Google Scholar] [CrossRef]
- Lin, M.T.; Occhialini, A.; Andralojc, P.J.; Parry, M.A.J.; Hanson, M.R. A faster Rubisco with potential to increase photosynthesis in crops. Nat. Cell Biol. 2014, 513, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Kromdijk, J.; Głowacka, K.; Leonelli, L.; Gabilly, S.T.; Iwai, M.; Niyogi, K.K.; Long, S.P. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 2016, 354, 857–861. [Google Scholar] [CrossRef]
- Singha, D.L.; Tuteja, N.; Boro, D.; Hazarika, G.N.; Singh, S. Heterologous expression of PDH47 confers drought tolerance in indica rice. Plant Cell. Tissue Organ Cult. 2017, 130, 577–589. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, R.; Li, R.; Yu, W.; Yang, M.; Sheng, J.; Shen, L. Enhanced drought tolerance in tomato plants by overexpression of SlMAPK1. Plant Cell. Tissue Organ Cult. 2018, 133, 27–38. [Google Scholar] [CrossRef]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Kang, C.; Song, X.; Yu, L.; Liu, D.; He, S.; Zhai, H.; Liu, Q. A ζ-carotene desaturase gene, IbZDS, increases β-carotene and lutein contents and enhances salt tolerance in transgenic sweetpotato. Plant Sci. 2017, 262, 39–51. [Google Scholar] [CrossRef]
- Liu, D.; He, S.; Song, X.; Zhai, H.; Liu, N.; Zhang, D.; Ren, Z.; Liu, Q. IbSIMT1, a novel salt-induced methyltransferase gene from Ipomoea batatas, is involved in salt tolerance. Plant Cell. Tissue Organ Cult. 2015, 120, 701–715. [Google Scholar] [CrossRef]
- Ambavaram, M.M.; Basu, S.; Krishnan, A.; Ramegowda, V.; Batlang, U.; Rahman, L.; Baisakh, N.; Pereira, A. Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat. Commun. 2014, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Hu, R.; Zhang, J.; Guo, H.-B.; Cheng, H.; Li, L.; Borland, A.M.; Qin, H.; Chen, J.-G.; Muchero, W.; et al. Overexpression of an Agave Phosphoenolpyruvate Carboxylase Improves Plant Growth and Stress Tolerance. Cells 2021, 10, 582. https://doi.org/10.3390/cells10030582
Liu D, Hu R, Zhang J, Guo H-B, Cheng H, Li L, Borland AM, Qin H, Chen J-G, Muchero W, et al. Overexpression of an Agave Phosphoenolpyruvate Carboxylase Improves Plant Growth and Stress Tolerance. Cells. 2021; 10(3):582. https://doi.org/10.3390/cells10030582
Chicago/Turabian StyleLiu, Degao, Rongbin Hu, Jin Zhang, Hao-Bo Guo, Hua Cheng, Linling Li, Anne M. Borland, Hong Qin, Jin-Gui Chen, Wellington Muchero, and et al. 2021. "Overexpression of an Agave Phosphoenolpyruvate Carboxylase Improves Plant Growth and Stress Tolerance" Cells 10, no. 3: 582. https://doi.org/10.3390/cells10030582
APA StyleLiu, D., Hu, R., Zhang, J., Guo, H.-B., Cheng, H., Li, L., Borland, A. M., Qin, H., Chen, J.-G., Muchero, W., Tuskan, G. A., & Yang, X. (2021). Overexpression of an Agave Phosphoenolpyruvate Carboxylase Improves Plant Growth and Stress Tolerance. Cells, 10(3), 582. https://doi.org/10.3390/cells10030582







