A Transcription Regulatory Sequence in the 5′ Untranslated Region of SARS-CoV-2 Is Vital for Virus Replication with an Altered Evolutionary Pattern against Human Inhibitory MicroRNAs
Abstract
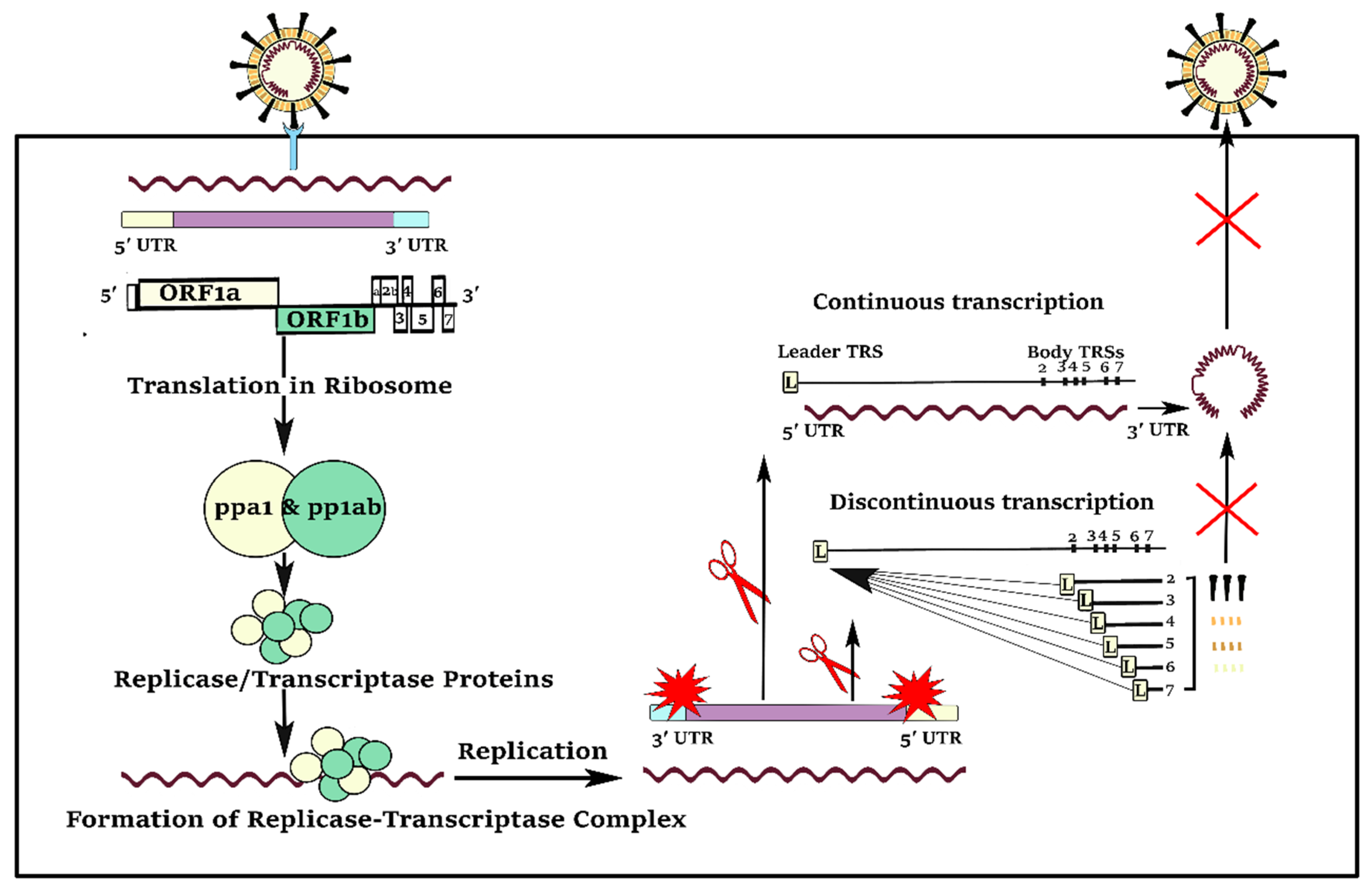
1. Background
2. Methods
2.1. Data Collection
2.2. Inhibitory MicroRNA Prediction
2.3. Sequence Alignment between 5′UTR of Coronavirus and MicroRNA Seed Regions and Phylogenetic Trees
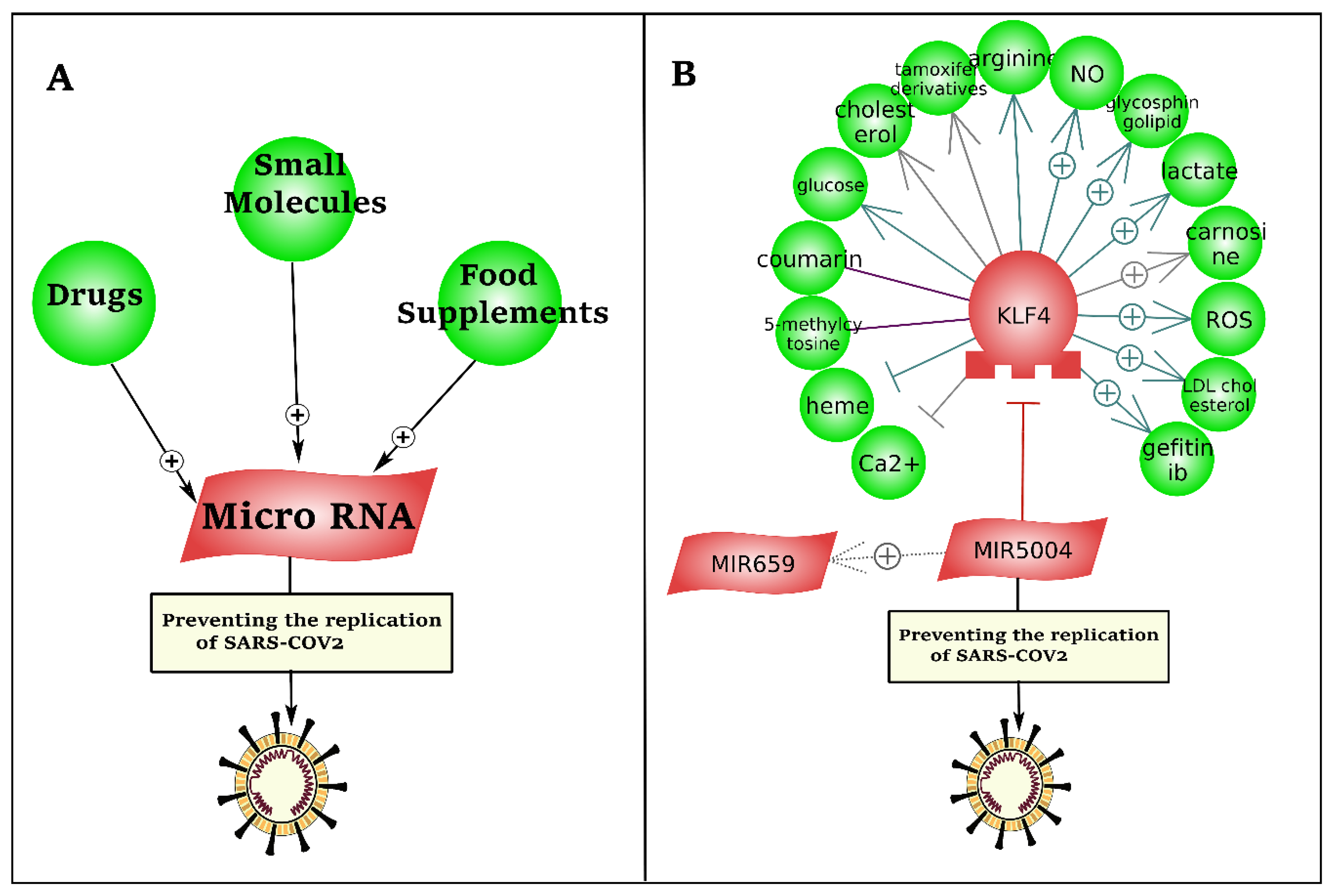
2.4. Literature-Mining Based Drug Repurposing
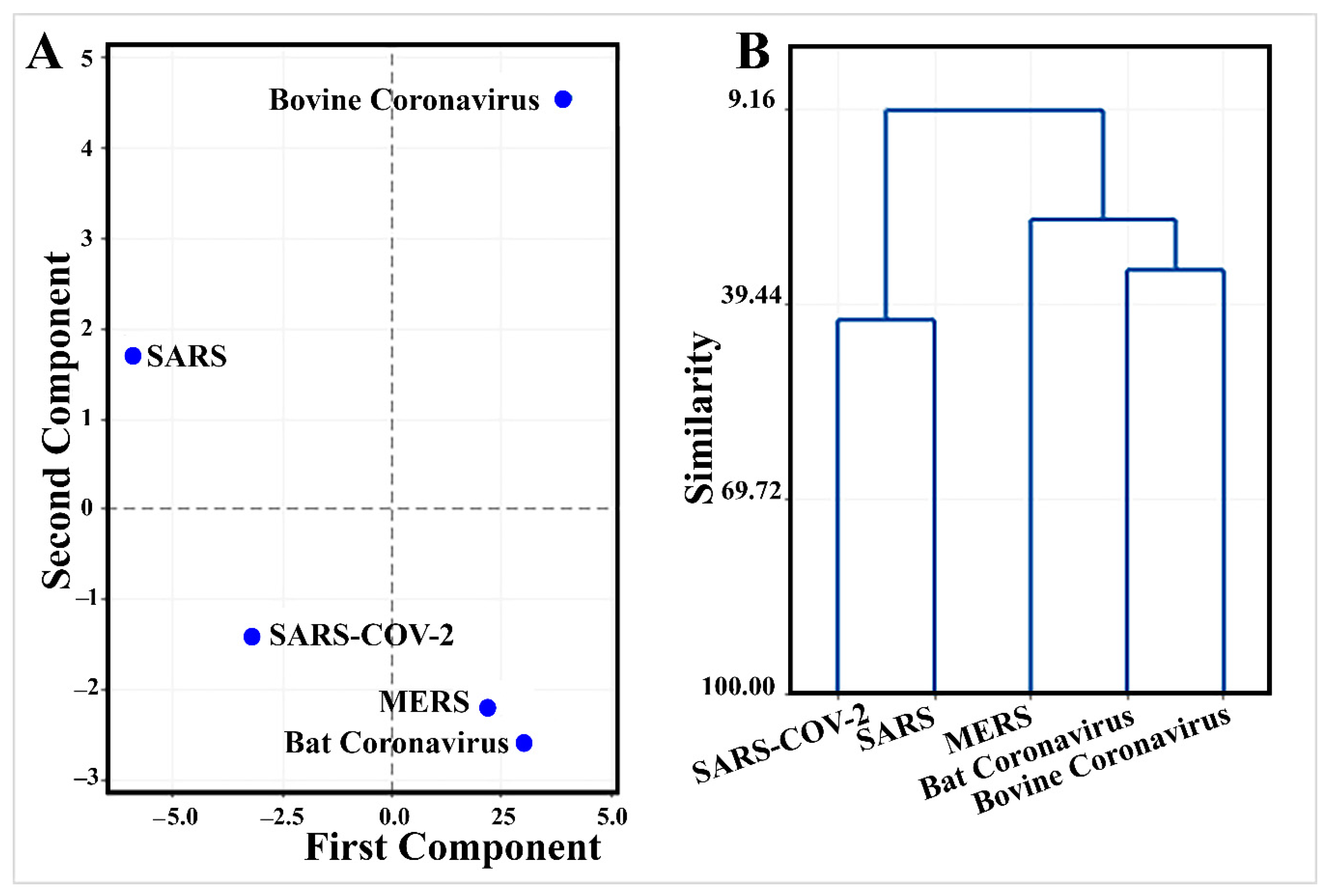
2.5. Multivariate Analysis
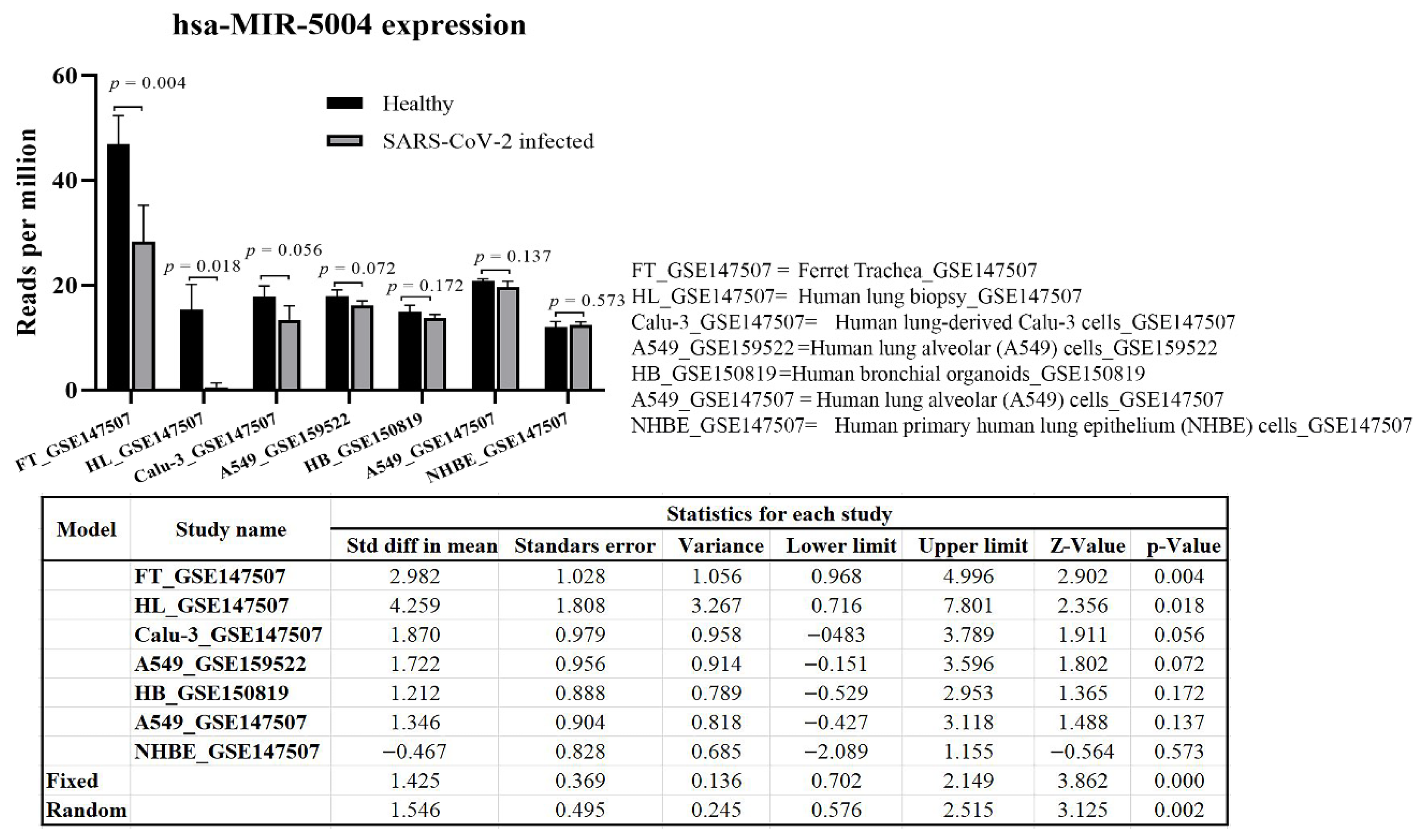
2.6. MIR-5004 Expression Analysis in Response to SARS-CoV-2 Infection: Meta-analysis Approach
2.7. Variant Discovery on Genomic Sequence of Hsa-MIR-5004-3p, 5′UTR Inhibitory MicroRNAs, as COVID-19 Risk Factors
3. Results
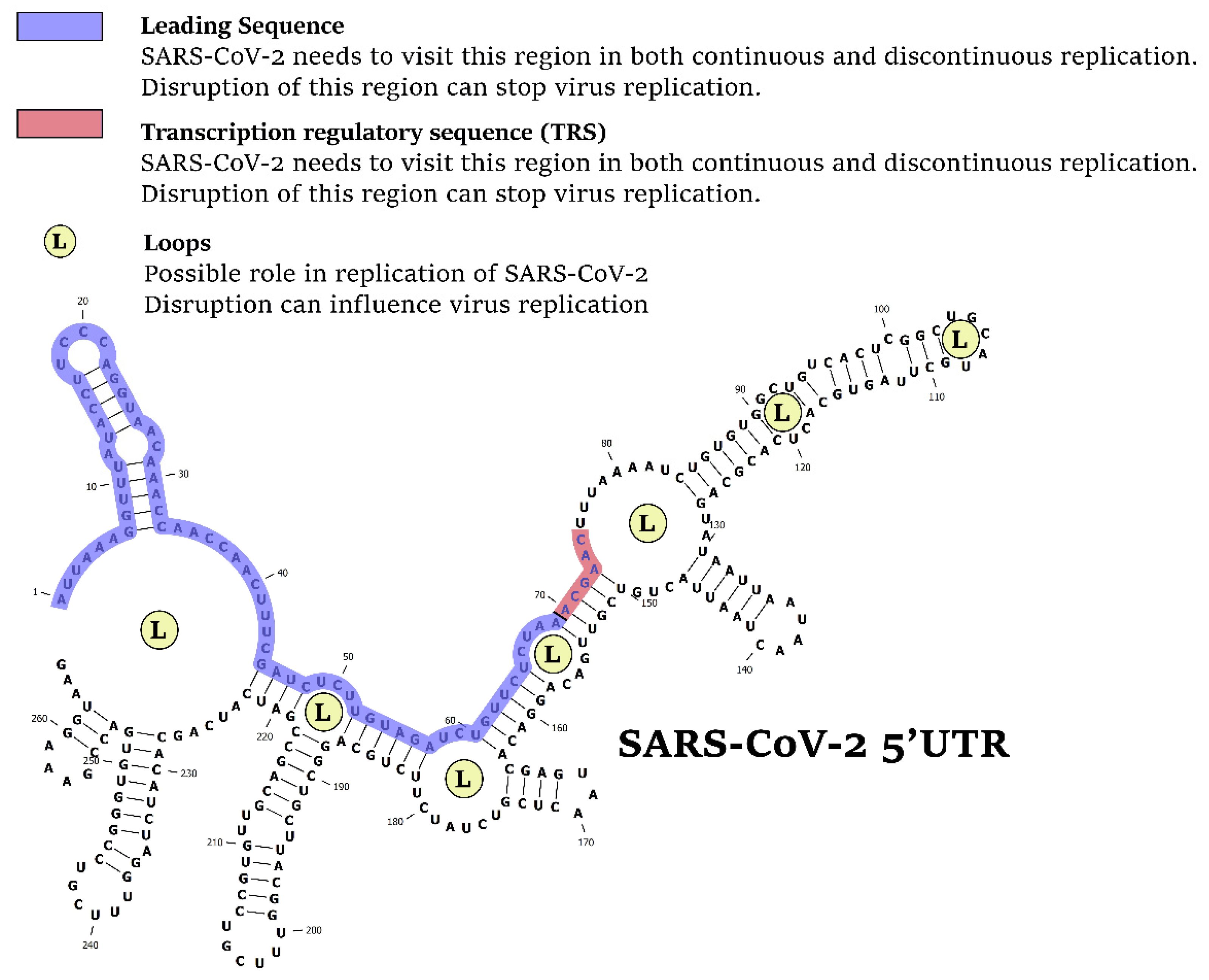
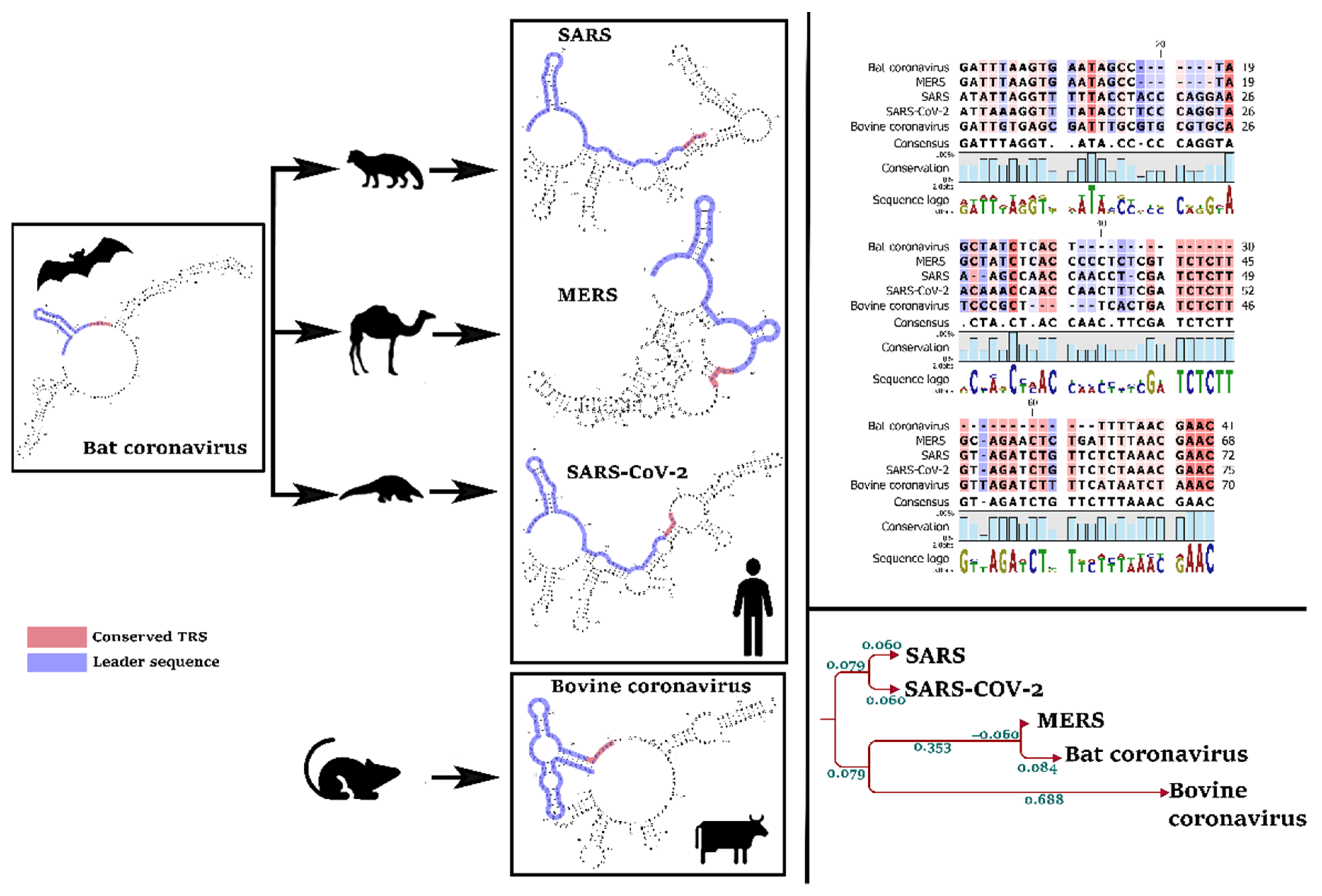
3.1. Comparative Analysis of the 5′UTR of Human Pathogenic and Non-Pathogenic Coronaviruses
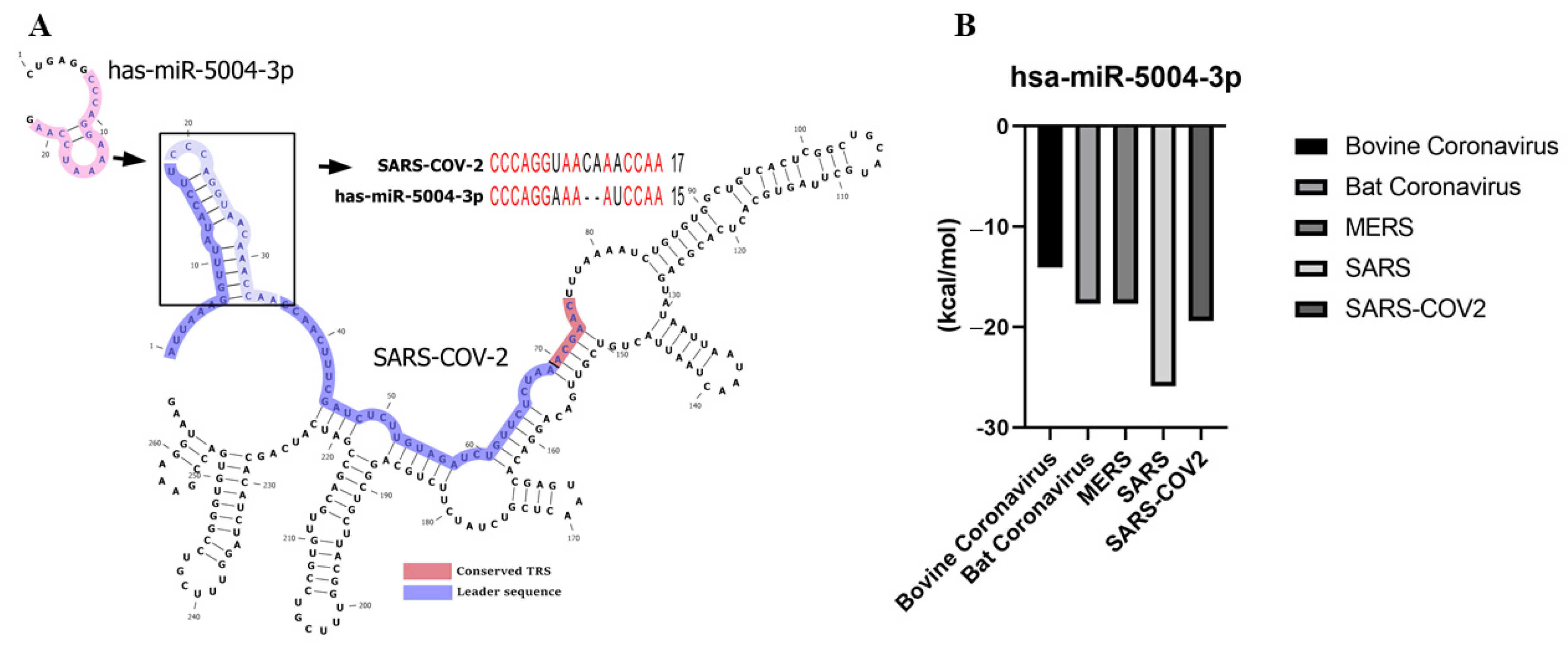
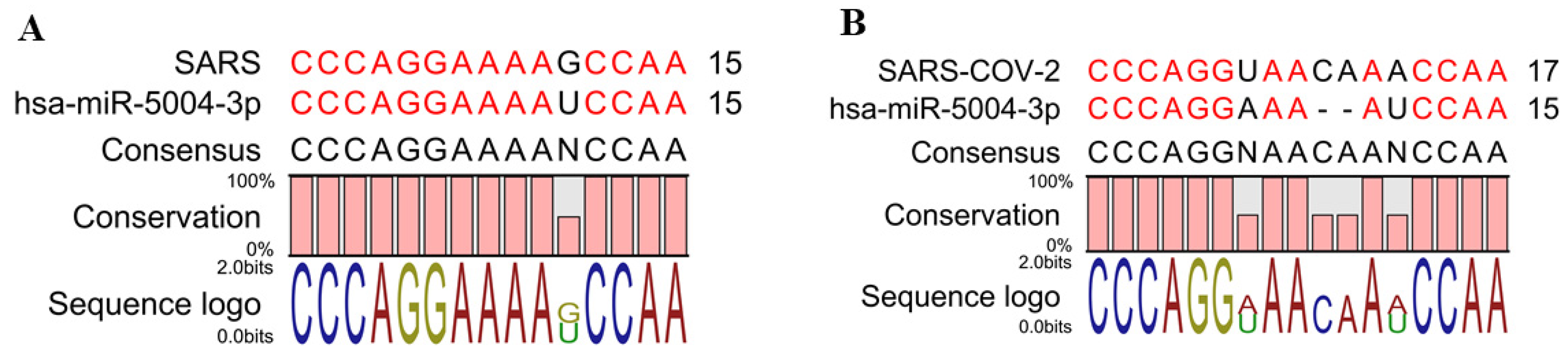
3.2. Identifying the MicroRNAs that Can Bind to the Leader Sequence and TRS of SARS-CoV-2 (5′UTR Inhibitory MicroRNAs)
3.3. The Leader Sequence of SARS-CoV-2 Has a Unique Pattern of MicroRNA Binding, Compared with SARS, MERS, Bat, and Bovine Coronaviruses
3.4. Drug Repurposing to Induce 5′UTR Inhibitory MicroRNAs
3.5. Significant Decline in Expression of MIR-5004 after SARS-COV-2 Infection
3.6. hsa-miR-5004-3p Genomic Variation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Yang, D.; Leibowitz, J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015, 206, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Sola, I.; Almazan, F.; Zuniga, S.; Enjuanes, L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef]
- Huang, C.; Lokugamage, K.G.; Rozovics, J.M.; Narayanan, K.; Semler, B.L.; Makino, S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: Viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011, 7, e1002433. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.; Bouma, P.; Williams, G.D.; Brian, D.A. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2003, 77, 6720–6730. [Google Scholar] [CrossRef] [PubMed]
- Alanazi, I.O.; Al Shehri, Z.S.; Ebrahimie, E.; Giahi, H.; Mohammadi-Dehcheshmeh, M. Non-coding and coding genomic variants distinguish prostate cancer, castration-resistant prostate cancer, familial prostate cancer, and metastatic castration-resistant prostate cancer from each other. Mol. Carcinog. 2019, 58, 862–874. [Google Scholar] [CrossRef] [PubMed]
- De Jong, V.; Zaldumbide, A.; Van Der Slik, A.; Persengiev, S.; Roep, B.; Koeleman, B. Post-transcriptional control of candidate risk genes for type 1 diabetes by rare genetic variants. Genes Immun. 2013, 14, 58–61. [Google Scholar] [CrossRef]
- Paul, S.; Bravo Vázquez, L.A.; Pérez Uribe, S.; Roxana Reyes-Pérez, P.; Sharma, A. Current status of microRNA-based therapeutic approaches in neurodegenerative disorders. Cells 2020, 9, 1698. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Bandyra, K.J.; Said, N.; Pfeiffer, V.; Górna, M.W.; Vogel, J.; Luisi, B.F. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell 2012, 47, 943–953. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, A.; Shankar, R.; Bagchi, S.; Grama, A.; Chaterji, S. MicroRNA target prediction using thermodynamic and sequence curves. BMC Genom. 2015, 16, 999. [Google Scholar] [CrossRef]
- Alanazi, I.O.; AlYahya, S.A.; Ebrahimie, E.; Mohammadi-Dehcheshmeh, M. Computational systems biology analysis of biomarkers in lung cancer; unravelling genomic regions which frequently encode biomarkers, enriched pathways, and new candidates. Gene 2018, 659, 29–36. [Google Scholar] [CrossRef]
- Novichkova, S.; Egorov, S.; Daraselia, N. MedScan, a natural language processing engine for MEDLINE abstracts. Bioinformatics 2003, 19, 1699–1706. [Google Scholar] [CrossRef]
- Bax, L.; Yu, L.-M.; Ikeda, N.; Moons, K.G. A systematic comparison of software dedicated to meta-analysis of causal studies. BMC Med. Res. Methodol. 2007, 7, 40. [Google Scholar] [CrossRef]
- Kulinskaya, E.; Morgenthaler, S.; Staudte, R.G. Meta Analysis: A Guide to Calibrating and Combining Statistical Evidence; John Wiley & Sons: Hoboken, NJ, USA, 2008; Volume 756. [Google Scholar]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Davydov, E.V.; Goode, D.L.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef] [PubMed]
- Fulzele, S.; Sahay, B.; Yusufu, I.; Lee, T.J.; Sharma, A.; Kolhe, R.; Isales, C.M. COVID-19 Virulence in Aged Patients Might Be Impacted by the Host Cellular MicroRNAs Abundance/Profile. Aging Dis. 2020, 11, 509. [Google Scholar] [CrossRef]
- Raman, S.; Brian, D.A. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2005, 79, 12434–12446. [Google Scholar] [CrossRef]
- Moens, U. Silencing viral microRNA as a novel antiviral therapy? BioMed Res. Int. 2009, 2009, 419539. [Google Scholar] [CrossRef]
- Tambyah, P.A.; Ching, C.S.; Sepramaniam, S.; Ali, J.M.; Armugam, A.; Jeyaseelan, K. microRNA expression in blood of dengue patients. Ann. Clin. Biochem. 2016, 53, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; McGoogan, J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Cruz-Rodrıguez, L.; Dilsiz, N.; Zıaratı, P.; Lambert, B.; Hochwımmer, B. A miRNA-peptide fusion as a vaccine candidate against the novel coronavirus (COVID-19). Exosomes as potential biomarkers of SARS-COV-2 in lung. J. Biosci. Biomed. Eng. 2020, 1, 1–11. [Google Scholar]
- Chauhan, N.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. COVID-19: Fighting the invisible enemy with microRNAs. Expert Rev. Anti-Infect. Ther. 2020, 1–9. [Google Scholar] [CrossRef]
- El-Nabi, S.H.; Elhiti, M.; El-Sheekh, M. A new approach for COVID-19 treatment by micro-RNA. Med. Hypotheses 2020, 143, 110203. [Google Scholar] [CrossRef]
- Hassab Elnabi, S.E. New strategies for treatment of COVID-19 and evolution of SARS-CoV-2 according to biodiversity and evolution theory. Egypt. J. Basic Appl. Sci. 2020, 7, 226–232. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, T.; Yin, Y.; Zhang, C.-Y.; Zhang, Y.-L. Dietary microRNA—A Novel Functional Component of Food. Adv. Nutr. 2019, 10, 711–721. [Google Scholar] [CrossRef]
- Munnink, B.B.O.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; Van Der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef]
- Mohammadi-Dehcheshmeh, M.; Niazi, A.; Ebrahimi, M.; Tahsili, M.; Nurollah, Z.; Ebrahimi Khaksefid, R.; Ebrahimi, M.; Ebrahimie, E. Unified transcriptomic signature of arbuscular mycorrhiza colonization in roots of Medicago truncatula by integration of machine learning, promoter analysis, and direct merging meta-analysis. Front. Plant Sci. 2018, 9, 1550. [Google Scholar] [CrossRef]
- Hosseinpour, B.; Bakhtiarizadeh, M.R.; Khosravi, P.; Ebrahimie, E. Predicting distinct organization of transcription factor binding sites on the promoter regions: A new genome-based approach to expand human embryonic stem cell regulatory network. Gene 2013, 531, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Kargarfard, F.; Sami, A.; Hemmatzadeh, F.; Ebrahimie, E. Identifying mutation positions in all segments of influenza genome enables better differentiation between pandemic and seasonal strains. Gene 2019, 697, 78–85. [Google Scholar] [CrossRef]
- Kargarfard, F.; Sami, A.; Ebrahimie, E. Knowledge discovery and sequence-based prediction of pandemic influenza using an integrated classification and association rule mining (CBA) algorithm. J. Biomed. Inform. 2015, 57, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Kargarfard, F.; Sami, A.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E. Novel approach for identification of influenza virus host range and zoonotic transmissible sequences by determination of host-related associative positions in viral genome segments. BMC Genom. 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Novikov, B.; Ebrahimie, E.; Spilman, A.; Ahsan, R.; Tahsili, M.R.; Najafi, M.; Navvabi, S.; Shariaty, F. The first report of the most important sequential differences between COVID-19 and MERS viruses by attribute weighting models, the importance of Nucleocapsid (N) protein. Bioinformatics 2020. [Google Scholar] [CrossRef]
| Entities | Number | Relations | Number |
|---|---|---|---|
| Small molecules (including drugs) | 1,053,259 | Binding | 1,123,702 |
| Protein | 138,106 | Biomarker | 120,448 |
| Cell process | 9771 | Cell expression | 1,213,035 |
| Cell Object | 607 | Chemical reaction | 58,888 |
| Cells | 4155 | Clinical trial | 109,386 |
| Clinical parameters | 5126 | Direct regulation | 766,707 |
| Complex | 998 | Expression | 832,784 |
| Diseases | 20,855 | Functional associations | 1,775,463 |
| Functional class | 5489 | Genetic change | 379,261 |
| Genetic Variant | 127,872 | Molsynthesis | 160,178 |
| Organ | 3839 | Moltransport | 251,347 |
| Treatments | 78 | Promoter binding | 44,619 |
| Tissue | 574 | Protein modification | 73,859 |
| Total number of entities | 1,370,729 | Quantitative change | 421,884 |
| Regulation | 5,193,796 | ||
| State change | 128,112 | ||
| MicroRNA effects | 57,743 | ||
| Total number of relations | 12,653,469 |
| Experiment ID | Sample ID (NCBI) | Organism | Tissue/Cell Line | SARS-CoV-2 Infected/Non-Infected | Total Number of Reads | SARS-CoV-2 Strain |
|---|---|---|---|---|---|---|
| GSE150819 | SRR11811019 | Human | Lung bronchial organoids | Non-infected | 32,214,210 | Non-infected (mock) |
| SRR11811020 | Human | Lung bronchial organoids | Non-infected | 32,443,162 | Non-infected (mock) | |
| SRR11811021 | Human | Lung bronchial organoids | Non-infected | 33,310,500 | Non-infected (mock) | |
| SRR11811022 | Human | Lung bronchial organoids | Infected | 31,662,278 | SARS-CoV-2/Hu/DP/Kng/19-020 | |
| SRR11811023 | Human | Lung bronchial organoids | Infected | 35,953,491 | SARS-CoV-2/Hu/DP/Kng/19-020 | |
| SRR11811024 | Human | Lung bronchial organoids | Infected | 32,416,198 | SARS-CoV-2/Hu/DP/Kng/19-020 | |
| GSE147507 | SRR11517725-28 | Human | human lung biopsies | Non-infected | 57,660,692 | Non-infected (mock) |
| SRR11517729-32 | Human | human lung biopsies | Non-infected | 40,524,836 | Non-infected (mock) | |
| SRR11517733-36 | Human | human lung biopsies | Infected | 10,561,476 | USA-WA1/2020 | |
| SRR11517737-40 | Human | human lung biopsies | Infected | 9,514,219 | USA-WA1/2020 | |
| SRR11412215-18 | Human | Lung epithelium NHBE cells | Non-infected | 17,003,573 | Non-infected (mock) | |
| SRR11412219-22 | Human | Lung epithelium NHBE cells | Non-infected | 16,311,121 | Non-infected (mock) | |
| SRR11412223-26 | Human | Lung epithelium NHBE cells | Non-infected | 24,286,949 | Non-infected (mock) | |
| SRR11412227-30 | Human | Lung epithelium NHBE cells | Infected | 15,032,096 | USA-WA1/2020 | |
| SRR11412231-34 | Human | Lung epithelium NHBE cells | Infected | 15,108,090 | USA-WA1/2020 | |
| SRR11412235-38 | Human | Lung epithelium NHBE cells | Infected | 44,210,735 | USA-WA1/2020 | |
| SRR11412239-42 | Human | Lung alveolar A549 cells | Non-infected | 27,013,945 | Non-infected (mock) | |
| SRR11412243-46 | Human | Lung alveolar A549 cells | Non-infected | 14,744,844 | Non-infected (mock) | |
| SRR11412247-50 | Human | Lung alveolar A549 cells | Non-infected | 11,683,707 | Non-infected (mock) | |
| SRR11412251-54 | Human | Lung alveolar A549 cells | Infected | 34,141,057 | USA-WA1/2020 | |
| SRR11412255-59 | Human | Lung alveolar A549 cells | Infected | 29,681,064 | USA-WA1/2020 | |
| SRR11412260-63 | Human | Lung alveolar A549 cells | Infected | 20,603,153 | USA-WA1/2020 | |
| SRR11517744 | Human | Lung-derived Calu-3 cells | Non-infected | 9,324,151 | Non-infected (mock) | |
| SRR11517745 | Human | Lung-derived Calu-3 cells | Non-infected | 17,436,078 | Non-infected (mock) | |
| SRR11517746 | Human | Lung-derived Calu-3 cells | Non-infected | 37,787,485 | Non-infected (mock) | |
| SRR11517747 | Human | Lung-derived Calu-3 cells | Infected | 23,623,325 | USA-WA1/2020 | |
| SRR11517748 | Human | Lung-derived Calu-3 cells | Infected | 13,583,713 | USA-WA1/2020 | |
| SRR11517749 | Human | Lung-derived Calu-3 cells | Infected | 28,688,015 | USA-WA1/2020 | |
| SRR11517699 | Ferret | Trachea | Non-infected | 328,105,259 | Non-infected (mock) | |
| SRR11517700 | Ferret | Trachea | Non-infected | 5,210,254 | Non-infected (mock) | |
| SRR11517701 | Ferret | Trachea | Non-infected | 4,746,327 | Non-infected (mock) | |
| SRR11517702 | Ferret | Trachea | Non-infected | 5,163,699 | Non-infected (mock) | |
| SRR11517703 | Ferret | Trachea | Infected | 9,169,859 | USA-WA1/2020 | |
| SRR11517707 | Ferret | Trachea | Infected | 14,124,547 | USA-WA1/2020 | |
| SRR11517711 | Ferret | Trachea | Infected | 12,933,325 | USA-WA1/2020 | |
| SRR11517715 | Ferret | Trachea | Infected | 14,644,347 | USA-WA1/2020 | |
| GSE159522 | SRR12828440-43 | Human | Lung alveolar A549 cells | Non-infected | 19,152,790 | Non-infected (mock) |
| SRR12828444-47 | Human | Lung alveolar A549 cells | Non-infected | 19,381,530 | Non-infected (mock) | |
| SRR12828448-51 | Human | Lung alveolar A549 cells | Non-infected | 16,483,541 | USA-WA1/2020 | |
| SRR12828428-31 | Human | Lung alveolar A549 cells | Infected | 17,644,925 | USA-WA1/2020 | |
| SRR12828432-35 | Human | Lung alveolar A549 cells | Infected | 19,504,193 | USA-WA1/2020 | |
| SRR12828436-39 | Human | Lung alveolar A549 cells | Infected | 19,491,861 | USA-WA1/2020 |
| MicroRNA | Organism | Thermodynamic Binding Energy against Leader Sequence (kcal/mol) | ||||
|---|---|---|---|---|---|---|
| SARS-COV-2 | SARS | MERS | Bat Coronavirus | Bovine Coronavirus | ||
| ptc-miR474b | Populus trichocarpa | −27.3 | −24.5 | −21.6 | −21.5 | −17 |
| ptc-miR474a | Populus trichocarpa | −27.3 | −22.2 | −22.8 | −22.2 | −18.1 |
| csa-let-7d | Ciona savignyi | −25.1 | −22.7 | −24.6 | −24.6 | −19 |
| cin-let-7d-5p | Ciona intestinalis | −25.1 | −22.7 | −24.6 | −24.6 | −19 |
| gga-miR-6608-3p | Gallus gallus | −25 | −23.6 | −30.1 | −14.6 | −17.1 |
| eca-miR-9080 | Equus caballus | −23.4 | −27.7 | −15.9 | −15.9 | −16.5 |
| csi-miR3953 | Citrus sinensis | −22.5 | −22.4 | −27.2 | −14.1 | −16.8 |
| ame-miR-3741 | Apis mellifera | −21.9 | −20.9 | −35.2 | −16 | −21.7 |
| cel-miR-8207-3p | Caenorhabditis elegans | −20.5 | −22 | −25.6 | −12.2 | −22.6 |
| ppy-miR-1273a | Pongo pygmaeus | −20.1 | −21.1 | −23.4 | −18.7 | −19.5 |
| hsa-miR-5004-3p | Homo sapiens | −19.4 | −25.9 | −17.7 | −17.7 | −14.1 |
| bta-miR-2284ab | Bos taurus | −19.3 | −21.6 | −16.8 | −19.9 | −13.8 |
| oan-miR-1395-5p | Ornithorhynchus anatinus | −19.3 | −27.8 | −17.5 | −15.3 | −13.4 |
| mdo-miR-137b-5p | Monodelphis domestica | −17.7 | −26.8 | −19.4 | −15.1 | −16.8 |
| dme-miR-4949-3p | Drosophila melanogaster | −17.7 | −17.4 | −13.2 | −12.6 | −24.4 |
| ssc-miR-9833-5p | Sus scrofa | −17.1 | −16.3 | −15.9 | −15.9 | −15.7 |
| ptc-miR6464 | Populus trichocarpa | −16.2 | −15.4 | −13.7 | −13.7 | −13.1 |
| mtr-miR2629g | Medicago truncatula | −15.1 | −21.7 | −13.1 | −13.1 | −14.1 |
| mtr-miR2629f | Medicago truncatula | −15.1 | −21.7 | −13.1 | −13.1 | −14.1 |
| mtr-miR2629e | Medicago truncatula | −15.1 | −21.7 | −13.1 | −13.1 | −14.1 |
| mtr-miR2629d | Medicago truncatula | −15.1 | −21.7 | −13.1 | −13.1 | −14.1 |
| mtr-miR2629c | Medicago truncatula | −15.1 | −21.7 | −13.1 | −13.1 | −14.1 |
| mtr-miR2629b | Medicago truncatula | −15.1 | −21.7 | −13.1 | −13.1 | −14.1 |
| mtr-miR2629a | Medicago truncatula | −15.1 | −21.7 | −13.1 | −13.1 | −14.1 |
| bmo-miR-3293 | Bombyx mori | −13.9 | −14.8 | −15.7 | −12.7 | −16 |
| dsi-miR-986-3p | Drosophila simulans | −12.4 | −16.5 | −25.1 | −25.1 | −19.7 |
| dme-miR-986-3p | Drosophila melanogaster | −12.4 | −16.5 | −25.1 | −25.1 | −19.7 |
| dsi-miR-986-3p | Drosophila simulans | −12.4 | −16.5 | −25.1 | −25.1 | −19.7 |
| dme-miR-986-3p | Drosophila melanogaster | −12.4 | −16.5 | −25.1 | −25.1 | −19.7 |
| mmu-miR-6957-3p | Mus musculus | −11.6 | −13.5 | −12.2 | −12.2 | −12.1 |
| ppc-miR-83-5p | Pristionchus pacificus | −11.4 | −14.7 | −10.9 | −11.4 | −17.6 |
| cme-miR1863 | Cucumis melo | −11.3 | −10.5 | −12.5 | −12.5 | −15.2 |
| cel-miR-2211-5p | Caenorhabditis elegans | −10.7 | −10.6 | −10.5 | −10.3 | −12.2 |
| ath-miR5638a | Arabidopsis thaliana | −9.8 | −7.8 | −10.9 | −10.9 | −16.6 |
| bdi-miR5065 | Brachypodium distachyon | −9.7 | −11.8 | −23.6 | −23.6 | −16.9 |
| bdi-miR5065 | Brachypodium distachyon | −9.7 | −11.8 | −23.6 | −23.6 | −16.9 |
| oan-miR-1421l-2-3p | Ornithorhynchus anatinus | −9.7 | −10.8 | −12.6 | −11.1 | −21.2 |
| mghv-miR-M1-2-3p | Mouse gammaherpesvirus 68 | −9.4 | −8.8 | −8.3 | −5.7 | −7.5 |
| dps-miR-2535-3p | Drosophila pseudoobscura | −9.3 | −10.6 | −17.1 | −17.1 | −19.6 |
| Average | −16.33 | −18.58 | −18.34 | −16.35 | −16.61 | |
| rsId | Chr. | Location | Ref | Alt. | MicroRNA | Gene Region | Translational Impact | GERP++ Score |
|---|---|---|---|---|---|---|---|---|
| rs369274154 | 6 | 33406128 | T | C | MIR5004 | 5UTR | ||
| rs371304188 | 6 | 33406147 | C | T | MIR5004 | 5UTR | ||
| rs375913209 | 6 | 33406168 | C | T | MIR5004 | 5UTR | ||
| Not assigned | 6 | 33406194 | A | C | MIR5004 | 5UTR | splice-disrupt | 4.77 |
| Not assigned | 6 | 33406194 | A | G | MIR5004 | 5UTR | splice-disrupt | 4.77 |
| Not assigned | 6 | 33406194 | A | T | MIR5004 | 5UTR | splice-disrupt | 4.77 |
| Not assigned | 6 | 33406195 | G | A | MIR5004 | 5UTR | splice-disrupt | 4.77 |
| Not assigned | 6 | 33406195 | G | C | MIR5004 | 5UTR | splice-disrupt | 4.77 |
| Not assigned | 6 | 33406195 | G | T | MIR5004 | 5UTR | splice-disrupt | 4.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammadi-Dehcheshmeh, M.; Moghbeli, S.M.; Rahimirad, S.; Alanazi, I.O.; Shehri, Z.S.A.; Ebrahimie, E. A Transcription Regulatory Sequence in the 5′ Untranslated Region of SARS-CoV-2 Is Vital for Virus Replication with an Altered Evolutionary Pattern against Human Inhibitory MicroRNAs. Cells 2021, 10, 319. https://doi.org/10.3390/cells10020319
Mohammadi-Dehcheshmeh M, Moghbeli SM, Rahimirad S, Alanazi IO, Shehri ZSA, Ebrahimie E. A Transcription Regulatory Sequence in the 5′ Untranslated Region of SARS-CoV-2 Is Vital for Virus Replication with an Altered Evolutionary Pattern against Human Inhibitory MicroRNAs. Cells. 2021; 10(2):319. https://doi.org/10.3390/cells10020319
Chicago/Turabian StyleMohammadi-Dehcheshmeh, Manijeh, Sadrollah Molaei Moghbeli, Samira Rahimirad, Ibrahim O. Alanazi, Zafer Saad Al Shehri, and Esmaeil Ebrahimie. 2021. "A Transcription Regulatory Sequence in the 5′ Untranslated Region of SARS-CoV-2 Is Vital for Virus Replication with an Altered Evolutionary Pattern against Human Inhibitory MicroRNAs" Cells 10, no. 2: 319. https://doi.org/10.3390/cells10020319
APA StyleMohammadi-Dehcheshmeh, M., Moghbeli, S. M., Rahimirad, S., Alanazi, I. O., Shehri, Z. S. A., & Ebrahimie, E. (2021). A Transcription Regulatory Sequence in the 5′ Untranslated Region of SARS-CoV-2 Is Vital for Virus Replication with an Altered Evolutionary Pattern against Human Inhibitory MicroRNAs. Cells, 10(2), 319. https://doi.org/10.3390/cells10020319







