Abstract
Our knowledge of the evolution and the role of untranslated region (UTR) in SARS-CoV-2 pathogenicity is very limited. Leader sequence, originated from UTR, is found at the 5′ ends of all encoded SARS-CoV-2 transcripts, highlighting its importance. Here, evolution of leader sequence was compared between human pathogenic and non-pathogenic coronaviruses. Then, profiling of microRNAs that can inactivate the key UTR regions of coronaviruses was carried out. A distinguished pattern of evolution in leader sequence of SARS-CoV-2 was found. Mining all available microRNA families against leader sequences of coronaviruses resulted in discovery of 39 microRNAs with a stable thermodynamic binding energy. Notably, SARS-CoV-2 had a lower binding stability against microRNAs. hsa-MIR-5004-3p was the only human microRNA able to target the leader sequence of SARS and to a lesser extent, also SARS-CoV-2. However, its binding stability decreased remarkably in SARS-COV-2. We found some plant microRNAs with low and stable binding energy against SARS-COV-2. Meta-analysis documented a significant (p < 0.01) decline in the expression of MIR-5004-3p after SARS-COV-2 infection in trachea, lung biopsy, and bronchial organoids as well as lung-derived Calu-3 and A549 cells. The paucity of the innate human inhibitory microRNAs to bind to leader sequence of SARS-CoV-2 can contribute to its high replication in infected human cells.
1. Background
Coronavirus genomes are single-stranded mRNAs, containing both coding and untranslated regions (UTRs). The 5′UTR and 3′UTR are crucial for coronavirus RNA replication, transcription, and dominating host systems biology [1]. However, their exact roles and their evolutions, specifically in the new SARS-CoV-2, are mainly unknown.
There are some 5′UTR regions that are vital for virus replication (Figure 1). A specific region of 70 nucleotides at the 5′ end of the genome, referred as the ‘leader’ sequence, has been found at the 5′ ends of all encoded transcripts, highlighting its importance. A conserved cis-acting element, named transcription regulatory sequence (TRS), immediately follows the leader sequence, representing a unique feature of coronaviruses.
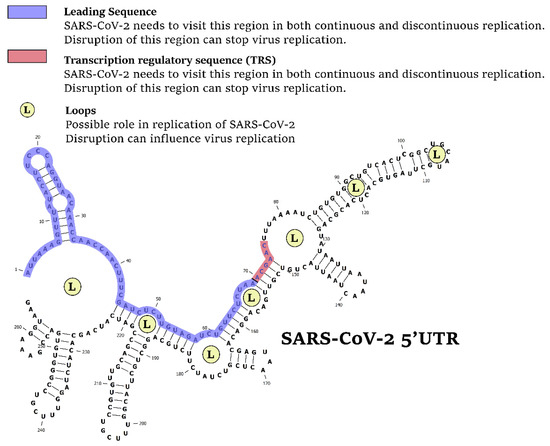
Figure 1.
The 5′UTR is the Achilles’ heel of SARS-CoV-2. Disruption of specific regions in 5′UTR, including leading sequence, transcription regulatory sequence, or loops (L) can stop the virus replication.
Compared to the other positive-strand RNA viruses, coronaviruses have a unique and complicated pattern of continuous and discontinuous RNA synthesis (Figure 2). Like the other positive-strand RNA viruses, continuous RNA synthesis happens for genome replication to yield multiple copies of the genomic RNA. In continuous RNA synthesis, a full-length complementary negative-strand RNA is utilized as the template for generation of progeny virus genomes [2]. In discontinuous transcription, during the synthesis of sub-genomic negative-strand RNAs, a premature termination and template switch occur to add copies of the leader sequence [2]. The presence of the 5′ leader sequence provides an efficient strategy for the efficient accumulation of coronaviruses mRNAs and proteins during infection because of the protection of viral mRNAs from endonucleolytic cleavage of the capped mRNAs [3]. Furthermore, the complement of the leader sequence supports initiation of positive-strand RNA synthesis, generating negative-strand sub-genomic RNAs as templates for further productions of positive-strand sub-genomic mRNAs [2].
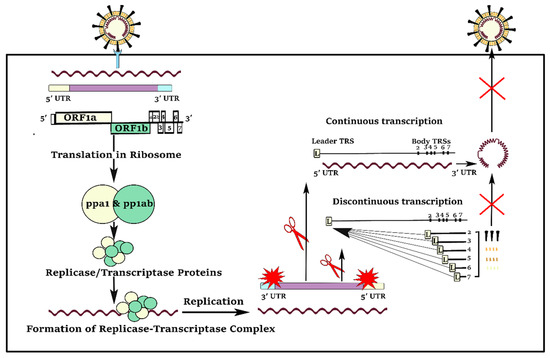
Figure 2.
In both continuous and discontinuous RNA synthesis patterns, SARS-CoV-2 virus segments need to visit the 5′UTR, particularly their transcription regulatory sequence (TRS) and leader sequence (L). The 5′UTR is a good target to reduce virus load in the host cell.
Whereas UTR is a non-coding region, another unique structure of coronaviruses is the existence of a short open reading frame (ORF) on one of the 5′UTR stem loops. In bovine coronavirus, maintenance of the short ORF is positively correlated with viral RNA accumulation [4]. The other non-coding region, 3′-UTR folds into a unique stem-loop two motif secondary structure that is required for virus viability.
UTRs are also important in the context of host microRNA interaction. MicroRNAs play their negative regulatory roles via sequence-specific interactions with the 3′ or 5′UTRs [5]. Consequently, sequence variation in the viral UTR regions can prevent the binding of human microRNAs, so preventing microRNA-based immunity [6,7]. The problem is more serious in case of SARS-CoV-2 which originated recently from bat, such that human microRNAs have not evolved to interact with their UTRs.
Here, we hypothesize that the TRS and leader sequences of 5′ UTRs are crucial for coronavirus RNA replication. Moreover, microRNAs can bind to the TRS and leader sequences of the 5′UTR to reduce SARS-CoV-2 replication. The first aim of this study was to unravel the evolution of the TRS and leader sequences in 5′UTR regions of SARS-CoV-2. The second aim was to identify inhibitory microRNAs, present in human and other organisms, that can bind to the key regulatory regions of SARS-CoV-2 sequences.
2. Methods
2.1. Data Collection
Complete genomes of SARS-COV-2, SARS, MERS, bat coronavirus, and bovine coronavirus, including complete sequences of non-coding regions, were downloaded from The National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov). The sequences were: Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1 (NC_045512), SARS coronavirus Tor2 (NC_004718), Tylonycteris bat coronavirus HKU4 isolate CZ01 (MH002337), Middle East respiratory syndrome-related coronavirus isolate NL13892 (MG987420), and bovine coronavirus isolate BCoV-ENT (NC_003045).
2.2. Inhibitory MicroRNA Prediction
Available microRNA information of human and other organisms, deposited in TargetScan [8], were downloaded. In total, the information of 9994 microRNAs, including sequences of seed region and mature sequences, were obtained. The seed region is a conserved sequence with the average length of 6–8 bp [9]. The seed region is critical for binding and hybridization of the microRNA to its target. Supplementary 1 represents the seed region, mature sequence, microRNA family, and the associated MiRBase IDs of the 9994 microRNAs.
Then, the reverse complement sequence of UTR regions of SARS-COV-2, SARS, MERS, and bovine coronavirus were searched against the known seed sequences to find the microRNAs with affinity binding to the specific regions in 5′UTR, TRS, and leader sequences by TargetScan.
Based on the MiRBase IDs, obtained from the TargetScan, fasta format of mature sequences of microRNAs were retrieved from the miRBase database [10]. Using the RNAhybrid v2.2 database, the binding probability of mature microRNAs against TRS and leader sequences of coronaviruses were evaluated through calculating of a minimum free energy hybridization [11]. The thermodynamic binding energy of −15 kcal/mol was used as the minimum cut-off, and binding energy of −25 kcal/mol pointed the stable binding [12].
2.3. Sequence Alignment between 5′UTR of Coronavirus and MicroRNA Seed Regions and Phylogenetic Trees
Multiple sequence alignment was performed using CLUSTALW algorithm by CLC Genomics Workbench 20 (QIGEN, Venlo, The Netherlands). Phylogenetic trees, based on the maximum likelihood phylogeny approach, were constructed using UPGMA method, nucleotide substitution model of Jukes Cantor, gamma distribution parameter of 1.0, and bootstrap of 1000.
2.4. Literature-Mining Based Drug Repurposing
Pathway Studio Database (Elsevier) was employed to find the drugs, food supplements, and chemicals (small molecules) with promoting effects on 5′ UTR inhibitory microRNAs, as described recently [13]. To perform literature mining, MedScan tool was used that employs the NLP algorithm to mine extract relations from biomedical texts, mainly PubMed [14]. In addition to the mined sentences of relations, Medscan also records the title of literature, authors, the year of publication, Medline (PubMed) reference number, and type of relation. The results are deposited in Mammalian + ChemEffect + DiseaseFx database of Pathway Studio which is enriched by a range of extra systems biology information like the subcellular location and protein class (such as receptor, ligand, transcription factor, small RNA, small molecule, etc.), from Gene Ontology Consortium, as well as KEGG pathways. The database updates using cloud technology by addition of new mined relationships and entities from recent publications. Statistics of Mammalian + ChemEffect + DiseaseFx database used for literature mining is presented at Table 1.

Table 1.
Statistics of Mammalian + ChemEffect + DiseaseFx database used for literature mining in this study (September 2020).
In short, a highly enriched database with more than one million chemicals, 138,000 proteins, and 12 million protein interactions were employed for drug repurposing.
2.5. Multivariate Analysis
Thermodynamic binding energy values (kcal/mol) between microRNAs and 5′UTR regions of coronaviruses were used as input for PCA and clustering analysis. Analysis was performed in MINITAB 18 (https://www.minitab.com). Graphs were visualized by GraphPad Prism 7 (GraphPad Software, Inc. California, CA, USA). Correlation matrix was used for PCA analysis. Clustering was performed based on Euclidean distance matrix and average linkage method.
2.6. MIR-5004 Expression Analysis in Response to SARS-CoV-2 Infection: Meta-analysis Approach
In a comprehensive study, the expression of MIR-5004 was evaluated in COVID-infected samples compared to non-infected ones in human and ferret. The raw RNA-seq reads in FASTQ format were downloaded from NCBI SRA (https://www.ncbi.nlm.nih.gov/sra) using CLC Genomics Workbench v.20.0 (QIAGEN, Venlo, The Netherlands). In total, 42 infected and non-infected samples of trachea, lung biopsy, bronchial organoids, lung-derived Calu-3 cells, lung alveolar A549 cells, and lung epithelium NHBE cells, from GSE150819, GSE147507, and GSE159522 were utilized (Table 2).

Table 2.
Samples (n = 42) used for MIR-5004 expression analysis against SARS-CoV-2 infection.
FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess quality raw. MIR-5004 genomic sequence, NR_049800, was downloaded from NCBI. Mapping of short reads to MIR-5004 reference genome was performed using CLC Genomics Workbench based on the following mapping parameters: Mismatch cost = 2, Insertion cost = 3, Deletion cost = 3, Length fraction = 0.1, Similarity fraction = 0.6, and Maximum number of hits for a read = 10. Mapped reads to MIR-5004 reference genome (mapped reads per million) were used as expression measurement.
Comprehensive Meta-analysis (CMA) software Version 3.3.070 (Biostat Inc., Englewood, NJ, USA) [15] was used for meta-analysis of MIR-5004 expression in response to SARS-CoV-2 infection in different studies and different tissue types (trachea, lung biopsy, bronchial organoids, Calu-3 cells, and A549 cells, and NHBE cells). Fixed standardized effects model was employed for meta-analysis [16].
2.7. Variant Discovery on Genomic Sequence of Hsa-MIR-5004-3p, 5′UTR Inhibitory MicroRNAs, as COVID-19 Risk Factors
The human genetic variation genetic variation database of NCBI (dbSNP database) [17] was employed as the main resource for variants gathering. The Pathway Studio tool was used for retrieving hsa-MIR-5004-3p genomic variants by mining more than two hundred million deposited variants in dbSNP and 1000 Genomes project. Genomic locations of variants were recorded (CDs, 3′UTR, 5′UTR, intergenic, or intronic variants). Then, translational impact of the identified hsa-MIR-5004-3p variants including missense, splice disrupt, CDs indel, nonsense, misstart, and non-stop were determined. Finally, the proportion of each type of variant was calculated.
To evaluate the possible functional impact of the identified hsa-MIR-5004-3p variants, the Genomic Evolutionary Rate Profiling (GERP)++ conservation score was used. GERP is an evolutionary conservation score which have a good correspondence with clinical significance and pathogenicity level [18]. GERP++ demonstrates the constrained elements in multiple alignments by quantifying substitution deficits. These deficits identify substitutions that would have happened if the element were neutral DNA but did not happen as the element has been experienced functional constraint. Low values of GERP++ score stand for low level of conservation and high values for high level of conservation.
3. Results
3.1. Comparative Analysis of the 5′UTR of Human Pathogenic and Non-Pathogenic Coronaviruses
Our primary analysis of reference sequences distinguished a pattern of evolution in the leader sequence and TRS of SARS-CoV-2, by comparison with MERS and the bovine coronavirus (Figure 3). Interestingly, the TRS sequence is identical in all coronaviruses that infect human (SARS-CoV-2, SARS, and MERS) which differs in non-human pathogens, such as bovine coronavirus (Figure 3). In other words, TRS can explain the host range of a coronavirus.
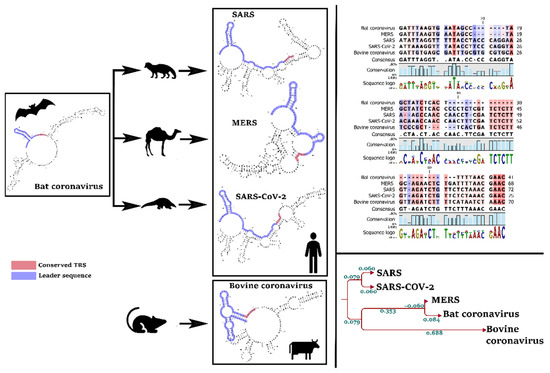
Figure 3.
Evolution of “leader sequence” and TRS (transcription regulatory sequence) in the 5′UTR of human pathogens (SARS, MERS, and SARS-CoV-2) and non-human pathogens (bovine). We found a significant evolution pattern in SARS-CoV-2 compared with the other coronaviruses.
3.2. Identifying the MicroRNAs that Can Bind to the Leader Sequence and TRS of SARS-CoV-2 (5′UTR Inhibitory MicroRNAs)
Mining microRNAs against the leader sequence of SARS-CoV-2 resulted in the discovery of 39 microRNAs with an acceptable thermodynamic binding energy (cut-off of less than minus 15 kcal/mol). Table 3 presents a list of microRNAs and their thermodynamic binding energy that potentially bind to at least one type of coronavirus (SARS-COV-2, SARS, MERS, bat coronavirus, or bovine coronavirus). ptc-MiR474b, ptc- MiR474a, csa-let-7d, cin-let-7d-5p, csi-miR3953, and gga-MiR-6608-3p were microRNAs with stable thermodynamic binding energy (lower than −22 kcal/mol) against leader sequence of SARS-CoV-2.

Table 3.
MicroRNAs potentially able to bind to the leader sequence of the coronavirus 5′ untranslated region (5′UTR) and their binding energy. Lower binding energy demonstrates higher binding stability between microRNA and leader sequence.
hsa-MIR-5004-3p was the only human microRNA able to target the leader sequence of SARS and SARS-CoV-2. However, its binding stability decreased remarkably in SARS-COV-2 (−19.4 kcal/mol), compared with SARS-COV-2 (−25.9 kcal/mol) (Table 3 and Figure 4). Notably, our analysis showed that the leader sequence of SARS-COV-2 is mutated (insertion type mutation, CA), thus escaping microRNA-RNA hybridization (Figure 5). Lack of innate human inhibitory microRNAs able to bind to SARS-COV-2 contributes to the high replication of SARS-CoV-2 in infected human cells.
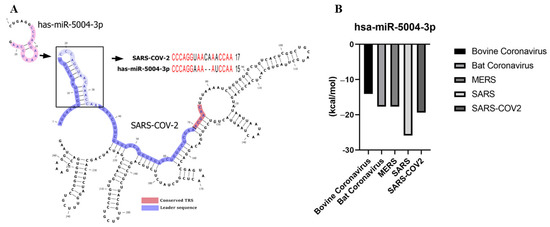
Figure 4.
(A) hsa-miR-5004-3p is the only human microRNA able to target the leader sequence of SARS-CoV-2. (B) Binding energy (Kcal/mol) between leader sequence of coronaviruses and hsa-miR-5004-3p. Lower binding energies (smaller values) demonstrate higher binding stability between microRNA and leader sequence.
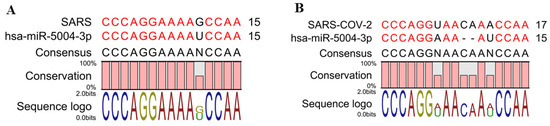
Figure 5.
A two-nucleotide insertion mutation in the leader sequence of SARS-COV-2 results in lower binding between the viral leader sequence and seed region of hsa-miR-5004-3p. (A) Alignment of hsa-miR-5004-3p with SARS. (B) Alignment of hsa-miR-5004-3p with SARS-CoV-2.
3.3. The Leader Sequence of SARS-CoV-2 Has a Unique Pattern of MicroRNA Binding, Compared with SARS, MERS, Bat, and Bovine Coronaviruses
Multivariate analysis of thermodynamic binding energy values of mined inhibitory microRNAs (Table 3) demonstrates a unique pattern of SARS-CoV-2 evolution (Figure 6).
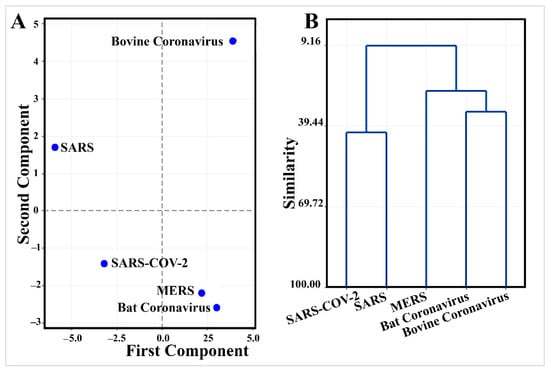
Figure 6.
Multivariate analysis of thermodynamic binding energy values of the leader sequence of coronaviruses against mined inhibitory microRNAs (Table 3) demonstrates a unique pattern for SARS-CoV-2 evolution. (A) Principal component analysis discriminates SARS-CoV-2 from other coronaviruses. (B) Clustering shows that SARS-CoV-2 has only 41.6% similarity to SARS in its pattern of binding to microRNAs.
PCA discriminated efficiently between SARS-CoV-2 and other types of coronaviruses where PCA1 and PCA2 described 71.9% of variation in the data (Supplementary 2). SARS-CoV-2 had negative values of both PCA1 and PCA2 (Figure 6). Interestingly, hsa-miR-5004-3p is one of the top five important microRNAs in PCA1 with an absolute coefficient > 0.2 (Supplementary 2). ptc-miR474b, cme-miR1863, bta-miR-2284ab, and bdi-miR5065 were the top microRNAs in PCA2. Consistent with this finding, in comparison with the other human pathogens (SARS and MERS), the leader sequence of SARS-COV-2 has a remarkably higher binding energy against microRNAs (−16.33 kcal/mol against 18.58 kcal/mol and −18.34 kcal/mol, respectively) (Table 3). Higher binding energy results in lower stability of microRNA-5′UTR binding and provides this opportunity for SARS-COV-2 to escape the inhibitory microRNAs.
Clustering shows that SARS-CoV-2 has only 41.6% similarity with SARS in its pattern of binding to microRNAs (Supplementary 2 and Figure 6).
3.4. Drug Repurposing to Induce 5′UTR Inhibitory MicroRNAs
As presented in Figure 7, we developed a literature mining-based drug repurposing approach to identify inhibitory microRNAs against the leader sequence of SARS-CoV-2. To this end, more than 1 million drugs and small molecules, and 12 million relations (binding, biomarker, expression, chemical reaction, promoter binding, microRNA effect, etc.) were mined by natural language processing (NLP).
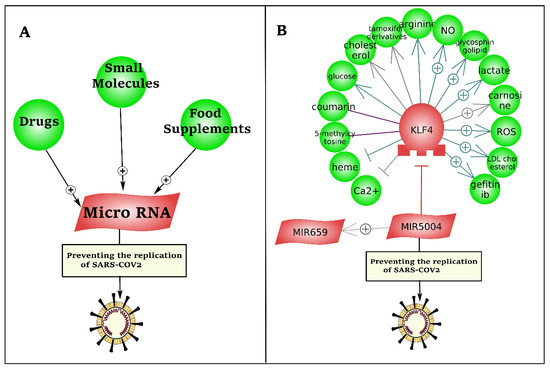
Figure 7.
Drug repurposing to induce inhibitory microRNAs against the leader sequence and TRS of the 5′UTR region of SARS-CoV-2. (A) The presented pipeline of literature mining-based drug discovery. (B) Upregulation of hsa-miR-5004-3p has potential to prevent SARS-CoV-2 replication via binding to the leader sequence in the 5′UTR of virus.
Literature mining-based drug repurposing could not identify any drug, small molecule, or food supplement with direct interaction with hsa-miR-5004-3p (Figure 7). However, downregulation of KLF4, by heme and calcium, sowed indirect potential to upregulate hsa-miR-5004-3p as an inhibitory microRNA against SARS-CoV-2. Heme is an iron-containing tetradentate ligand.
3.5. Significant Decline in Expression of MIR-5004 after SARS-COV-2 Infection
Meta-analysis of MIR-5004 expression in a range of tissues and cells (42 samples), including trachea, lung biopsy, lung-derived Calu-3 cells, and lung alveolar A549 cells, highlighted a significant (p < 0.01) decline in expression in of MIR-5004 after SARS-COV-2 infection (Figure 8). Interestingly, the decline in expression of MIR-5004-3p was more significant in trachea and lung tissues than in cell lines. Noticeably, the decline in expression of human inhibitory microRNAs with increasing age, and in a range of diseases such as diabetes and obesity, has been observed [19] that can contribute to higher mortality of SARS-CoV-2 in elderly patients.
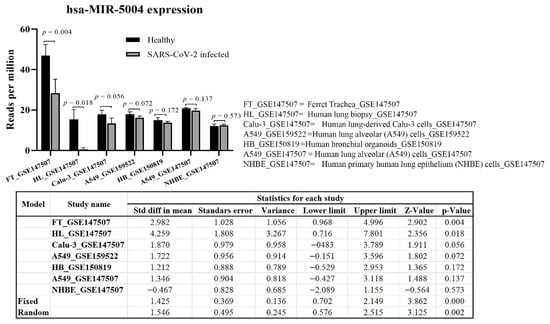
Figure 8.
hsa-MIR-5004 expression response to SARS-CoV-2 infection. Meta-analysis revealed a significant (p < 0.01) decline in expression of hsa-MIR-5004 after SARS-COV-2 infection. hsa-MIR-5004 expression was studied in 42 samples of trachea, lung biopsy, lung-derived Calu-3 cells, and lung alveolar A549 cells before and after SARS-CoV-2 infection.
3.6. hsa-miR-5004-3p Genomic Variation
Impaired 5′UTR inhibitory microRNAs in the human genome could account for the high rate of virus replication in human cells. Consequently, mutations in genomic sequences in the 5′UTR inhibitory microRNAs may be considered as a risk factor of COVID-19 infection. Mining more than two hundred million deposited variants in dbSNP, using the Pathway Studio tool (Elsevier), resulted in discovery of 9 variants in hsa-MIR-5004 (Table 4). Six of these variants were splice-disrupted mutations with possible regulatory functions. Table 4 presents the potentially damaging variants according to their GERP++ conservation scores. The GERP++ conservation scores vary from −12.3 to 6.17. Larger values demonstrate higher conservation. Splice-disrupted mutations in MIR5004 have high GERP++ scores.

Table 4.
hsa-miR-5004-3p genomic variation. High GERP++ conservation score is associated with higher functional impact.
4. Discussion
UTRs, particularly in the 5′UTR, are of high translational importance. The leader sequence and TRS in the 5′ non-coding part of SARS-CoV-2 can be considered as the Achilles’ heel of SARS-CoV-2. Leader sequence is located at the 5′ ends of all encoded transcripts, highlighting its potential significance. The TRS can explain the host range and pathogenicity of a coronavirus. UTRs are potential sites for antiviral drugs to bind and inhibit the virus replication. There is no report on disruption of 5′UTR in SARS-CoV-2 but, in bovine coronavirus, it has been found that disruption of either stem-loop III or stem-loop IV of the 5′UTR stops virus RNA replication, suggesting that these regions function as cis-acting elements [4,20]. On the other hand, microRNAs that bind to SARS-CoV-2 UTRs can be induced by drugs or food supplements to reduce virus replication. Enhancing host microRNA defense machinery against the 5′UTR region of a virus can help in prevention of SARS-CoV-2 infection. The above-mentioned strategies represent potentially achievable treatments against COVID-19 infection. In this study, we presented a model of literature mining-based drug discovery to induce inhibitory microRNAs against leader sequence of SARS-CoV-2.
Some host cell microRNAs are important components of the host immune defense against viral infection as they destroy the viral RNA [21]. In this study, we found that hsa-miR-5004-3p is a unique human microRNA with the ability to target the leader sequence of SARS and SARS-CoV-2. Our comprehensive meta-analysis also documented a significant (p < 0.01) decline in the expression of MIR-5004-3p after SARS-COV-2 infection in trachea, lung, and bronchial organoids as well as in lung-derived Calu-3 and A549 cells. Interestingly, a decreased level of hsa-miR-5004-3p has been reported in the whole blood of patients during dengue virus (DENV) infection. hsa-miR-5004-3p was undetectable in early DENV infection, but expression was high in most of the healthy controls and recovered dengue patients. This finding demonstrated suppression of hsa-miR-5004-3p during the early phases of DENV infection and its importance in patient recovery [22]. Like COVID-19, DENV is a rampant arboviral illness worldwide [22] with high viral transmission success and immune evasion.
We found a significant trend in the 5′UTR of SARS-CoV-2 to escape from binding of hsa-miR-5004-3p by insertion-type mutation. Such mutations decrease microRNA-5′UTR binding stability and allow SARS-CoV-2 to escape the available human microRNA immunity system. We suggest that the lack of innate human inhibitory microRNAs for SARS-CoV-2 contributes to its high replication in the infected human cells. On the other hand, mining of two hundred million deposited human genomic variants led us to discover splice-disrupted mutations in the genomic structure of hsa-miR-5004-3p. These mutations can negatively affect hsa-miR-5004-3p function in preventing SARS-CoV-2 replication.
The abundance of some human inhibitory microRNAs against SARS-CoV-2 is associated with age [19]. It has been discussed that higher COVID-19 virulence in the aged patients [23] is potentially related to the decline in microRNAs in elderly people [19]. In addition to age, some of the human microRNAs downregulate in coronary artery disease, kidney disease, colorectal cancer, osteosarcoma, prostate cancer, obesity/diabetes, myocardial injury, hepatocellular carcinoma, non-small cell lung cancer, gastrointestinal tumors, and colorectal cancer [19]. Like age, the mentioned conditions can also contribute to lower abundance of human inhibitory microRNAs and consequently, higher severity and mortality of COVID-19. This hypothesis needs to be examined in future studies.
The 5′UTR inhibitory microRNAs, such as hsa-miR-5004-3p or those of plant origin such as ptc-miR474 and csi-miR3953, have promising potential for developing new therapies. MicroRNA vaccine, nanoparticles, synthesized microRNAs, microRNA exosome, and dietary microRNAs can be considered in this context. MicroRNA-peptide fusion is also suggested as potential vaccine candidates [24]. In addition to hsa-miR-5004-3p, plant microRNAs with very low binding energy (less than −22 kcal/mol) against leader sequence of 5′UTR of SARS-CoV-2, such as ptc-miR474b and ptc-miR474a (Populus trichocarpa) and csi-miR3953 (Citrus sinensis) are possible candidates for inclusion. Nanoformulations of the SARS-CoV-2 inhibitory microRNAs can successfully and safely deliver microRNAs to lung cells, to reduce viral replication in the host cell and suppress the viral load [25]. MicroRNA-based nanoparticles can also be utilized in the form of nanovaccines to prevent SARS-CoV-2 infection. Compared with traditional vaccines, nanovaccines have many potential benefits, including: (1) Specific targeting to infection sites, (2) minimal off-target effects, and (3) the possibility of nasal spray/drop formulation for direct activation of the immune system in the respiratory tract [25]. Due to shortness of microRNA sequences (18–25 nucleotides), synthesizing of inhibitory microRNAs are readily conducted. It has been suggested that mixtures of synthesized microRNAs, called multiple microRNA cocktail, targeting different coding (such as S gene) and UTR regions of SARS-CoV-2 can be considered as new COVID-19 treatment strategy [19,26]. This synthesized mixture may be delivered to the target host cells by liposomes like exosome (extracellular vesicle), polymer-based carriers, or nanoparticles [26]. High potential of microRNA exosomes as biomarkers of infection and recovery in COVID 19 has been suggested [24]. On the other hand, the combination of a mRNA vaccine with the immune promoting agents, such as inhibitory microRNAs, can increase its efficiency and protection, even in the case of new SARS-CoV-2 mutations.
Interestingly, expression of microRNAs, complementary to the 3′UTR of viruses, is the main protecting strategy in plants that degrades the viral RNA or blocks its translation by ribosomes [26]. Plants are a rich source for microRNAs, with remarkable therapeutic and preventive roles in many diseases [27]. Recently, it has been suggested that microRNAs in food can be absorbed by the human circulatory system [28]. The absorbed microRNA, named dietary microRNA, can regulate gene expression and biological processes in mammalian cells [28]. Dietary microRNA, as a novel functional feature of food [28], opens a new potential avenue for safe and accessible COVID19 protection and treatment. Further research on plant dietary microRNAs may lead to a safe treatment strategy against COVID19 by oral use of plant microRNAs or whole plants containing SARS-CoV-2 inhibitory microRNAs.
Sequence variation in the non-coding viral genomic regions may predispose people to develop more severe disease. It should be noted that the genetic signature of the pathogenesis severity in the non-coding regions of SARS-CoV-2 is still unknown. A recent report on human–mink–human transmission cycle [29] highlights the necessity to uncover all functional mutations, including the ones that occur in UTR regions. The availability of 212,346 (at 21 November 2020) SARS-CoV-2 genomic sequences, including 159,057 sequences of the full genome and high coverage, in GISAID (https://www.gisaid.org/) [30] and NCBI provides the chance of pattern recognition in 5′UTR sequences of SARS-CoV-2, particularly against host microRNA inhibitory machinery, by machine learning models. Models and statistics such as decision tree classification based on association rule mining and deep learning [31,32,33,34,35,36] that have been used for eukaryotic promoter and UTR analysis, can be examined for UTR analysis of SARS-CoV-2. It should be noted that some of the sequences that have been deposited in GISAID as full genomes have incomplete or low-quality sequencing in 3′UTR and 5′UTR regions. Data cleaning of 5′UTR and 3′UTR regions is a major task before analysis as many of the announced SARS-CoV-2 full genomes with complete coding sequences have incomplete, short, or low-quality UTR sequences. This problem casts doubt on the reliability of the currently identified SARS-CoV-2 sequences and UTR mutations.
5. Conclusions
Non-coding regions are crucial for SARS-CoV-2 replication, transcription, and domination of host systems biology. Unravelling the reasons underpinning SARS-CoV-2 success in dominating human cells and its high transmission rate is crucial for future research. The present study is a significant step towards unravelling the evolution of the 5′UTR in SARS-CoV-2, discovering the key regions, and utilizing the UTRs for lowering virus load in the infected cells. We have shown that human hsa-miR-5004-3p and several plant microRNAs are good microRNA candidates to target the leader sequence of SARS-CoV-2 at 5′UTR region.
In this study, an evolutionary pattern in the 5′UTR of SARS-CoV-2 from SARS was discovered whereby SARS-CoV-2 tries to escape hsa-miR-5004-3p binding by the generation of insertion-type mutations. The lack of a human inhibitory microRNAs to target the 5′ UTR of SARS-CoV-2 can contribute in immunity evasion and pathogenesis in SARS-CoV-2. Decline in expression of human inhibitory microRNAs with increasing age, and in a range of diseases such as diabetes and obesity, can also contribute to higher mortality of SARS-CoV-2 in elderly patients and in individuals suffering from these diseases.
We also developed a literature mining-based drug repurposing strategy to induce inhibitory microRNAs that are potentially active against the leader sequence of SARS-CoV-2. Activation of inhibitory machinery microRNAs by drug repurposing and food supplements are easily achievable treatment strategies against COVID-19. Additionally, the microRNAs identified can be utilized for other therapeutic strategies such as development of microRNA vaccines, nanoparticles, synthesized microRNAs, microRNA exosome, and dietary microRNA.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/10/2/319/s1, Supplementary 1: Seed region of 9994 microRNAs families in targetscan database, Supplementary 2: Multivariate analysis.
Author Contributions
Conceptualization, M.M.-D., S.M.M., S.R. and E.E.; Methodology, M.M.-D., S.M.M., S.R. and E.E.; Software, S.R. and E.E.; Validation, S.R. and E.E.; Formal Analysis, M.M.-D., S.R., I.O.A. and E.E.; Investigation, M.M.-D., S.M.M., S.R. and E.E.; Resources, I.O.A., Z.S.A.S. and E.E. Data Curation, S.M.M., S.R. and E.E.; Writing—Original Draft Preparation, M.M.-D., S.M.M., S.R. and E.E.; Writing—Review & Editing, I.O.A., Z.S.A.S. and E.E.; Visualization, M.M.-D.; Supervision, E.E.; Project Administration, E.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We would like to greatly thank Genomics Research Platform, College of Science, Health and Engineering of La Trobe University as well as School of Animal and Veterinary Sciences of The University of Adelaide for providing the computational resources to perform this study. This research was supported by use of the Nectar Research Cloud, a collaborative Australian research platform supported by the National Collaborative Research Infrastructure Strategy (NCRIS). Additionally, this work was supported by resources provided by the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia. We thank Jeremy Timmis, Emeritus Professor of Genetics at The University of Adelaide, for editing of the revised manuscript.
Conflicts of Interest
The authors declare that there is no conflict of interest.
References
- Yang, D.; Leibowitz, J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015, 206, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Sola, I.; Almazan, F.; Zuniga, S.; Enjuanes, L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef]
- Huang, C.; Lokugamage, K.G.; Rozovics, J.M.; Narayanan, K.; Semler, B.L.; Makino, S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: Viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011, 7, e1002433. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.; Bouma, P.; Williams, G.D.; Brian, D.A. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2003, 77, 6720–6730. [Google Scholar] [CrossRef] [PubMed]
- Alanazi, I.O.; Al Shehri, Z.S.; Ebrahimie, E.; Giahi, H.; Mohammadi-Dehcheshmeh, M. Non-coding and coding genomic variants distinguish prostate cancer, castration-resistant prostate cancer, familial prostate cancer, and metastatic castration-resistant prostate cancer from each other. Mol. Carcinog. 2019, 58, 862–874. [Google Scholar] [CrossRef] [PubMed]
- De Jong, V.; Zaldumbide, A.; Van Der Slik, A.; Persengiev, S.; Roep, B.; Koeleman, B. Post-transcriptional control of candidate risk genes for type 1 diabetes by rare genetic variants. Genes Immun. 2013, 14, 58–61. [Google Scholar] [CrossRef]
- Paul, S.; Bravo Vázquez, L.A.; Pérez Uribe, S.; Roxana Reyes-Pérez, P.; Sharma, A. Current status of microRNA-based therapeutic approaches in neurodegenerative disorders. Cells 2020, 9, 1698. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Bandyra, K.J.; Said, N.; Pfeiffer, V.; Górna, M.W.; Vogel, J.; Luisi, B.F. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell 2012, 47, 943–953. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, A.; Shankar, R.; Bagchi, S.; Grama, A.; Chaterji, S. MicroRNA target prediction using thermodynamic and sequence curves. BMC Genom. 2015, 16, 999. [Google Scholar] [CrossRef]
- Alanazi, I.O.; AlYahya, S.A.; Ebrahimie, E.; Mohammadi-Dehcheshmeh, M. Computational systems biology analysis of biomarkers in lung cancer; unravelling genomic regions which frequently encode biomarkers, enriched pathways, and new candidates. Gene 2018, 659, 29–36. [Google Scholar] [CrossRef]
- Novichkova, S.; Egorov, S.; Daraselia, N. MedScan, a natural language processing engine for MEDLINE abstracts. Bioinformatics 2003, 19, 1699–1706. [Google Scholar] [CrossRef]
- Bax, L.; Yu, L.-M.; Ikeda, N.; Moons, K.G. A systematic comparison of software dedicated to meta-analysis of causal studies. BMC Med. Res. Methodol. 2007, 7, 40. [Google Scholar] [CrossRef]
- Kulinskaya, E.; Morgenthaler, S.; Staudte, R.G. Meta Analysis: A Guide to Calibrating and Combining Statistical Evidence; John Wiley & Sons: Hoboken, NJ, USA, 2008; Volume 756. [Google Scholar]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Davydov, E.V.; Goode, D.L.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef] [PubMed]
- Fulzele, S.; Sahay, B.; Yusufu, I.; Lee, T.J.; Sharma, A.; Kolhe, R.; Isales, C.M. COVID-19 Virulence in Aged Patients Might Be Impacted by the Host Cellular MicroRNAs Abundance/Profile. Aging Dis. 2020, 11, 509. [Google Scholar] [CrossRef]
- Raman, S.; Brian, D.A. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2005, 79, 12434–12446. [Google Scholar] [CrossRef]
- Moens, U. Silencing viral microRNA as a novel antiviral therapy? BioMed Res. Int. 2009, 2009, 419539. [Google Scholar] [CrossRef]
- Tambyah, P.A.; Ching, C.S.; Sepramaniam, S.; Ali, J.M.; Armugam, A.; Jeyaseelan, K. microRNA expression in blood of dengue patients. Ann. Clin. Biochem. 2016, 53, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; McGoogan, J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Cruz-Rodrıguez, L.; Dilsiz, N.; Zıaratı, P.; Lambert, B.; Hochwımmer, B. A miRNA-peptide fusion as a vaccine candidate against the novel coronavirus (COVID-19). Exosomes as potential biomarkers of SARS-COV-2 in lung. J. Biosci. Biomed. Eng. 2020, 1, 1–11. [Google Scholar]
- Chauhan, N.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. COVID-19: Fighting the invisible enemy with microRNAs. Expert Rev. Anti-Infect. Ther. 2020, 1–9. [Google Scholar] [CrossRef]
- El-Nabi, S.H.; Elhiti, M.; El-Sheekh, M. A new approach for COVID-19 treatment by micro-RNA. Med. Hypotheses 2020, 143, 110203. [Google Scholar] [CrossRef]
- Hassab Elnabi, S.E. New strategies for treatment of COVID-19 and evolution of SARS-CoV-2 according to biodiversity and evolution theory. Egypt. J. Basic Appl. Sci. 2020, 7, 226–232. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, T.; Yin, Y.; Zhang, C.-Y.; Zhang, Y.-L. Dietary microRNA—A Novel Functional Component of Food. Adv. Nutr. 2019, 10, 711–721. [Google Scholar] [CrossRef]
- Munnink, B.B.O.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; Van Der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef]
- Mohammadi-Dehcheshmeh, M.; Niazi, A.; Ebrahimi, M.; Tahsili, M.; Nurollah, Z.; Ebrahimi Khaksefid, R.; Ebrahimi, M.; Ebrahimie, E. Unified transcriptomic signature of arbuscular mycorrhiza colonization in roots of Medicago truncatula by integration of machine learning, promoter analysis, and direct merging meta-analysis. Front. Plant Sci. 2018, 9, 1550. [Google Scholar] [CrossRef]
- Hosseinpour, B.; Bakhtiarizadeh, M.R.; Khosravi, P.; Ebrahimie, E. Predicting distinct organization of transcription factor binding sites on the promoter regions: A new genome-based approach to expand human embryonic stem cell regulatory network. Gene 2013, 531, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Kargarfard, F.; Sami, A.; Hemmatzadeh, F.; Ebrahimie, E. Identifying mutation positions in all segments of influenza genome enables better differentiation between pandemic and seasonal strains. Gene 2019, 697, 78–85. [Google Scholar] [CrossRef]
- Kargarfard, F.; Sami, A.; Ebrahimie, E. Knowledge discovery and sequence-based prediction of pandemic influenza using an integrated classification and association rule mining (CBA) algorithm. J. Biomed. Inform. 2015, 57, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Kargarfard, F.; Sami, A.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E. Novel approach for identification of influenza virus host range and zoonotic transmissible sequences by determination of host-related associative positions in viral genome segments. BMC Genom. 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Novikov, B.; Ebrahimie, E.; Spilman, A.; Ahsan, R.; Tahsili, M.R.; Najafi, M.; Navvabi, S.; Shariaty, F. The first report of the most important sequential differences between COVID-19 and MERS viruses by attribute weighting models, the importance of Nucleocapsid (N) protein. Bioinformatics 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).