Pin1 Regulates IL-5 Induced Eosinophil Polarization and Migration
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Mice
2.3. Human Eos Preparation
2.4. Immunostaining, Immunoblots and Intracellular Flow Cytometry
2.5. Recombinant TAT Proteins
2.6. Pull Down Assay
2.7. Cell Polarization and Migration Assay
2.8. Eos Differentiation from Wild Type and Pin1 KO Bone Marrow
2.9. Pin1 Activity Assay
2.10. Statistics
3. Results
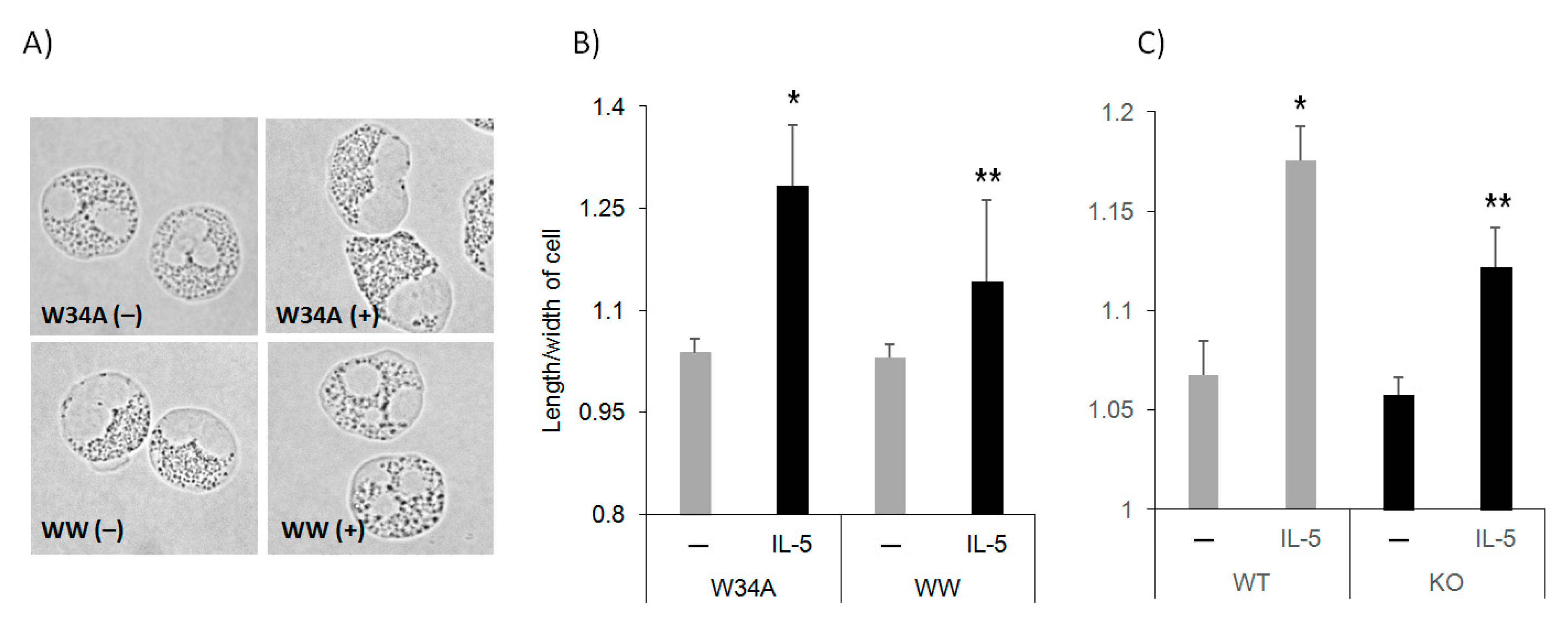
3.1. Pin1 Is Required for IL-5 Induced Eos Polarization
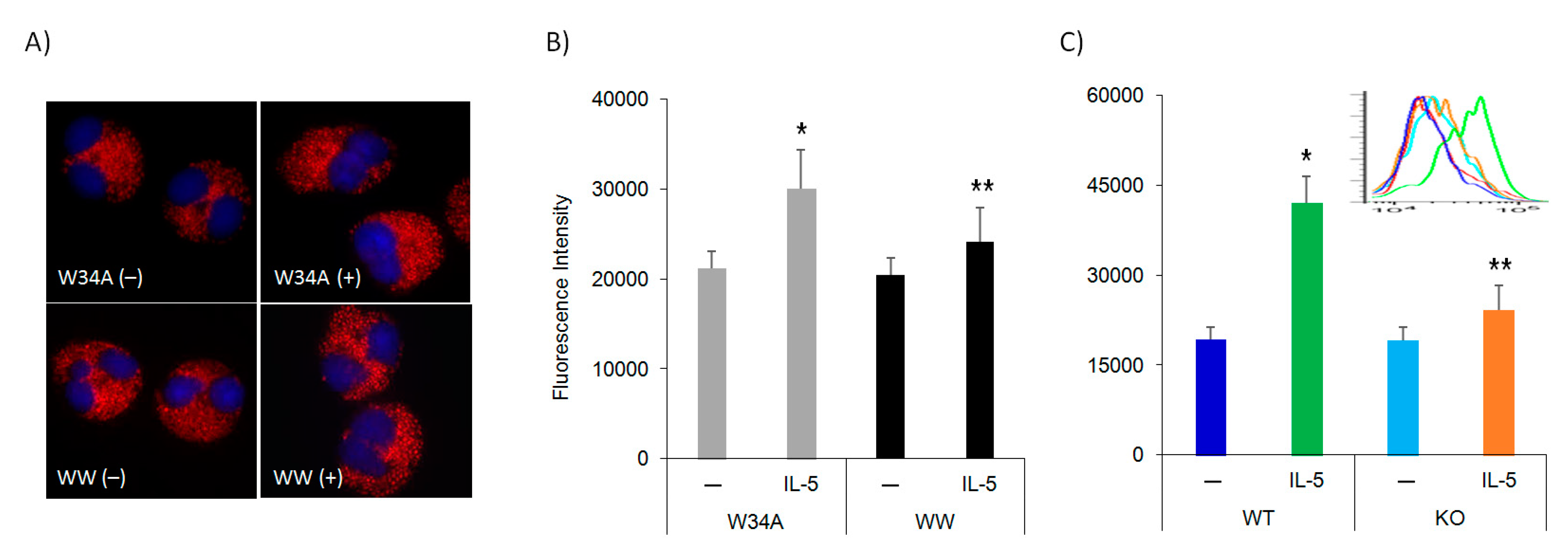
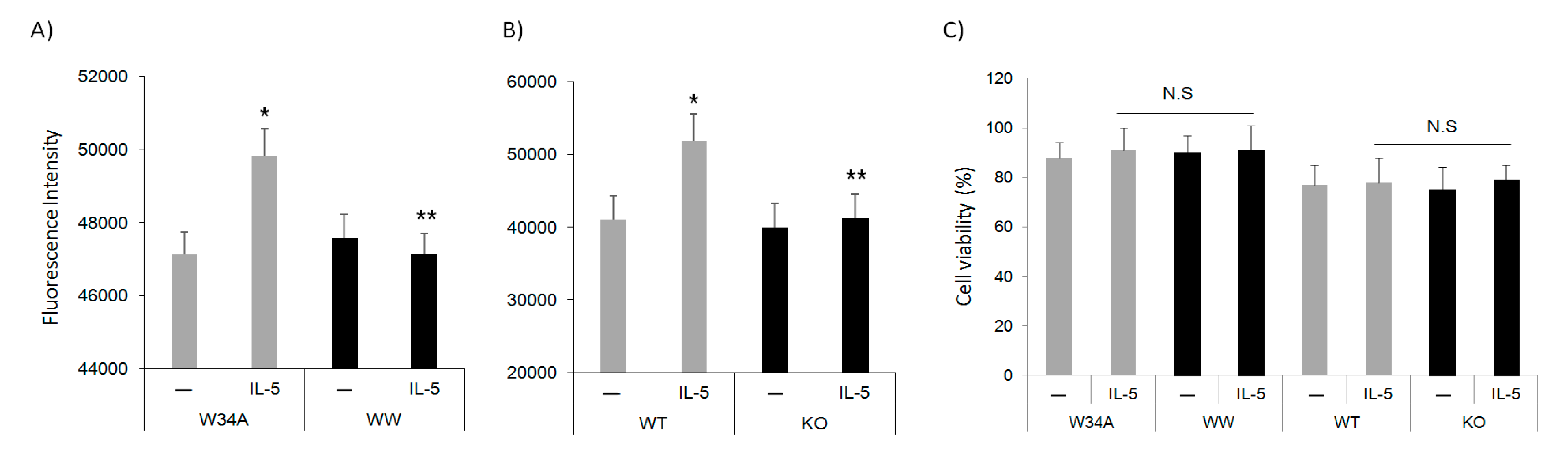
3.2. IL-5 Induction of F-Actin Polymerization Is Pin1 Dependent
3.3. Pin1 Is Required for IL5-Induced Cell Migration
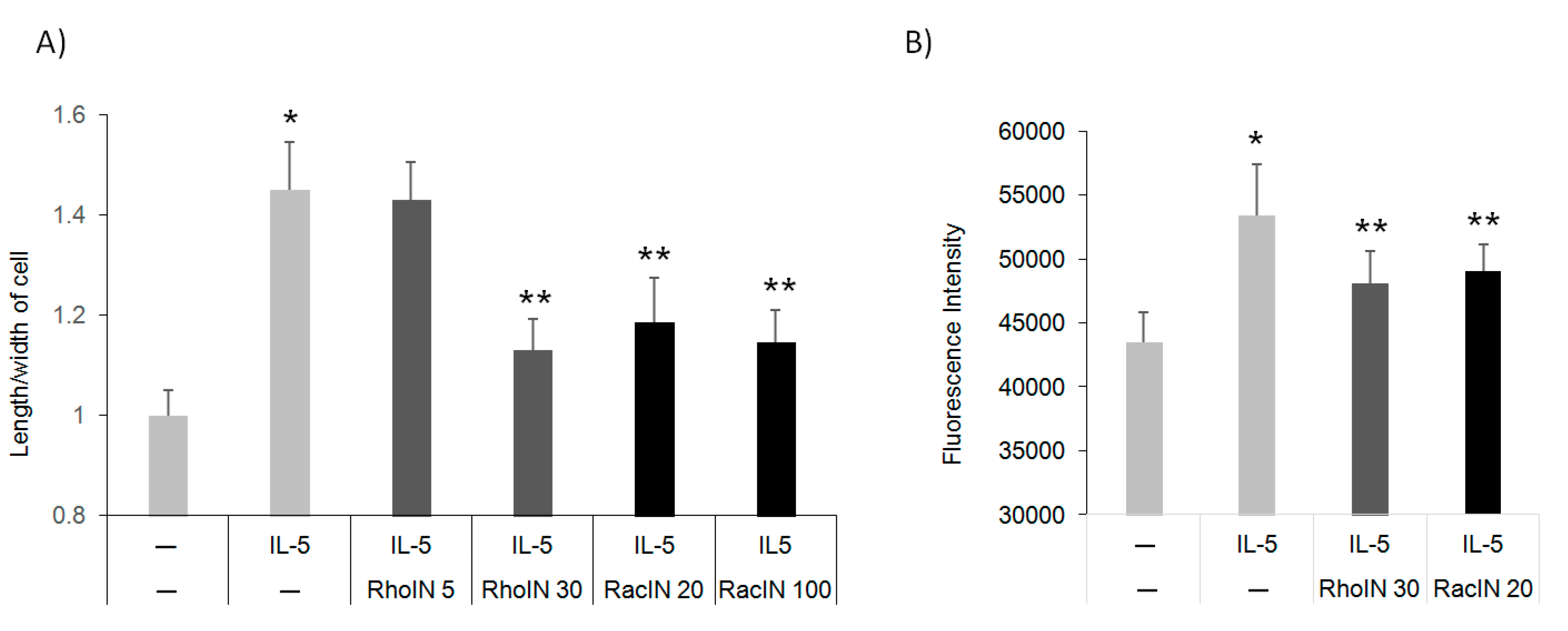
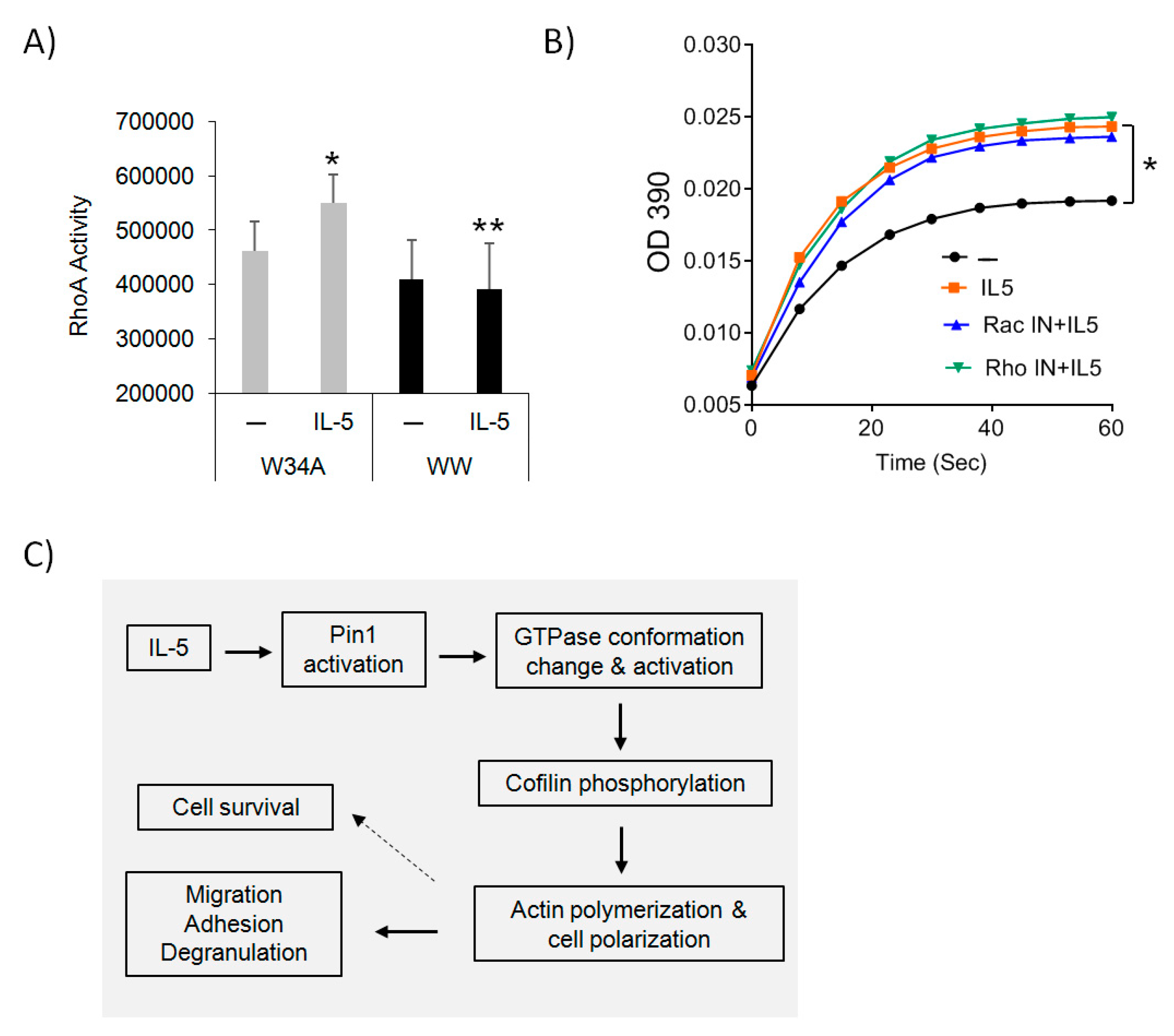
3.4. The Effect of Rho Inhibitors on Eos Polarization and Migration
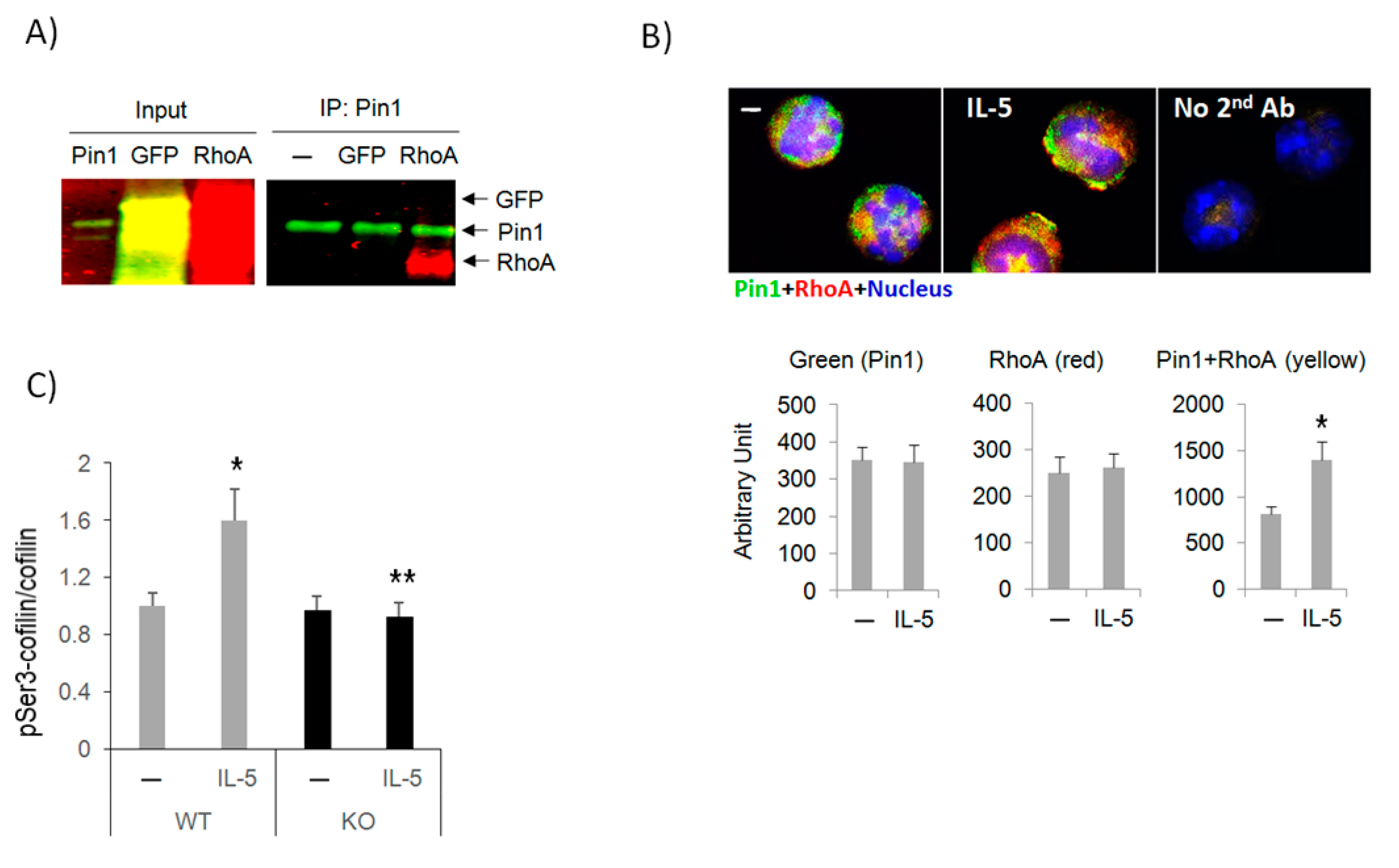
3.5. Pin1 Interacts with Rho GTPase and Regulates Downstream Signaling
3.6. Pin1 Regulates Rho GTPase Activity
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rothenberg, M.E.; Hogan, S.P. The eosinophil. Annu. Rev. Immunol. 2006, 24, 147–174. [Google Scholar] [CrossRef] [PubMed]
- Han, S.T.; Mosher, D.F. IL-5 induces suspended eosinophils to undergo unique global reorganization associated with priming. Am. J. Respir. Cell Mol. Biol. 2014, 50, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.N.; Choi, M.K.; Park, C.S.; Chung, I.Y. A parallel signal-transduction pathway for eotaxin- and interleukin-5-induced eosinophil shape change. Immunology 2003, 108, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.D.; Ridley, A.J. Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 2018, 217, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.F.; Sahai, E. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 2009, 26, 273–287. [Google Scholar] [CrossRef]
- Tang, Y.; Olufemi, L.; Wang, M.T.; Nie, D. Role of Rho GTPases in breast cancer. Front. Biosci. 2008, 13, 759–776. [Google Scholar] [CrossRef][Green Version]
- Campellone, K.G.; Welch, M.D. A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010, 11, 237–251. [Google Scholar] [CrossRef]
- Jaffe, A.B.; Hall, A. Rho GTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005, 21, 247–269. [Google Scholar] [CrossRef]
- Wilkerson, E.M.; Johansson, M.W.; Hebert, A.S.; Westphall, M.S.; Mathur, S.K.; Jarjour, N.N.; Schwantes, E.A.; Mosher, D.F.; Coon, J.J. The Peripheral Blood Eosinophil Proteome. J. Proteome Res. 2016, 15, 1524–1533. [Google Scholar] [CrossRef]
- Esnault, S.; Rosenthal, L.A.; Shen, Z.J.; Sedgwick, J.B.; Szakaly, R.J.; Sorkness, R.L.; Malter, J.S. A critical role for Pin1 in allergic pulmonary eosinophilia in rats. J. Allergy Clin. Immunol. 2007, 120, 1082–1088. [Google Scholar] [CrossRef]
- Shen, Z.J.; Hu, J.; Kashi, V.; Bochkov, Y.A.; Gern, J.E.; Malter, J.S. TLR-7 Stress Signaling in Differentiating and Mature Eosinophils Is Mediated by the Prolyl Isomerase Pin1. J. Immunol. 2018, 201, 3503–3513. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.J.; Esnault, S.; Rosenthal, L.A.; Szakaly, R.J.; Sorkness, R.L.; Westmark, P.R.; Sandor, M.; Malter, J.S. Pin1 regulates TGF-beta1 production by activated human and murine eosinophils and contributes to allergic lung fibrosis. J. Clin. Investig. 2008, 118, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.J.; Esnault, S.; Malter, J.S. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat. Immunol. 2005, 6, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.J.; Hu, J.; Kashi, V.P.; Kelly, E.A.; Denlinger, L.C.; Lutchman, K.; McDonald, J.G.; Jarjour, N.N.; Malter, J.S. Epstein-Barr Virus-induced Gene 2 Mediates Allergen-induced Leukocyte Migration into Airways. Am. J. Respir. Crit. Care Med. 2017, 195, 1576–1585. [Google Scholar] [CrossRef]
- Shen, Z.J.; Esnault, S.; Schinzel, A.; Borner, C.; Malter, J.S. The peptidyl-prolyl isomerase Pin1 facilitates cytokine-induced survival of eosinophils by suppressing Bax activation. Nat. Immunol. 2009, 10, 257–265. [Google Scholar] [CrossRef]
- Lu, K.P.; Zhou, X.Z. The prolyl isomerase PIN1: A pivotal new twist in phosphorylation signalling and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 904–916. [Google Scholar] [CrossRef]
- Hansel, T.T.; De Vries, I.J.; Iff, T.; Rihs, S.; Wandzilak, M.; Betz, S.; Blaser, K.; Walker, C. An improved immunomagnetic procedure for the isolation of highly purified human blood eosinophils. J. Immunol. Methods 1991, 145, 105–110. [Google Scholar] [CrossRef]
- Dyer, K.D.; Moser, J.M.; Czapiga, M.; Siegel, S.J.; Percopo, C.M.; Rosenberg, H.F. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J. Immunol. 2008, 181, 4004–4009. [Google Scholar] [CrossRef]
- Hennig, L.; Christner, C.; Kipping, M.; Schelbert, B.; Rücknagel, K.P.; Grabley, S.; Küllertz, G.; Fischer, G. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry 1998, 37, 5953–5960. [Google Scholar] [CrossRef]
- Rosenberg, H.F.; Dyer, K.D.; Foster, P.S. Eosinophils: Changing perspectives in health and disease. Nat. Rev. Immunol. 2013, 13, 9–22. [Google Scholar] [CrossRef]
- Tirnauer, J.S. Coupled zones of f-actin and microtubule movement in polarized cells. Dev. Cell 2002, 3, 152–153. [Google Scholar] [CrossRef]
- Wickramarachchi, D.C.; Theofilopoulos, A.N.; Kono, D.H. Immune pathology associated with altered actin cytoskeleton regulation. Autoimmunity 2010, 43, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Lacy, P.; Willetts, L.; Kim, J.D.; Lo, A.N.; Lam, B.; Maclean, E.I.; Moqbel, R.; Rothenberg, M.E.; Zimmermann, N. Agonist activation of f-actin-mediated eosinophil shape change and mediator release is dependent on Rac2. Int. Arch. Allergy Immunol. 2011, 156, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Mosher, D.F.; Wilkerson, E.M.; Turton, K.B.; Hebert, A.S.; Coon, J.J. Proteomics of Eosinophil Activation. Front. Med. (Lausanne). 2017, 4, 159. [Google Scholar] [CrossRef]
- Tong, J.; Li, L.; Ballermann, B.; Wang, Z. Phosphorylation and Activation of RhoA by ERK in Response to Epidermal Growth Factor Stimulation. PLoS ONE 2016, 11, e0147103. [Google Scholar] [CrossRef]
- Oleinik, N.V.; Helke, K.L.; Kistner-Griffin, E.; Krupenko, N.I.; Krupenko, S.A. Rho GTPases RhoA and Rac1 mediate effects of dietary folate on metastatic potential of A549 cancer cells through the control of cofilin phosphorylation. J. Biol. Chem. 2014, 289, 26383–26394. [Google Scholar] [CrossRef]
- Kuhn, T.B.; Meberg, P.J.; Brown, M.D.; Bernstein, B.W.; Minamide, L.S.; Jensen, J.R.; Okada, K.; Soda, E.A.; Bamburg, J.R. Regulating actin dynamics in neuronal growth cones by ADF/cofilin and rho family GTPases. J. Neurobiol. 2000, 44, 126–144. [Google Scholar] [CrossRef]
- Shamri, R.; Young, K.M.; Weller, P.F. Rho and Rac, but not ROCK, are required for secretion of human and mouse eosinophil-associated RNases. Clin. Exp. Allergy 2019, 49, 190–198. [Google Scholar] [CrossRef]
- Wang, L.; Yang, L.; Filippi, M.D.; Williams, D.A.; Zheng, Y. Genetic deletion of Cdc42GAP reveals a role of Cdc42 in erythropoiesis and hematopoietic stem/progenitor cell survival, adhesion, and engraftment. Blood 2006, 107, 98–105. [Google Scholar] [CrossRef]
- Wang, L.; Yang, L.; Burns, K.; Kuan, C.Y.; Zheng, Y. Cdc42GAP regulates c-Jun N-terminal kinase (JNK)-mediated apoptosis and cell number during mammalian perinatal growth. Proc. Natl. Acad. Sci. USA 2005, 102, 13484–13489. [Google Scholar] [CrossRef]
- Coughlin, J.J.; Odemuyiwa, S.O.; Davidson, C.E.; Moqbel, R. Differential expression and activation of Rab27A in human eosinophils: Relationship to blood eosinophilia. Biochem. Biophys. Res. Commun. 2008, 373, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J.; Hall, A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 1992, 70, 389–399. [Google Scholar] [CrossRef]
- Ridley, A.J.; Paterson, H.F.; Johnston, C.L.; Diekmann, D.; Hall, A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 1992, 70, 401–410. [Google Scholar] [CrossRef]
- Nobes, C.D.; Hall, A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995, 81, 53–62. [Google Scholar] [CrossRef]
- Matsuura, I.; Chiang, K.N.; Lai, C.Y.; He, D.; Wang, G.; Ramkumar, R.; Uchida, T.; Ryo, A.; Lu, K.; Liu, F. Pin1 promotes transforming growth factor-beta-induced migration and invasion. J. Biol. Chem. 2010, 285, 1754–1764. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Z.-J.; Hu, J.; O’Neal, M.A.; Malter, J.S. Pin1 Regulates IL-5 Induced Eosinophil Polarization and Migration. Cells 2021, 10, 211. https://doi.org/10.3390/cells10020211
Shen Z-J, Hu J, O’Neal MA, Malter JS. Pin1 Regulates IL-5 Induced Eosinophil Polarization and Migration. Cells. 2021; 10(2):211. https://doi.org/10.3390/cells10020211
Chicago/Turabian StyleShen, Zhong-Jian, Jie Hu, Melissa A. O’Neal, and James S. Malter. 2021. "Pin1 Regulates IL-5 Induced Eosinophil Polarization and Migration" Cells 10, no. 2: 211. https://doi.org/10.3390/cells10020211
APA StyleShen, Z.-J., Hu, J., O’Neal, M. A., & Malter, J. S. (2021). Pin1 Regulates IL-5 Induced Eosinophil Polarization and Migration. Cells, 10(2), 211. https://doi.org/10.3390/cells10020211





