Inflammation-Related Risk Loci in Genome-Wide Association Studies of Coronary Artery Disease
Abstract
:1. Introduction
2. Methodological Background
2.1. GWAS in a Nutshell
2.2. Challenges and Limitations of GWAS
3. Genetic Variants in CAD Linked to Inflammation
3.1. IL6R
3.2. ARHGEF26
3.3. IL5
3.4. SVEP1
3.5. CXCL12
3.6. SH2B3/LNK
3.7. PECAM1
3.8. Further CAD Risk Loci Associated with Inflammation
3.9. The Drop-by-Drop Start of a Great Wave?
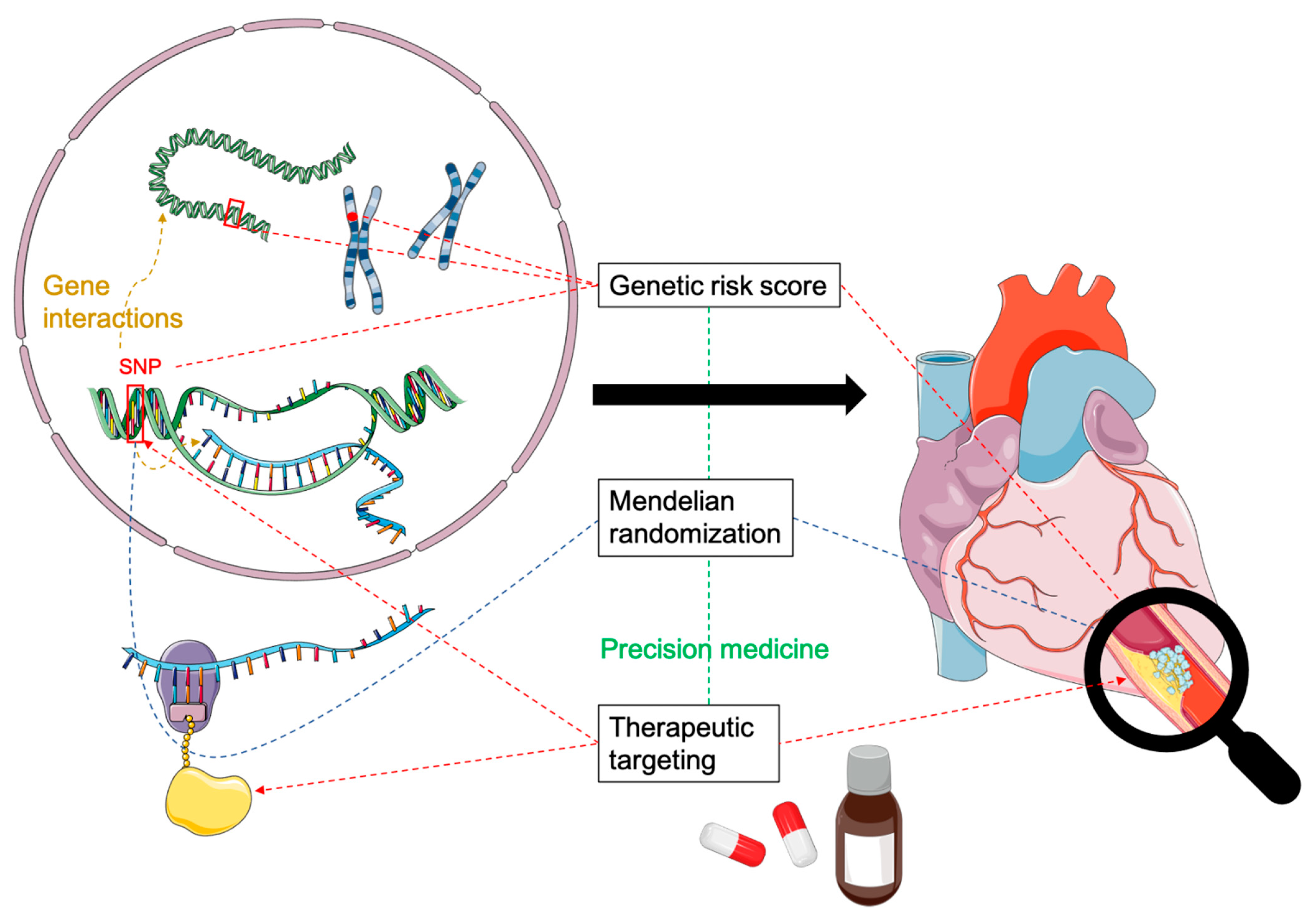
4. Clinical Applications of GWAS Findings
4.1. Polygenic Risk Scores
4.2. Precision Medicine
5. Conclusions
Funding
Conflicts of Interest
Abbreviations
| ARHGEF26 | Rho Guanine Nucleotide Exchange Factor 26 |
| ASSAIL-MI | ASSessing the Effect of Anti-IL-6 Treatment in Myocardial Infarction |
| C1S | Complement Component 1 Subcomponent S |
| C2 | Complement Component 2 |
| CAD | Coronary Artery Disease |
| CANTOS | Canakinumab Antiinflammatory Thrombosis Outcome Study |
| CD | Cluster of Differentiation |
| CFTR | Cystic Fibrosis Transmembrane Conductance Regulator |
| CRE | cis-Regulatory Elements |
| CRISPR-Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats- CRISPR-associated protein 9 |
| CXCL1 | C-X-C Motif Chemokine Ligand 1 |
| CXCL12 | C-X-C Motif Chemokine 12 |
| CXCR4 | C-X-C Chemokine Receptor Type 4 |
| DHX58 | DExH-Box Helicase 58 |
| eQTL | Expression Quantitative Trait Locus |
| GUCY1A1 | Guanylate Cyclase 1 Soluble Subunit Alpha 1 |
| GWAS | Genome-Wide Association Study |
| GxE | Gene-Environment Interaction |
| HDAC9 | Histone Deacetylase 9 |
| IKK | IκB Kinase |
| IL | Interleukin |
| IL6R | Interleukin-6 Receptor |
| ITGB5 | Integrin Beta 5 |
| JCAD/KIAA1462 | Junctional Cadherin 5 Associated |
| MRAS | Muscle RAS Oncogene Homolog |
| mRNA | Messenger Ribonucleic Acid |
| NLRP3 | NOD-, LRR- And Pyrin Domain-Containing Protein 3 |
| PCSK9 | Proprotein Convertase Subtilisin/Kexin Type 9 |
| PECAM-1 | Platelet Endothelial Cell Adhesion Molecule 1 |
| PLG | Plasminogen |
| pQTL | Protein Quantitative Trait Locus |
| PRKCE | Protein Kinase C Epsilon |
| PRS | Polygenic Risk Score |
| PRXL2A/FAM213A | Peroxiredoxin Like 2A |
| RCT | Randomized, Controlled Trial |
| SGEF | Src Homology 3 Domain-Containing Guanine Nucleotide Exchange Factor |
| SH2B3/LNK | SH2B Adapter Protein 3/Lymphocyte Adapter Protein |
| SNP | Single Nucleotide Polymorphism |
| SVEP1 | Sushi, Von Willebrand Factor Type A, EGF and Pentraxin Domain-Containing Protein 1 |
| TLR-4 | Toll-Like-Receptor 4 |
| TNF | Tumor Necrosis Factor |
| TRIM | Tripartite Motif Containing |
References
- Ridker, P.M.; Everett, B.M.; Thuren, T.; MacFadyen, J.G.; Chang, W.H.; Ballantyne, C.; Fonseca, F.; Nicolau, J.; Koenig, W.; Anker, S.D.; et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N. Engl. J. Med. 2017, 377, 1119–1131. [Google Scholar] [CrossRef]
- Tardif, J.C.; Kouz, S.; Waters, D.D.; Bertrand, O.F.; Diaz, R.; Maggioni, A.P.; Pinto, F.J.; Ibrahim, R.; Gamra, H.; Kiwan, G.S.; et al. Efficacy and Safety of Low-Dose Colchicine after Myocardial Infarction. N. Engl. J. Med. 2019, 381, 2497–2505. [Google Scholar] [CrossRef] [PubMed]
- Nidorf, S.M.; Fiolet, A.T.L.; Mosterd, A.; Eikelboom, J.W.; Schut, A.; Opstal, T.S.J.; The, S.H.K.; Xu, X.F.; Ireland, M.A.; Lenderink, T.; et al. Colchicine in Patients with Chronic Coronary Disease. N. Engl. J. Med. 2020, 383, 1838–1847. [Google Scholar] [CrossRef]
- Ridker, P.M.; Everett, B.M.; Pradhan, A.; MacFadyen, J.G.; Solomon, D.H.; Zaharris, E.; Mam, V.; Hasan, A.; Rosenberg, Y.; Iturriaga, E.; et al. Low-Dose Methotrexate for the Prevention of Atherosclerotic Events. N. Engl. J. Med. 2019, 380, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Hall, K.T.; Kessler, T.; Buring, J.E.; Passow, D.; Sesso, H.D.; Zee, R.Y.L.; Ridker, P.M.; Chasman, D.I.; Schunkert, H. Genetic variation at the coronary artery disease risk locus GUCY1A3 modifies cardiovascular disease prevention effects of aspirin. Eur. Heart J. 2019, 40, 3385–3392. [Google Scholar] [CrossRef] [PubMed]
- Stranger, B.E.; Forrest, M.S.; Dunning, M.; Ingle, C.E.; Beazley, C.; Thorne, N.; Redon, R.; Bird, C.P.; de Grassi, A.; Lee, C.; et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007, 315, 848–853. [Google Scholar] [CrossRef] [Green Version]
- Pe’er, I.; Yelensky, R.; Altshuler, D.; Daly, M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008, 32, 381–385. [Google Scholar] [CrossRef] [PubMed]
- International HapMap Consortium; Frazer, K.A.; Ballinger, D.G.; Cox, D.R.; Hinds, D.A.; Stuve, L.L.; Gibbs, R.A.; Belmont, J.W.; Boudreau, A.; Hardenbol, P.; et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007, 449, 851–861. [Google Scholar]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [PubMed] [Green Version]
- Erdmann, J.; Kessler, T.; Munoz Venegas, L.; Schunkert, H. A decade of genome-wide association studies for coronary artery disease: The challenges ahead. Cardiovasc. Res. 2018, 114, 1241–1257. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harismendy, O.; Notani, D.; Song, X.; Rahim, N.G.; Tanasa, B.; Heintzman, N.; Ren, B.; Fu, X.D.; Topol, E.J.; Rosenfeld, M.G.; et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 2011, 470, 264–268. [Google Scholar] [CrossRef] [Green Version]
- Folkersen, L.; van’t Hooft, F.; Chernogubova, E.; Agardh, H.E.; Hansson, G.K.; Hedin, U.; Liska, J.; Syvanen, A.C.; Paulsson-Berne, G.; Franco-Cereceda, A.; et al. Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease. Circ. Cardiovasc. Genet. 2010, 3, 365–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franzen, O.; Ermel, R.; Cohain, A.; Akers, N.K.; Di Narzo, A.; Talukdar, H.A.; Foroughi-Asl, H.; Giambartolomei, C.; Fullard, J.F.; Sukhavasi, K.; et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 2016, 353, 827–830. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, K.; Umicevic Mirkov, M.; de Leeuw, C.A.; van den Heuvel, M.P.; Posthuma, D. Genetic mapping of cell type specificity for complex traits. Nat Commun 2019, 10, 3222. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Talukdar, H.A.; Koplev, S.; Giannarelli, C.; Ivert, T.; Gan, L.M.; Ruusalepp, A.; Schadt, E.E.; Kovacic, J.C.; Lusis, A.J.; et al. Contribution of Gene Regulatory Networks to Heritability of Coronary Artery Disease. J. Am. Coll. Cardiol. 2019, 73, 2946–2957. [Google Scholar] [CrossRef]
- von Scheidt, M.; Zhao, Y.; Kurt, Z.; Pan, C.; Zeng, L.; Yang, X.; Schunkert, H.; Lusis, A.J. Applications and Limitations of Mouse Models for Understanding Human Atherosclerosis. Cell Metab. 2017, 25, 248–261. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, M.D.; Chen-Plotkin, A.S. The Post-GWAS Era: From Association to Function. Am. J. Hum. Genet. 2018, 102, 717–730. [Google Scholar] [CrossRef] [Green Version]
- Lichou, F.; Trynka, G. Functional studies of GWAS variants are gaining momentum. Nat Commun 2020, 11, 6283. [Google Scholar] [CrossRef]
- Kessler, T.; Schunkert, H. Genomic Strategies Toward Identification of Novel Therapeutic Targets. Handb. Exp. Pharmacol. 2020, 1–34. [Google Scholar] [CrossRef]
- CARDIoGRAMplusC4D Consortium; Deloukas, P.; Kanoni, S.; Willenborg, C.; Farrall, M.; Assimes, T.L.; Thompson, J.R.; Ingelsson, E.; Saleheen, D.; Erdmann, J.; et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013, 45, 25–33. [Google Scholar] [CrossRef]
- Zuniga, M.C.; Raghuraman, G.; Hitchner, E.; Weyand, C.; Robinson, W.; Zhou, W. PKC-epsilon and TLR4 synergistically regulate resistin-mediated inflammation in human macrophages. Atherosclerosis 2017, 259, 51–59. [Google Scholar] [CrossRef] [Green Version]
- van der Harst, P.; Verweij, N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018, 122, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, A.K.; Yu, C.; Mann, E.A.; Schridde, A.; Finnemann, S.C.; Mowat, A.M. Expression and characterization of alphavbeta5 integrin on intestinal macrophages. Eur. J. Immunol. 2018, 48, 1181–1187. [Google Scholar] [CrossRef]
- Verweij, N.; Eppinga, R.N.; Hagemeijer, Y.; van der Harst, P. Identification of 15 novel risk loci for coronary artery disease and genetic risk of recurrent events, atrial fibrillation and heart failure. Sci. Rep. 2017, 7, 2761. [Google Scholar] [CrossRef]
- Klarin, D.; Zhu, Q.M.; Emdin, C.A.; Chaffin, M.; Horner, S.; McMillan, B.J.; Leed, A.; Weale, M.E.; Spencer, C.C.A.; Aguet, F.; et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat. Genet. 2017, 49, 1392–1397. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.P.; Goel, A.; Butterworth, A.S.; Kanoni, S.; Webb, T.R.; Marouli, E.; Zeng, L.; Ntalla, I.; Lai, F.Y.; Hopewell, J.C.; et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017, 49, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, J.; Grosshennig, A.; Braund, P.S.; Konig, I.R.; Hengstenberg, C.; Hall, A.S.; Linsel-Nitschke, P.; Kathiresan, S.; Wright, B.; Tregouet, D.A.; et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat. Genet. 2009, 41, 280–282. [Google Scholar] [CrossRef]
- Schunkert, H.; Konig, I.R.; Kathiresan, S.; Reilly, M.P.; Assimes, T.L.; Holm, H.; Preuss, M.; Stewart, A.F.; Barbalic, M.; Gieger, C.; et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011, 43, 333–338. [Google Scholar] [CrossRef] [PubMed]
- van Buul, J.D.; Allingham, M.J.; Samson, T.; Meller, J.; Boulter, E.; Garcia-Mata, R.; Burridge, K. RhoG regulates endothelial apical cup assembly downstream from ICAM1 engagement and is involved in leukocyte trans-endothelial migration. J. Cell Biol. 2007, 178, 1279–1293. [Google Scholar] [CrossRef] [Green Version]
- IBC 50K CAD Consortium. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet 2011, 7, e1002260. [Google Scholar]
- Webb, T.R.; Erdmann, J.; Stirrups, K.E.; Stitziel, N.O.; Masca, N.G.; Jansen, H.; Kanoni, S.; Nelson, C.P.; Ferrario, P.G.; Konig, I.R.; et al. Systematic Evaluation of Pleiotropy Identifies 6 Further Loci Associated with Coronary Artery Disease. J. Am. Coll. Cardiol. 2017, 69, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.K.; Strickland, S. A critical role for plasminogen in inflammation. J. Exp. Med. 2020, 217, e20191865. [Google Scholar] [CrossRef] [PubMed]
- Asare, Y.; Campbell-James, T.A.; Bokov, Y.; Yu, L.L.; Prestel, M.; El Bounkari, O.; Roth, S.; Megens, R.T.A.; Straub, T.; Thomas, K.; et al. Histone Deacetylase 9 Activates IKK to Regulate Atherosclerotic Plaque Vulnerability. Circ. Res. 2020, 127, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.P.; Chmiel, J.F. Inflammation and its genesis in cystic fibrosis. Pediatr. Pulmonol. 2015, 50 (Suppl. 40), S39–S56. [Google Scholar] [CrossRef]
- Winkler, M.J.; Muller, P.; Sharifi, A.M.; Wobst, J.; Winter, H.; Mokry, M.; Ma, L.; van der Laan, S.W.; Pang, S.; Miritsch, B.; et al. Functional investigation of the coronary artery disease gene SVEP1. Basic Res. Cardiol. 2020, 115, 67. [Google Scholar] [CrossRef]
- Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators; Stitziel, N.O.; Stirrups, K.E.; Masca, N.G.; Erdmann, J.; Ferrario, P.G.; Konig, I.R.; Weeke, P.E.; Webb, T.R.; Auer, P.L.; et al. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N. Engl. J. Med. 2016, 374, 1134–1144. [Google Scholar]
- Samani, N.J.; Erdmann, J.; Hall, A.S.; Hengstenberg, C.; Mangino, M.; Mayer, B.; Dixon, R.J.; Meitinger, T.; Braund, P.; Wichmann, H.E.; et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007, 357, 443–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Xu, Y.; Liu, P.; Zhang, S.; Liu, H.; Slavin, S.; Kumar, S.; Koroleva, M.; Luo, J.; Wu, X.; et al. The novel coronary artery disease risk gene JCAD/KIAA1462 promotes endothelial dysfunction and atherosclerosis. Eur. Heart J. 2019, 40, 2398–2408. [Google Scholar] [CrossRef]
- Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011, 43, 339–344. [Google Scholar] [CrossRef]
- Erdmann, J.; Willenborg, C.; Nahrstaedt, J.; Preuss, M.; Konig, I.R.; Baumert, J.; Linsel-Nitschke, P.; Gieger, C.; Tennstedt, S.; Belcredi, P.; et al. Genome-wide association study identifies a new locus for coronary artery disease on chromosome 10p11.23. Eur. Heart J. 2011, 32, 158–168. [Google Scholar] [CrossRef] [Green Version]
- Oh, C.K.; Ha, M.; Han, M.E.; Heo, H.J.; Myung, K.; Lee, Y.; Oh, S.O.; Kim, Y.H. FAM213A is linked to prognostic significance in acute myeloid leukemia through regulation of oxidative stress and myelopoiesis. Hematol. Oncol. 2020, 38, 381–389. [Google Scholar] [CrossRef]
- Ozato, K.; Shin, D.M.; Chang, T.H.; Morse, H.C., 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 2008, 8, 849–860. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.L.; Huang, C.C.; Wu, M.T.; Hsu, W.M.; Chuang, J.H. Innate immune sensor laboratory of genetics and physiology 2 suppresses tumor cell growth and functions as a prognostic marker in neuroblastoma. Cancer Sci. 2018, 109, 3494–3502. [Google Scholar] [CrossRef]
- Howson, J.M.M.; Zhao, W.; Barnes, D.R.; Ho, W.K.; Young, R.; Paul, D.S.; Waite, L.L.; Freitag, D.F.; Fauman, E.B.; Salfati, E.L.; et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat. Genet. 2017, 49, 1113–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium; Swerdlow, D.I.; Holmes, M.V.; Kuchenbaecker, K.B.; Engmann, J.E.; Shah, T.; Sofat, R.; Guo, Y.; Chung, C.; Peasey, A.; et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: A mendelian randomisation analysis. Lancet 2012, 379, 1214–1224. [Google Scholar]
- Reiss, A.B.; Siegart, N.M.; De Leon, J. Interleukin-6 in atherosclerosis: Atherogenic or atheroprotective? Clin. Lipidol. 2017, 12, 14–23. [Google Scholar]
- Kleveland, O.; Kunszt, G.; Bratlie, M.; Ueland, T.; Broch, K.; Holte, E.; Michelsen, A.E.; Bendz, B.; Amundsen, B.H.; Espevik, T.; et al. Effect of a single dose of the interleukin-6 receptor antagonist tocilizumab on inflammation and troponin T release in patients with non-ST-elevation myocardial infarction: A double-blind, randomized, placebo-controlled phase 2 trial. Eur. Heart J. 2016, 37, 2406–2413. [Google Scholar] [CrossRef] [PubMed]
- Anstensrud, A.K. Il-6 inhibition in acute ST-elevation myocardial infarction. Results Immunol. 2014, 4, 8–13. [Google Scholar]
- Sager, H.B.; Heidt, T.; Hulsmans, M.; Dutta, P.; Courties, G.; Sebas, M.; Wojtkiewicz, G.R.; Tricot, B.; Iwamoto, Y.; Sun, Y.; et al. Targeting Interleukin-1beta Reduces Leukocyte Production After Acute Myocardial Infarction. Circulation 2015, 132, 1880–1890. [Google Scholar] [CrossRef]
- Cahill, P.A.; Redmond, E.M. Vascular endothelium - Gatekeeper of vessel health. Atherosclerosis 2016, 248, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Sager, H.B.; Dutta, P.; Dahlman, J.E.; Hulsmans, M.; Courties, G.; Sun, Y.; Heidt, T.; Vinegoni, C.; Borodovsky, A.; Fitzgerald, K.; et al. RNAi targeting multiple cell adhesion molecules reduces immune cell recruitment and vascular inflammation after myocardial infarction. Sci. Transl. Med. 2016, 8, 342ra80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sager, H.B.; Nahrendorf, M. Inflammation: A trigger for acute coronary syndrome. Q. J. Nucl. Med. Mol. Imaging 2016, 60, 185–193. [Google Scholar] [PubMed]
- van Rijssel, J.; Kroon, J.; Hoogenboezem, M.; van Alphen, F.P.; de Jong, R.J.; Kostadinova, E.; Geerts, D.; Hordijk, P.L.; van Buul, J.D. The Rho-guanine nucleotide exchange factor Trio controls leukocyte transendothelial migration by promoting docking structure formation. Mol. Biol. Cell 2012, 23, 2831–2844. [Google Scholar] [CrossRef]
- Samson, T.; van Buul, J.D.; Kroon, J.; Welch, C.; Bakker, E.N.; Matlung, H.L.; van den Berg, T.K.; Sharek, L.; Doerschuk, C.; Hahn, K.; et al. The guanine-nucleotide exchange factor SGEF plays a crucial role in the formation of atherosclerosis. PLoS ONE 2013, 8, e55202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder, C.J.; Hartvigsen, K.; Chang, M.K.; Miller, M.; Broide, D.; Palinski, W.; Curtiss, L.K.; Corr, M.; Witztum, J.L. IL-5 links adaptive and natural immunity specific for epitopes of oxidized LDL and protects from atherosclerosis. J. Clin. Investig. 2004, 114, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Newland, S.A.; Mohanta, S.; Clement, M.; Taleb, S.; Walker, J.A.; Nus, M.; Sage, A.P.; Yin, C.; Hu, D.; Kitt, L.L.; et al. Type-2 innate lymphoid cells control the development of atherosclerosis in mice. Nat Commun 2017, 8, 15781. [Google Scholar] [CrossRef]
- Knutsson, A.; Bjorkbacka, H.; Duner, P.; Engstrom, G.; Binder, C.J.; Nilsson, A.H.; Nilsson, J. Associations of Interleukin-5 With Plaque Development and Cardiovascular Events. JACC Basic Transl Sci 2019, 4, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Berglund, G.; Elmstahl, S.; Janzon, L.; Larsson, S.A. The Malmo Diet and Cancer Study. Design and feasibility. J. Intern. Med. 1993, 233, 45–51. [Google Scholar] [CrossRef]
- Sato-Nishiuchi, R.; Nakano, I.; Ozawa, A.; Sato, Y.; Takeichi, M.; Kiyozumi, D.; Yamazaki, K.; Yasunaga, T.; Futaki, S.; Sekiguchi, K. Polydom/SVEP1 is a ligand for integrin alpha9beta1. J. Biol. Chem. 2012, 287, 25615–25630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morooka, N.; Futaki, S.; Sato-Nishiuchi, R.; Nishino, M.; Totani, Y.; Shimono, C.; Nakano, I.; Nakajima, H.; Mochizuki, N.; Sekiguchi, K. Polydom Is an Extracellular Matrix Protein Involved in Lymphatic Vessel Remodeling. Circ. Res. 2017, 120, 1276–1288. [Google Scholar] [CrossRef]
- Farouk, S.S.; Rader, D.J.; Reilly, M.P.; Mehta, N.N. CXCL12: A new player in coronary disease identified through human genetics. Trends Cardiovasc. Med. 2010, 20, 204–209. [Google Scholar] [CrossRef] [Green Version]
- Heidt, T.; Sager, H.B.; Courties, G.; Dutta, P.; Iwamoto, Y.; Zaltsman, A.; von Zur Muhlen, C.; Bode, C.; Fricchione, G.L.; Denninger, J.; et al. Chronic variable stress activates hematopoietic stem cells. Nat. Med. 2014, 20, 754–758. [Google Scholar] [CrossRef] [Green Version]
- Mehta, N.N.; Li, M.; William, D.; Khera, A.V.; DerOhannessian, S.; Qu, L.; Ferguson, J.F.; McLaughlin, C.; Shaikh, L.H.; Shah, R.; et al. The novel atherosclerosis locus at 10q11 regulates plasma CXCL12 levels. Eur. Heart J. 2011, 32, 963–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doring, Y.; van der Vorst, E.P.C.; Duchene, J.; Jansen, Y.; Gencer, S.; Bidzhekov, K.; Atzler, D.; Santovito, D.; Rader, D.J.; Saleheen, D.; et al. CXCL12 Derived from Endothelial Cells Promotes Atherosclerosis to Drive Coronary Artery Disease. Circulation 2019, 139, 1338–1340. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.H.; He, L.H.; Yu, X.H.; Zhao, Z.W.; Wang, G.; Zou, J.; Wen, F.J.; Zhou, L.; Wan, X.J.; Zhang, D.W.; et al. CXCL12 promotes atherosclerosis by downregulating ABCA1 expression via the CXCR4/GSK3beta/beta-catenin(T120)/TCF21 pathway. J. Lipid Res. 2019, 60, 2020–2033. [Google Scholar] [CrossRef]
- Zernecke, A.; Bot, I.; Djalali-Talab, Y.; Shagdarsuren, E.; Bidzhekov, K.; Meiler, S.; Krohn, R.; Schober, A.; Sperandio, M.; Soehnlein, O.; et al. Protective role of CXC receptor 4/CXC ligand 12 unveils the importance of neutrophils in atherosclerosis. Circ. Res. 2008, 102, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Doring, Y.; Noels, H.; van der Vorst, E.P.C.; Neideck, C.; Egea, V.; Drechsler, M.; Mandl, M.; Pawig, L.; Jansen, Y.; Schroder, K.; et al. Vascular CXCR4 Limits Atherosclerosis by Maintaining Arterial Integrity: Evidence from Mouse and Human Studies. Circulation 2017, 136, 388–403. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dembowsky, K.; Chevalier, E.; Stuve, P.; Korf-Klingebiel, M.; Lochner, M.; Napp, L.C.; Frank, H.; Brinkmann, E.; Kanwischer, A.; et al. C-X-C Motif Chemokine Receptor 4 Blockade Promotes Tissue Repair After Myocardial Infarction by Enhancing Regulatory T Cell Mobilization and Immune-Regulatory Function. Circulation 2019, 139, 1798–1812. [Google Scholar] [CrossRef]
- Mori, T.; Iwasaki, Y.; Seki, Y.; Iseki, M.; Katayama, H.; Yamamoto, K.; Takatsu, K.; Takaki, S. Lnk/Sh2b3 controls the production and function of dendritic cells and regulates the induction of IFN-gamma-producing T cells. J. Immunol. 2014, 193, 1728–1736. [Google Scholar] [CrossRef] [Green Version]
- Boulday, G.; Coulon, F.; Fraser, C.C.; Soulillou, J.P.; Charreau, B. Transcriptional up-regulation of the signaling regulatory protein LNK in activated endothelial cells. Transplantation 2002, 74, 1352–1354. [Google Scholar] [CrossRef] [PubMed]
- Hunt, K.A.; Zhernakova, A.; Turner, G.; Heap, G.A.; Franke, L.; Bruinenberg, M.; Romanos, J.; Dinesen, L.C.; Ryan, A.W.; Panesar, D.; et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008, 40, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Clayton, D.G.; Concannon, P.; Akolkar, B.; Cooper, J.D.; Erlich, H.A.; Julier, C.; Morahan, G.; Nerup, J.; Nierras, C.; et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009, 41, 703–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, D.; Ehret, G.B.; Rice, K.; Verwoert, G.C.; Launer, L.J.; Dehghan, A.; Glazer, N.L.; Morrison, A.C.; Johnson, A.D.; Aspelund, T.; et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009, 41, 677–687. [Google Scholar] [CrossRef] [Green Version]
- Newton-Cheh, C.; Johnson, T.; Gateva, V.; Tobin, M.D.; Bochud, M.; Coin, L.; Najjar, S.S.; Zhao, J.H.; Heath, S.C.; Eyheramendy, S.; et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009, 41, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Gudbjartsson, D.F.; Bjornsdottir, U.S.; Halapi, E.; Helgadottir, A.; Sulem, P.; Jonsdottir, G.M.; Thorleifsson, G.; Helgadottir, H.; Steinthorsdottir, V.; Stefansson, H.; et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009, 41, 342–347. [Google Scholar] [CrossRef]
- Soranzo, N.; Spector, T.D.; Mangino, M.; Kuhnel, B.; Rendon, A.; Teumer, A.; Willenborg, C.; Wright, B.; Chen, L.; Li, M.; et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009, 41, 1182–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takizawa, H.; Nishimura, S.; Takayama, N.; Oda, A.; Nishikii, H.; Morita, Y.; Kakinuma, S.; Yamazaki, S.; Okamura, S.; Tamura, N.; et al. Lnk regulates integrin alphaIIbbeta3 outside-in signaling in mouse platelets, leading to stabilization of thrombus development in vivo. J. Clin. Investig. 2010, 120, 179–190. [Google Scholar] [CrossRef]
- Wang, W.; Tang, Y.; Wang, Y.; Tascau, L.; Balcerek, J.; Tong, W.; Levine, R.L.; Welch, C.; Tall, A.R.; Wang, N. LNK/SH2B3 Loss of Function Promotes Atherosclerosis and Thrombosis. Circ. Res. 2016, 119, e91–e103. [Google Scholar] [CrossRef] [Green Version]
- Flister, M.J.; Hoffman, M.J.; Lemke, A.; Prisco, S.Z.; Rudemiller, N.; O’Meara, C.C.; Tsaih, S.W.; Moreno, C.; Geurts, A.M.; Lazar, J.; et al. SH2B3 Is a Genetic Determinant of Cardiac Inflammation and Fibrosis. Circ. Cardiovasc. Genet. 2015, 8, 294–304. [Google Scholar] [CrossRef] [Green Version]
- Privratsky, J.R.; Paddock, C.M.; Florey, O.; Newman, D.K.; Muller, W.A.; Newman, P.J. Relative contribution of PECAM-1 adhesion and signaling to the maintenance of vascular integrity. J. Cell Sci. 2011, 124, 1477–1485. [Google Scholar] [CrossRef] [Green Version]
- Serebruany, V.L.; Murugesan, S.R.; Pothula, A.; Semaan, H.; Gurbel, P.A. Soluble PECAM-1, but not P-selectin, nor osteonectin identify acute myocardial infarction in patients presenting with chest pain. Cardiology 1999, 91, 50–55. [Google Scholar] [CrossRef]
- Harry, B.L.; Sanders, J.M.; Feaver, R.E.; Lansey, M.; Deem, T.L.; Zarbock, A.; Bruce, A.C.; Pryor, A.W.; Gelfand, B.D.; Blackman, B.R.; et al. Endothelial cell PECAM-1 promotes atherosclerotic lesions in areas of disturbed flow in ApoE-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 2003–2008. [Google Scholar] [CrossRef]
- Goel, R.; Schrank, B.R.; Arora, S.; Boylan, B.; Fleming, B.; Miura, H.; Newman, P.J.; Molthen, R.C.; Newman, D.K. Site-specific effects of PECAM-1 on atherosclerosis in LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1996–2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, Y.; Satoh, T.; Tamura, T.; Wei, P.; Bilasy, S.E.; Edamatsu, H.; Aiba, A.; Katagiri, K.; Kinashi, T.; Nakao, K.; et al. The M-Ras-RA-GEF-2-Rap1 pathway mediates tumor necrosis factor-alpha dependent regulation of integrin activation in splenocytes. Mol. Biol. Cell 2007, 18, 2949–2959. [Google Scholar] [CrossRef]
- Barthel, D.; Schindler, S.; Zipfel, P.F. Plasminogen is a complement inhibitor. J. Biol. Chem. 2012, 287, 18831–18842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, G.; Mehta, V.; Al Haj Zen, A.; Akoumianakis, I.; Goel, A.; Rashbrook, V.S.; Trelfa, L.; Donovan, L.; Drydale, E.; Chuaiphichai, S.; et al. A key role for the novel coronary artery disease gene JCAD in atherosclerosis via shear stress mechanotransduction. Cardiovasc. Res. 2020, 116, 1863–1874. [Google Scholar] [CrossRef]
- Bjorkegren, J.L.M.; Kovacic, J.C.; Dudley, J.T.; Schadt, E.E. Genome-wide significant loci: How important are they? Systems genetics to understand heritability of coronary artery disease and other common complex disorders. J. Am. Coll. Cardiol. 2015, 65, 830–845. [Google Scholar] [PubMed] [Green Version]
- Vilne, B.; Schunkert, H. Integrating Genes Affecting Coronary Artery Disease in Functional Networks by Multi-OMICs Approach. Front Cardiovasc Med 2018, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Civelek, M.; Lusis, A.J. Systems genetics approaches to understand complex traits. Nat Rev Genet 2014, 15, 34–48. [Google Scholar] [CrossRef] [Green Version]
- Braenne, I.; Civelek, M.; Vilne, B.; Di Narzo, A.; Johnson, A.D.; Zhao, Y.; Reiz, B.; Codoni, V.; Webb, T.R.; Foroughi Asl, H.; et al. Prediction of Causal Candidate Genes in Coronary Artery Disease Loci. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 2207–2217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lempiainen, H.; Braenne, I.; Michoel, T.; Tragante, V.; Vilne, B.; Webb, T.R.; Kyriakou, T.; Eichner, J.; Zeng, L.; Willenborg, C.; et al. Network analysis of coronary artery disease risk genes elucidates disease mechanisms and druggable targets. Sci. Rep. 2018, 8, 3434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef]
- Tikkanen, E.; Gustafsson, S.; Ingelsson, E. Associations of Fitness, Physical Activity, Strength, and Genetic Risk with Cardiovascular Disease: Longitudinal Analyses in the UK Biobank Study. Circulation 2018, 137, 2583–2591. [Google Scholar] [CrossRef]
- Ntalla, I.; Kanoni, S.; Zeng, L.; Giannakopoulou, O.; Danesh, J.; Watkins, H.; Samani, N.J.; Deloukas, P.; Schunkert, H.; Group, U.K.B.C.C.C.W. Genetic Risk Score for Coronary Disease Identifies Predispositions to Cardiovascular and Noncardiovascular Diseases. J. Am. Coll. Cardiol. 2019, 73, 2932–2942. [Google Scholar] [CrossRef]
- Mega, J.L.; Stitziel, N.O.; Smith, J.G.; Chasman, D.I.; Caulfield, M.; Devlin, J.J.; Nordio, F.; Hyde, C.; Cannon, C.P.; Sacks, F.; et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: An analysis of primary and secondary prevention trials. Lancet 2015, 385, 2264–2271. [Google Scholar] [CrossRef] [Green Version]
- Schunkert, H.; Samani, N.J. Statin treatment: Can genetics sharpen the focus? Lancet 2015, 385, 2227–2229. [Google Scholar] [CrossRef]
- Damask, A.; Steg, P.G.; Schwartz, G.G.; Szarek, M.; Hagstrom, E.; Badimon, L.; Chapman, M.J.; Boileau, C.; Tsimikas, S.; Ginsberg, H.N.; et al. Patients with High Genome-Wide Polygenic Risk Scores for Coronary Artery Disease May Receive Greater Clinical Benefit from Alirocumab Treatment in the ODYSSEY OUTCOMES Trial. Circulation 2020, 141, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Marston, N.A.; Kamanu, F.K.; Nordio, F.; Gurmu, Y.; Roselli, C.; Sever, P.S.; Pedersen, T.R.; Keech, A.C.; Wang, H.; Lira Pineda, A.; et al. Predicting Benefit from Evolocumab Therapy in Patients with Atherosclerotic Disease Using a Genetic Risk Score: Results From the FOURIER Trial. Circulation 2020, 141, 616–623. [Google Scholar] [CrossRef]
| Locus | Lead SNP | Inflammation-Related Gene | Gene Function(s) | GWAS References |
|---|---|---|---|---|
| 1q21.3 | rs4845625 | IL6R | IL-6 signaling, pro-inflammatory immune responses | [21] |
| 2p21 | rs582384 | PRKCE | Cardiac muscle contraction, cardioprotection, macrophage and dendritic cell activation (TLR-4 signaling) [22] | [23] |
| 3q21.2 | rs142695226 | ITGB5 | αvβ5 integrin component, apoptotic cell phagocytosis [24] | [25,26,27] |
| 3q22.3 | rs2306374 | MRAS | Signal transduction in cell growth and differentiation, oncogene | [28,29] |
| 3q25.2 | rs12493885 | ARHGEF26 | Macropinocytosis, leukocyte transendothelial migration [30] | [25,26,27] |
| 5q31.1 | rs2706399 | IL5 | Eosinophil biology | [31] |
| 6p21.33 | rs3130683 | C2 | Stimulation of phagocytes (complement component) | [32] |
| 6q26 | rs4252120 | PLG | Fibrinolysis, degradation of extracellular matrices, leukocyte migration [33] | [21] |
| 7p21.1 | rs2023938 | HDAC9 | Histone deacetylase, IKK activation, inflammatory responses in macrophages and endothelial cells [34] | [21] |
| 7q31.2 | rs975722 | CFTR | Cystic fibrosis-related chloride channel, respiratory and intestinal immune responses [35] | [23] |
| 9q31.3 | rs111245230 | SVEP1 | Leukocyte transendothelial migration [36] | [37] |
| 10q11.21 | rs1746048 | CXCL12 | Lymphocyte chemotaxis, angiogenesis | [29,38] |
| 10p11.23 | rs2505083 | JCAD/KIAA1462 | Leukocyte endothelium adhesion [39] | [40,41] |
| 10q23.1 | rs17680741 | PRXL2A/FAM213A | Myelopoiesis, negative regulation of p53 tumor suppressor gene [42] | [23] |
| 11p15.4 | rs11601507 | TRIM5, TRIM22, TRIM6 | Innate immune responses, viral infections [43] | [23] |
| 12p13.31 | rs11838267 | C1S | Stimulation of phagocytes (complement component) | [23] |
| 12q24.12 | rs3184504 | SH2B3/LNK | Cytokine signaling | [29] |
| 17q21.2 | rs2074158 | DHX58 | Antiviral signaling, suppression of tumor cell growth [44] | [23] |
| 17q23.3 | rs1867624 | PECAM1 | Endothelium intercellular junctions | [45] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mauersberger, C.; Schunkert, H.; Sager, H.B. Inflammation-Related Risk Loci in Genome-Wide Association Studies of Coronary Artery Disease. Cells 2021, 10, 440. https://doi.org/10.3390/cells10020440
Mauersberger C, Schunkert H, Sager HB. Inflammation-Related Risk Loci in Genome-Wide Association Studies of Coronary Artery Disease. Cells. 2021; 10(2):440. https://doi.org/10.3390/cells10020440
Chicago/Turabian StyleMauersberger, Carina, Heribert Schunkert, and Hendrik B. Sager. 2021. "Inflammation-Related Risk Loci in Genome-Wide Association Studies of Coronary Artery Disease" Cells 10, no. 2: 440. https://doi.org/10.3390/cells10020440
APA StyleMauersberger, C., Schunkert, H., & Sager, H. B. (2021). Inflammation-Related Risk Loci in Genome-Wide Association Studies of Coronary Artery Disease. Cells, 10(2), 440. https://doi.org/10.3390/cells10020440






