TFEB Biology and Agonists at a Glance
Abstract
1. Introduction
2. Autophagy and the Lysosome
3. TFEB as a Drug Target
3.1. The Structure of TFEB
3.2. TFEB Function
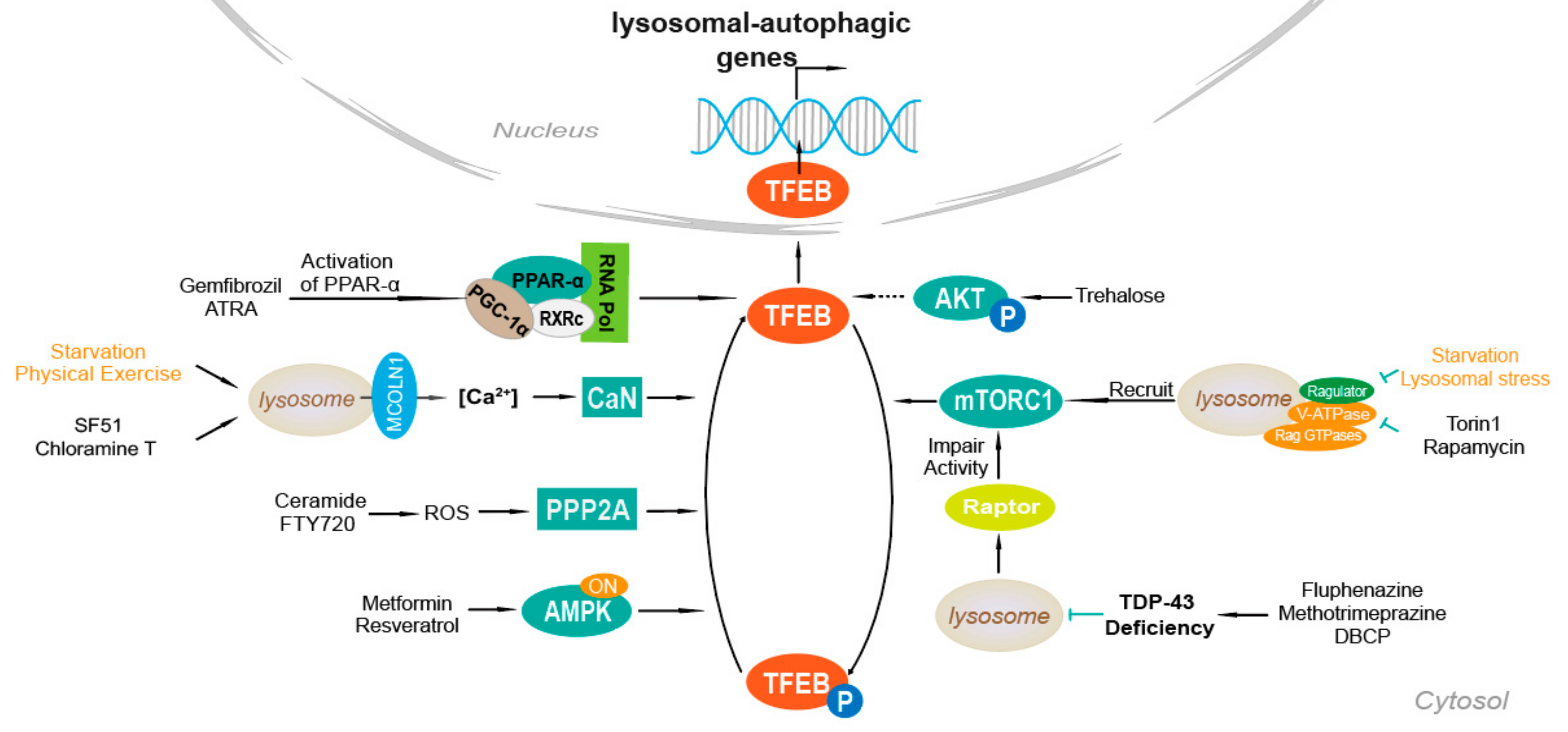
4. Mechanisms of TFEB Activation
4.1. Ca2+-Dependent Mechanism
4.2. AKT
4.3. mTOR
4.4. PPAR-α Activation
4.5. TDP-43 Loss of Function
4.6. AMPK, FLCN, ERK
4.7. PP2A Stimulation
5. TFEB and Diseases
6. Methods to Screen TFEB Agonists
7. TFEB Agonists
7.1. Direct TFEB Agonists
7.1.1. Resveratrol
7.1.2. Curcumin Analog C1
7.1.3. Progestin R5020
7.1.4. Potential TFEB Agonists
7.2. Indirect TFEB Agonists
7.2.1. Torin1
7.2.2. Rapamycin
7.2.3. 3,4-Dimethoxychalcone
7.2.4. Fisetin
8. Clinical Trials and Preclinical Trials of TFEB Agonists
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays Biochem. 2017, 61, 609–624. [Google Scholar]
- Yoshimori, T. Autophagy: A regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 2004, 313, 453–458. [Google Scholar] [CrossRef]
- Chao, X.; Wang, S.; Zhao, K.; Li, Y.; Williams, J.A.; Li, T.; Chavan, H.; Krishnamurthy, P.; He, X.C.; Li, L.; et al. Impaired TFEB-Mediated Lysosome Biogenesis and Autophagy Promote Chronic Ethanol-Induced Liver Injury and Steatosis in Mice. Gastroenterology 2018, 155, 865–879.e12. [Google Scholar] [CrossRef]
- Sha, Y.; Rao, L.; Settembre, C.; Ballabio, A.; Eissa, N.T. STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 2017, 36, 2544–2552. [Google Scholar]
- Wu, C.; Wang, Q.; Xu, D.; Li, M.; Zeng, X. Sirolimus for patients with connective tissue disease-related refractory thrombocytopenia: A single-arm, open-label clinical trial. Rheumatology 2020, keaa645. [Google Scholar] [CrossRef]
- Yang, B.; Ding, L.; Chen, Y.; Shi, J. Augmenting Tumor-Starvation Therapy by Cancer Cell Autophagy Inhibition. Adv. Sci. 2020, 7, 1902847. [Google Scholar] [CrossRef]
- Matoba, K.; Kotani, T.; Tsutsumi, A.; Tsuji, T.; Mori, T.; Noshiro, D.; Sugita, Y.; Nomura, N.; Iwata, S.; Ohsumi, Y.; et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 2020, 27, 1185–1193. [Google Scholar] [CrossRef]
- Maeda, S.; Yamamoto, H.; Kinch, L.N.; Garza, C.M.; Takahashi, S.; Otomo, C.; Grishin, N.V.; Forli, S.; Mizushima, N.; Otomo, T. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 2020, 27, 1194–1201. [Google Scholar] [CrossRef]
- Chen, F.; Amgalan, D.; Kitsis, R.N.; Pessin, J.E.; Feng, D. ATG16L1 autophagy pathway regulates BAX protein levels and programmed cell death. J. Biol. Chem. 2020, 295, 15045–15053. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, C.; Wu, Y.; Chen, B.; Zhang, W.; Zhang, J. Autophagy Protects the Blood-Brain Barrier Through Regulating the Dynamic of Claudin-5 in Short-Term Starvation. Front. Physiol. 2019, 10, 2. [Google Scholar] [CrossRef]
- Liu, K.; Sun, X.; Chen, W.; Sun, Y. Autophagy: A double-edged sword for neuronal survival after cerebral ischemia. Neural Regen. Res. 2014, 9, 1210–1216. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Liu, T.; He, W.; Chen, Y.; Chen, X.; Li, X.; Zhou, W.; Yi, J.; Ren, Z. Antitumor effect of matrine in human hepatoma G2 cells by inducing apoptosis and autophagy. World J. Gastroenterol. 2010, 16, 4281–4290. [Google Scholar] [CrossRef]
- Ge, J.; Liu, Y.; Guo, X.; Gu, L.; Ma, Z.G.; Zhu, Y.P. Resveratrol Induces Apoptosis and Autophagy in T-cell Acute Lymphoblastic Leukemia Cells by Inhibiting Akt/mTOR and Activating p38-MAPK. Biomed. Environ. Sci. 2013, 26, 902–911. [Google Scholar]
- Cai, D.; Han, R.; Liu, J.; Zhang, X.; Lian, D.; Lai, X.; Chen, Y. Puerarin inhibits hyperglycemia-induced NLRP3 inflammasome activation in endothelial cells via regulating autophagy process. Chin. J. Pharm. Toxicol. 2019, 33, 795. [Google Scholar]
- Yang, Y.; Wen, F.; Dang, L.; Fan, Y.; Liu, N.; Wu, K.; Zhao, S. Insulin enhances apoptosis induced by cisplatin in human esophageal squamous cell carcinoma EC9706 cells related to inhibition of autophagy. Chin. Med. J. 2014, 127, 353–358. [Google Scholar]
- Zhu, M.; Deng, G.; Tan, P.; Xing, C.; Guan, C.; Jiang, C.; Zhang, Y.; Ning, B.; Li, C.; Yin, B.; et al. Beclin 2 negatively regulates innate immune signaling and tumor development. J. Clin. Investig. 2020, 130, 5349–5369. [Google Scholar] [CrossRef]
- Tong, H.; Yin, H.; Hossain, M.A.; Wang, Y.; Wu, F.; Dong, X.; Gao, S.; Zhan, K.; He, W. Starvation-induced autophagy promotes the invasion and migration of human bladder cancer cells via TGF-beta1/Smad3-mediated epithelial-mesenchymal transition activation. J. Cell. Biochem. 2019, 120, 5118–5127. [Google Scholar]
- Yang, A.; Herter-Sprie, G.; Zhang, H.; Lin, E.Y.; Biancur, D.; Wang, X.; Deng, J.; Hai, J.; Yang, S.; Wong, K.-K.; et al. Autophagy Sustains Pancreatic Cancer Growth through Both Cell-Autonomous and Nonautonomous Mechanisms. Cancer Discov. 2018, 8, 276–287. [Google Scholar] [CrossRef]
- Vera-Ramirez, L.; Vodnala, S.K.; Nini, R.; Hunter, K.W.; Green, J.E. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Jung, H.S.; Chung, K.W.; Kim, J.W.; Kim, J.; Komatsu, M.; Tanaka, K.; Nguyen, Y.H.; Kang, T.M.; Yoon, K.-H.; Kim, J.-W.; et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008, 8, 418–424. [Google Scholar]
- Han, D.; Yang, B.; Olson, L.K.; Greenstein, A.; Baek, S.-H.; Claycombe, K.J.; Goudreau, J.L.; Yu, S.-W.; Kim, E.-K. Activation of autophagy through modulation of 5′-AMP-activated protein kinase protects pancreatic beta-cells from high glucose. Biochem. J. 2010, 425, 541–551. [Google Scholar]
- Kuma, A.; Mizushima, N. Physiological role of autophagy as an intracellular recycling system: With an emphasis on nutrient metabolism. Semin. Cell Dev. Biol. 2010, 21, 683–690. [Google Scholar] [CrossRef]
- Reiners, J.J.; Agostinis, P.; Berg, K.; Oleinick, N.L.; Kessel, D. Assessing autophagy in the context of photodynamic therapy. Autophagy 2010, 6, 7–18. [Google Scholar] [CrossRef]
- Haoxing, X.; Dejian, R. Lysosomal physiology. Annu. Rev. Physiol. 2015, 77, 57–80. [Google Scholar]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef]
- Gerasimenko, J.V.; Gerasimenko, O.V.; Petersen, O.H. Membrane repair Ca2+-elicited lysosomal exocytosis. Curr. Biol. 2001, 11, R971–R974. [Google Scholar]
- Blott, E.J.; Griffiths, G.M. Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 2002, 3, 122–131. [Google Scholar]
- Samie, M.A.; Xu, H. Lysosomal exocytosis and lipid storage disorders. J. Lipid Res. 2014, 55, 995–1009. [Google Scholar] [CrossRef]
- Wanner, C.; Arad, M.; Baron, R.; Burlina, A.P.; Elliott, P.M.; Feldt-Rasmussen, U.; Fomin, V.V.; Germain, D.; Hughes, A.D.; Jovanovic, A.; et al. European expert consensus statement on therapeutic goals in Fabry disease. Mol. Genet. Metab. 2018, 124, 189–203. [Google Scholar] [CrossRef]
- Carcel-Trullols, J.; Kovacs, A.D.; Pearce, D.A. Cell biology of the NCL proteins: What they do and don’t do. Biochim. Biophys. Acta 2015, 1852, 2242–2255. [Google Scholar]
- Pitcairn, C.; Wani, W.Y.; Mazzulli, J.R. Dysregulation of the autophagic-lysosomal pathway in Gaucher and Parkinson’s disease. Neurobiol. Dis. 2018, 122, 72–82. [Google Scholar] [CrossRef]
- Kazuyoshi, S.; Yoshiya, T. Recent topics on new biological agents for treatment of rheumatoid arthritis. Nihon Naika Gakkai Zasshi J. Jpn. Soc. Intern. Med. 2008, 97, 2418–2423. [Google Scholar]
- Foghsgaard, L.; Wissing, D.; Mauch, D.; Lademann, U.; Bastholm, L.; Boes, M.; Elling, F.; Leist, M.; Jäättelä, M. Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 2001, 153, 999–1010. [Google Scholar]
- Carr, C.S.; Sharp, P.A. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol. Cell. Biol. 1990, 10, 4384–4388. [Google Scholar] [CrossRef]
- Steingrimsson, E.; Copeland, N.G.; Jenkins, N.A. Melanocytes and the Microphthalmia transcription factor network. Annu. Rev. Genet. 2004, 38, 365–411. [Google Scholar]
- Zhang, X.; Yu, L.; Xu, H. Lysosome calcium in ROS regulation of autophagy. Autophagy 2016, 12, 1954–1955. [Google Scholar] [CrossRef]
- Sardiello, M.; Palmieri, M.; Di Ronza, A.; Medina, D.L.; Valenza, M.; Gennarino, V.A.; Di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R.S.; et al. A Gene Network Regulating Lysosomal Biogenesis and Function. Science 2009, 325, 473–477. [Google Scholar] [CrossRef]
- Bajaj, L.; Lotfi, P.; Pal, R.; di Ronza, A.; Sharma, J.; Sardiello, M. Lysosome biogenesis in health and disease. J. Neurochem. 2018, 148, 573–589. [Google Scholar] [CrossRef]
- Rodriguez, A.; Webster, P.; Ortego, J.; Andrews, N.W. Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J. Cell Biol. 1997, 137, 93–104. [Google Scholar]
- Palmieri, M.; Impey, S.; Kang, H.; Di Ronza, A.; Pelz, C.; Sardiello, M.; Ballabio, A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011, 20, 3852–3866. [Google Scholar] [CrossRef]
- Settembre, C.; Ballabio, A. TFEB regulates autophagy: An integrated coordination of cellular degradation and recycling processes. Autophagy 2011, 7, 1379–1381. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Zhou, Z.; Park, J.E.; Wang, L.; Wu, S.; Sun, X.; Lu, L.; Wang, T.; Lin, Q.; et al. Importance of TFEB acetylation in control of its transcriptional activity and lysosomal function in response to histone deacetylase inhibitors. Autophagy 2018, 14, 1–17. [Google Scholar] [CrossRef]
- Miller, A.J.; Levy, C.; Davis, I.J.; Razin, E.; Fisher, D.E. Sumoylation of MITF and Its Related Family Members TFE3 and TFEB. J. Biol. Chem. 2005, 280, 146–155. [Google Scholar] [CrossRef]
- Young, N.P.; Kamireddy, A.; Van Nostrand, J.L.; Eichner, L.J.; Shokhirev, M.N.; Dayn, Y.; Shaw, R.J. AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes Dev. 2016, 30, 535–552. [Google Scholar] [CrossRef]
- Ferron, M.; Settembre, C.; Shimazu, J.; Lacombe, J.; Kato, S.; Rawlings, D.J.; Ballabio, A.; Karsenty, G. A RANKL-PKC-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013, 27, 955–969. [Google Scholar] [CrossRef]
- Sun, J.; Lu, H.; Liang, W.; Zhao, G.; Ren, L.; Hu, D.; Chang, Z.; Liu, Y.; Garcia-Barrio, M.T.; Zhang, J.; et al. Endothelial TFEB (Transcription Factor EB) Improves Glucose Tolerance via Upregulation of IRS (Insulin Receptor Substrate) 1 and IRS2. Arter. Thromb. Vasc. Biol. 2020, 120315310. [Google Scholar] [CrossRef]
- Murano, T.; Najibi, M.; Paulus, G.L.C.; Adiliaghdam, F.; Valencia-Guerrero, A.; Selig, M.; Wang, X.; Jeffrey, K.; Xavier, R.J.; Xavier, R.J.; et al. Transcription factor TFEB cell-autonomously modulates susceptibility to intestinal epithelial cell injury in vivo. Sci. Rep. 2017, 7, 13938. [Google Scholar] [CrossRef]
- Rizzuto, R.; Pozzan, T. Microdomains of Intracellular Ca2+: Molecular Determinants and Functional Consequences. Physiol. Rev. 2006, 86, 369–408. [Google Scholar] [CrossRef]
- Tong, Y.; Song, F. Intracellular calcium signaling regulates autophagy via calcineurin-mediated TFEB dephosphorylation. Autophagy 2015, 11, 1192–1195. [Google Scholar]
- Palmieri, M.; Pal, R.; Sardiello, M. AKT modulates the autophagy-lysosome pathway via TFEB. Cell Cycle 2017, 16, 1237–1238. [Google Scholar] [CrossRef]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Rosato, A.S.; Prezioso, C.; Forrester, A.; et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef]
- Settembre, C.; Zoncu, R.; Medina, D.L.; Vetrini, F.; Erdin, S.; Erdin, S.; Huynh, T.; Ferron, M.; Karsenty, G.; Vellard, M.C.; et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012, 31, 1095–1108. [Google Scholar] [CrossRef]
- Ghosh, A.; Jana, M.; Modi, K.; Gonzales, F.J.; Sims, K.B.; Berry-Kravis, E.; Pahan, K. Activation of peroxisome proliferator-activated receptor alpha induces lysosomal biogenesis in brain cells: Implications for lysosomal storage disorders. J. Biol. Chem. 2015, 290, 10309–10324. [Google Scholar]
- Xia, Q.; Wang, H.; Hao, Z.; Fu, C.; Hu, Q.; Gao, F.; Ren, H.; Chen, D.; Han, J.; Ying, Z.; et al. TDP-43 loss of function increases TFEB activity and blocks autophagosome–lysosome fusion. EMBO J. 2015, 35, 121–142. [Google Scholar] [CrossRef]
- Brown, D.G.; Shorter, J.; Wobst, H.J. Emerging small-molecule therapeutic approaches for amyotrophic lateral sclerosis and frontotemporal dementia. Bioorgan. Med. Chem. Lett. 2020, 30, 126942. [Google Scholar] [CrossRef]
- Collodet, C.; Foretz, M.; Deak, M.; Bultot, L.; Metairon, S.; Viollet, B.; Lefebvre, G.; Raymond, F.; Parisi, A.; Civiletto, G.; et al. AMPK promotes induction of the tumor suppressor FLCN through activation of TFEB independently of mTOR. FASEB J. 2019, 33, 12374–12391. [Google Scholar] [CrossRef]
- El-Houjeiri, L.; Possik, E.; Vijayaraghavan, T.; Paquette, M.; Martina, J.A.; Kazan, J.M.; Ma, E.H.; Jones, R.; Blanchette, P.; Puertollano, R.; et al. The Transcription Factors TFEB and TFE3 Link the FLCN-AMPK Signaling Axis to Innate Immune Response and Pathogen Resistance. Cell Rep. 2019, 26, 3613–3628. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef]
- Wang, J.; Whiteman, M.W.; Lian, H.; Wang, G.; Singh, A.; Huang, D.; Denmark, T. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J. Biol. Chem. 2009, 284, 21412–21424. [Google Scholar] [CrossRef]
- O’Connor, C.M.; Perl, A.; Leonard, D.; Sangodkar, J.; Narla, G. Therapeutic targeting of PP2A. Int. J. Biochem. Cell Biol. 2018, 96, 182–193. [Google Scholar] [CrossRef]
- Martina, J.A.; Puertollano, R. Protein phosphatase 2A stimulates activation of TFEB and TFE3 transcription factors in response to oxidative stress. J. Biol. Chem. 2018, 293, 12525–12534. [Google Scholar] [CrossRef]
- Angelini, C.; Semplicini, C. Enzyme Replacement Therapy for Pompe Disease. Curr. Neurol. Neurosci. Rep. 2011, 12, 70–75. [Google Scholar] [CrossRef]
- Spampanato, C.; Feeney, E.; Li, L.; Cardone, M.; Lim, J.-A.; Annunziata, F.; Zare, H.; Polishchuk, R.; Puertollano, R.; Parenti, G.; et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 2013, 5, 691–706. [Google Scholar] [CrossRef]
- Feeney, E.J.; Spampanato, C.; Puertollano, R.; Ballabio, A.; Parenti, G.; Raben, N. What else is in store for autophagy? Exocytosis of autolysosomes as a mechanism of TFEB-mediated cellular clearance in Pompe disease. Autophagy 2013, 9, 1117–1118. [Google Scholar] [CrossRef][Green Version]
- Tsunemi, T.; Ashe, T.D.; Morrison, B.E.; Soriano, K.R.; Au, J.; Vazquez Roque, R.A.; Lazarowski, E.R.; Damian, V.A.; Masliah, E.; La Spada, A.R. PGC-1alpha rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 2012, 4, 142ra97. [Google Scholar]
- La Spada, A.R. PPARGC1A/PGC-1alpha, TFEB and enhanced proteostasis in Huntington disease: Defining regulatory linkages between energy production and protein-organelle quality control. Autophagy 2012, 8, 1845–1847. [Google Scholar]
- Hartmann, A.; Hunot, S.; Hirsch, E.C. Inflammation and dopaminergic neuronal loss in Parkinson’s disease: A complex matter. Exp. Neurol. 2003, 184, 561–564. [Google Scholar]
- Singh, P.K.; Kotia, V.; Ghosh, D.; Mohite, G.M.; Kumar, A.; Maji, S.K. Curcumin Modulates α-Synuclein Aggregation and Toxicity. ACS Chem. Neurosci. 2013, 4, 393–407. [Google Scholar] [CrossRef]
- Zhang, Y.-D.; Zhao, J.-J. TFEB Participates in the Aβ-Induced Pathogenesis of Alzheimer’s Disease by Regulating the Autophagy-Lysosome Pathway. DNA Cell Biol. 2015, 34, 661–668. [Google Scholar]
- Klionsky, D.J. The autophagosome is overrated! Autophagy 2011, 7, 353–354. [Google Scholar] [CrossRef]
- Najibi, M.; Labed, S.A.; Visvikis, O.; Irazoqui, E.J. An Evolutionarily Conserved PLC-PKD-TFEB Pathway for Host Defense. Cell Rep. 2016, 15, 1728–1742. [Google Scholar] [CrossRef]
- Bao, J.; Zheng, L.; Zhang, Q.; Li, X.; Zhang, X.; Li, Z.; Bai, X.; Zhang, Z.; Hou, W.; Zhao, X.; et al. Deacetylation of TFEB promotes fibrillar Abeta degradation by upregulating lysosomal biogenesis in microglia. Protein Cell 2016, 7, 417–433. [Google Scholar]
- Castillo, K.; Valenzuela, V.; Matus, S.; Nassif, M.; Onate, M.G.; Fuentealba, Y.; Encina, G.; Irrazabal, T.; Parsons, G.; Court, A.F.; et al. Measurement of autophagy flux in the nervous system in vivo. Cell Death Dis. 2013, 4, e917. [Google Scholar] [CrossRef]
- Wang, A.L.; Boulton, M.E.; Dunn, J.W.A.; Rao, H.V.; Cai, J.; Lukas, T.J.; Neufeld, A.H.; A Dunn, W. Using LC3 to Monitor Autophagy Flux in the Retinal Pigment Epithelium. Autophagy 2009, 5, 1190–1193. [Google Scholar] [CrossRef][Green Version]
- Dai, Y.; Li, K.; Wu, W.; Wu, K.; Yi, H.; Li, W.; Xiao, Y.; Zhong, Y.; Cao, Y.; Tian, L. Steroid hormone 20-hydroxyecdysone induces the transcription and complex assembly of V-ATPases to facilitate autophagy in Bombyx mori. Insect Biochem. Mol. Biol. 2020, 116, 103255. [Google Scholar] [CrossRef]
- Kosacka, J.; Nowicki, M.; Blüher, M.; Baum, P.; Stockinger, M.; Toyka, K.; Klöting, I.; Stumvoll, M.; Serke, H.; Bechmann, I.; et al. Increased autophagy in peripheral nerves may protect Wistar Ottawa Karlsburg W rats against neuropathy. Exp. Neurol. 2013, 250, 125–135. [Google Scholar] [CrossRef]
- Wang, C.; Niederstrasser, H.; Douglas, P.M.; Lin, R.; Jaramillo, J.; Li, Y.; Oswald, N.W.; Zhou, A.; McMillan, E.A.; Mendiratta, S.; et al. Small-molecule TFEB pathway agonists that ameliorate metabolic syndrome in mice and extend C. elegans lifespan. Nat. Commun. 2017, 8, 2270. [Google Scholar] [CrossRef]
- Zhou, X.; Yang, J.; Zhou, M.; Zhang, Y.; Liu, Y.; Hou, P.; Zeng, X.; Yi, L.; Mi, M. Resveratrol attenuates endothelial oxidative injury by inducing autophagy via the activation of transcription factor EB. Nutr. Metab. 2019, 16, 1–12. [Google Scholar] [CrossRef]
- Omidian, M.; Abdolahi, M.; Daneshzad, E.; Sedighiyan, M.; Aghasi, M.; Abdollahi, H.; Omidian, P.; Dabiri, S.; Mahmoudi, M.; Hadavi, S.; et al. The Effects of Resveratrol on Oxidative Stress Markers: A Systematic Review and Meta-Analysis of Randomized Clinical Trials. Endocr. Metab. Immune Disord. Drug Targets (Former. Curr. Drug Targets Immune Endocr. Metab. Disord.) 2020, 20, 718–727. [Google Scholar] [CrossRef]
- Song, J.-X.; Sun, Y.-R.; Peluso, I.; Zeng, Y.; Yu, X.; Lu, J.-H.; Xu, Z.; Wang, M.-Z.; Liu, L.-F.; Huang, Y.-Y.; et al. A novel curcumin analog binds to and activates TFEB in vitro and in vivo independent of MTOR inhibition. Autophagy 2016, 12, 1372–1389. [Google Scholar] [CrossRef]
- Song, J.X.; Malampati, S.; Zeng, Y.; Durairajan, S.S.K.; Yang, C.-B.; Tong, B.C.-K.; Iyaswamy, A.; Shang, W.-B.; Sreenivasmurthy, S.G.; Zhu, Z.; et al. A small molecule transcription factor EB activator ameliorates beta-amyloid precursor protein and Tau pathology in Alzheimer’s disease models. Aging Cell 2020, 19, e13069. [Google Scholar]
- Tan, S.; Bajalovic, N.; Wong, E.S.P.; Lin, V.C.L. Ligand-activated progesterone receptor B activates transcription factor EB to promote autophagy in human breast cancer cells. Exp. Cell Res. 2019, 382, 111433. [Google Scholar] [CrossRef]
- Settembre, C.; De Cegli, R.; Mansueto, G.; Saha, P.K.; Vetrini, F.; Visvikis, O.; Huynh, T.; Carissimo, A.; Palmer, D.; Klisch, T.J.; et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013, 15, 647–658. [Google Scholar]
- Thoreen, C.C.; Kang, S.A.; Chang, J.W.; Liu, Q.; Zhang, J.; Gao, Y.; Reichling, L.J.; Sim, T.; Sabatini, D.M.; Gray, N.S. An ATP-competitive Mammalian Target of Rapamycin Inhibitor Reveals Rapamycin-resistant Functions of mTORC1. J. Biol. Chem. 2009, 284, 8023–8032. [Google Scholar] [CrossRef]
- Cheng, N.-T.; Guo, A.; Cui, Y.-P. Intra-articular injection of Torin 1 reduces degeneration of articular cartilage in a rabbit osteoarthritis model. Bone Jt. Res. 2016, 5, 218–224. [Google Scholar] [CrossRef]
- Chen, G.; Xie, W.; Nah, J.; Sauvat, A.; Liu, P.; Pietrocola, F.; Sica, V.; Carmona-Gutierrez, D.; Zimmermann, A.; Pendl, T.; et al. 3,4-Dimethoxychalcone induces autophagy through activation of the transcription factors TFE 3 and TFEB. EMBO Mol. Med. 2019, 11, e10469. [Google Scholar] [CrossRef]
- Kim, S.; Choi, K.J.; Cho, S.-J.; Yun, S.-M.; Jeon, J.-P.; Koh, Y.H.; Song, J.; Johnson, G.V.W.; Jo, C. Fisetin stimulates autophagic degradation of phosphorylated tau via the activation of TFEB and Nrf2 transcription factors. Sci. Rep. 2016, 6, 24933. [Google Scholar] [CrossRef]
- Abe, K.; Yano, T.; Tanno, M.; Miki, T.; Kuno, A.; Sato, T.; Kouzu, H.; Nakata, K.; Ohwada, W.; Kimura, Y.; et al. mTORC1 inhibition attenuates necroptosis through RIP1 inhibition-mediated TFEB activation. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2019, 1865, 165552. [Google Scholar] [CrossRef]
- Cui, H.; Cheng, Y.; He, Y.; Cheng, W.; Zhao, W.; Zhao, H.; Zhou, F.H.; Wang, L.; Dong, J.; Cai, S. The AKT inhibitor MK2206 suppresses airway inflammation and the pro-remodeling pathway in a TDI-induced asthma mouse model. Mol. Med. Rep. 2020, 22, 3723–3734. [Google Scholar] [CrossRef]
- Peña-Llopis, S.; Vega-Rubin-De-Celis, S.; Schwartz, J.C.; Wolff, N.C.; Tran, T.A.T.; Zou, L.; Xie, X.-J.; Corey, D.R.; Brugarolas, J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011, 30, 3242–3258. [Google Scholar] [CrossRef]
- Peña-Llopis, S.; Brugarolas, J. TFEB, a novel mTORC1 effector implicated in lysosome biogenesis, endocytosis and autophagy. Cell Cycle 2011, 10, 3987–3988. [Google Scholar] [CrossRef]
- Martina, J.A.; Puertollano, R. Rag GTPases mediate amino acid–dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 2013, 200, 475–491. [Google Scholar] [CrossRef]
- Roczniak-Ferguson, A.; Petit, C.S.; Froehlich, F.; Qian, S.; Ky, J.; Angarola, B.; Walther, T.C.; Ferguson, S.M. The Transcription Factor TFEB Links mTORC1 Signaling to Transcriptional Control of Lysosome Homeostasis. Sci. Signal. 2012, 5, ra42. [Google Scholar] [CrossRef]
- Morran, D.C.; Wu, J.; Jamieson, N.B.; Mrowinska, A.; Kalna, G.; Karim, S.A.; Au, A.Y.M.; Scarlett, C.J.; Chang, D.K.; Pajak, M.Z.; et al. Targeting mTOR dependency in pancreatic cancer. Gut 2014, 63, 1481–1489. [Google Scholar] [CrossRef]
- Kepp, O.; Chen, G.; Carmona-Gutierrez, D.; Madeo, F.; Kroemer, G. A discovery platform for the identification of caloric restriction mimetics with broad health-improving effects. Autophagy 2019, 16, 188–189. [Google Scholar] [CrossRef]
- Lara, P.N.; Longmate, J.; Mack, P.C.; Kelly, K.; Socinski, M.A.; Salgia, R.; Gitlitz, B.J.; Li, T.; Koczywas, M.; Reckamp, K.L.; et al. Phase II Study of the AKT Inhibitor MK-2206 plus Erlotinib in Patients with Advanced Non–Small Cell Lung Cancer Who Previously Progressed on Erlotinib. Clin. Cancer Res. 2015, 21, 4321–4326. [Google Scholar] [CrossRef]
- Palmieri, M.; Pal, R.; Nelvagal, H.R.; Lotfi, P.; Stinnett, G.R.; Seymour, M.L.; Chaudhury, A.; Bajaj, L.; Bondar, V.V.; Bremner, L.; et al. Corrigendum: mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 2017, 8, 15793. [Google Scholar] [CrossRef]
- Patel, K.R.; Scott, E.; Brown, V.A.; Gescher, A.J.; Steward, W.P.; Brown, A.K. Clinical trials of resveratrol. Ann. N. Y. Acad. Sci. 2011, 1215, 161–169. [Google Scholar] [CrossRef]
| Method | Function | Detection Indicator | Reference |
|---|---|---|---|
| Nuclear translocation | When TFEB is activated, it is displaced into the nucleus, so it is possible to determine whether TFEB is activated by observing the nucleus translocation. | DAPI, GFP, FITC, EB | Najibi, et al., 2016 [71] |
| Gene expression (qPCR) | Fluorescence chemicals were used to measure the total amount of products after each PCR cycle | CT value | Bao, et al., 2016 [72] |
| Promoter activation | The initiation of transcription is the key stage of gene expression, and promoter activation increases the level of gene expression. | Target protein | Ghosh, et al., 2015 [53] |
| Autophagy flux | Observing the accumulation of autophagosomes can reflect the induction of autophagosomes and the reduction of autophagosome consumption | mRFP-GFP-LC3, LC3-I, LC3-II | Castillo, et al., 2013 [73] Wang, et al., 2009 [74] |
| Protein expression (WB) | TFEB is activated and the amount of target protein produced increases. | Target protein | Dai, et al., 2020 [75] |
| Electrophysiology | Membrane potential changes as substances cross cell membranes, and TFEB changes are observed by recording the electrical activity of cells in the body. | Transmembrane current, Action potential | Kosacka, et al., 2013 [76] |
| A UPS-enabled high-throughput screen | Trace, fast, sensitive, and accurate screening the TFEB agonists. | PH, LAMP1 | Wang, et al., 2017 [77] |
| TFEB Agonists | Structure | Mechanism | Clinical Trials and Preclinical Trials | Reference |
|---|---|---|---|---|
| Resveratrol (D) | 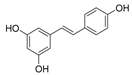 | How RSV regulates autophagy has not been discovered. | Clinical trials | Zhou, et al., 2019 [78] Elnaz, et al., 2020 [79] |
| Curcumin analog C1 (D) | 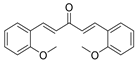 | C1 binds specifically to TFEB, facilitating TFEB entry into the nucleus. | Preclinical trials | Song, et al., 2016 [80] Song, et al., 2020 [81] |
| Progestin R5020 (D) |  | The interaction between PRB and TFEB increased autophagy in McF-7 breast cancer cells. | Preclinical trials | Tan, et al., 2019 [82] |
| Potential TFEB agonists (D) | A class of compounds related to Ca2+ that are structurally diverse | Ca2+ dependent mechanisms | Approved drug digoxin, ikarugamycin, alexidine dihydrochloride | Wang, et al., 2017 [77] Settembre, et al., 2013 [83] |
| Torin1 (In) | 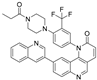 | Inhibits mTOR | Preclinical trials | Thoreen, et al., 2009 [84] Cheng, et al., 2016 [85] |
| 3,4-Dimet-hoxychalc-one (In) |  | Inhibits mTOR | Preclinical trials | Chen, et al., 2019 [86] |
| Fisetin (In) |  | Inhibits mTOR and is related to ALP | Preclinical trials | Kim, et al., 2016 [87] |
| Rapamycin (In) | 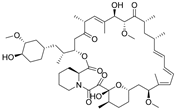 | Inhibits mTOR | Approved drug: Sirolimus | Abe, et al., 2019 [88] Cui, et al., 2020 [89] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Dai, Y.; Liu, S.; Fan, Y.; Ding, Z.; Li, D. TFEB Biology and Agonists at a Glance. Cells 2021, 10, 333. https://doi.org/10.3390/cells10020333
Chen M, Dai Y, Liu S, Fan Y, Ding Z, Li D. TFEB Biology and Agonists at a Glance. Cells. 2021; 10(2):333. https://doi.org/10.3390/cells10020333
Chicago/Turabian StyleChen, Mingyue, Yashuang Dai, Siyu Liu, Yuxin Fan, Zongxian Ding, and Dan Li. 2021. "TFEB Biology and Agonists at a Glance" Cells 10, no. 2: 333. https://doi.org/10.3390/cells10020333
APA StyleChen, M., Dai, Y., Liu, S., Fan, Y., Ding, Z., & Li, D. (2021). TFEB Biology and Agonists at a Glance. Cells, 10(2), 333. https://doi.org/10.3390/cells10020333






