Double-Stranded RNA Structural Elements Holding the Key to Translational Regulation in Cancer: The Case of Editing in RNA-Binding Motif Protein 8A
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA Interference
2.3. RNA Extraction, cDNA Synthesis, and RT-qPCR
2.4. Protein Extraction, Western Blotting, and Cell Fractionation
2.5. Adar2 Cloning, Sequencing, and Transfection
2.6. Immunohistochemistry
2.7. Analysis of Mitochondrial Ribosomal Protein L3 Alternative Splicing
2.8. Editing Quantification
2.9. Generation of the RBM8A 3′UTR and Mutant Luciferase Reporters
2.10. Dual-Luciferase Assay
2.11. RNA Pull-Down
2.12. RNA Analysis
2.13. Statistical Analysis
3. Results
3.1. Mesothelioma Cells Are More Sensitive to RBM8A Deficiency Compared to Mesothelial Cells
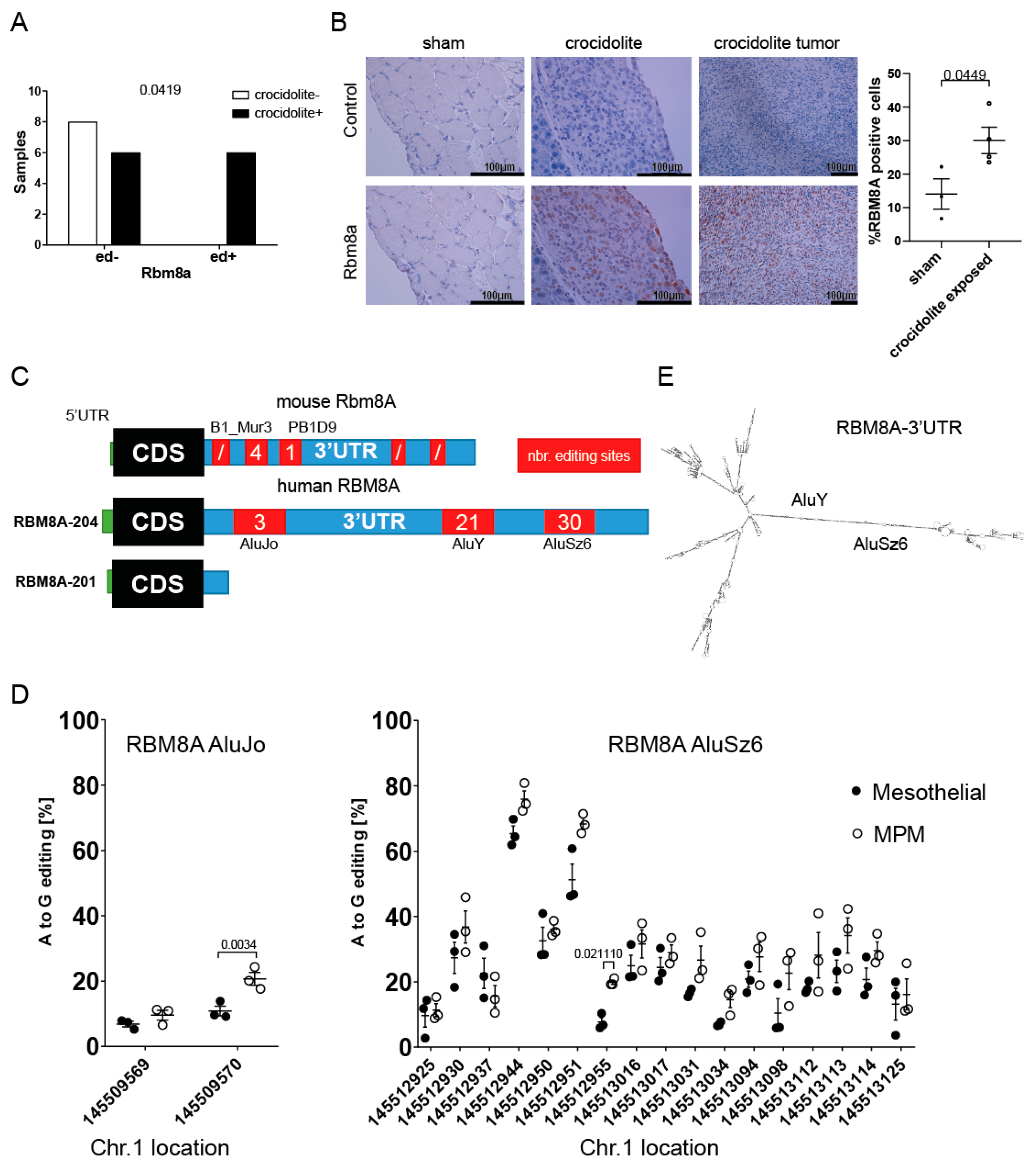
3.2. RBM8A mRNA Has Higher Editing Levels in Mesothelioma Compared to Mesothelial Cells
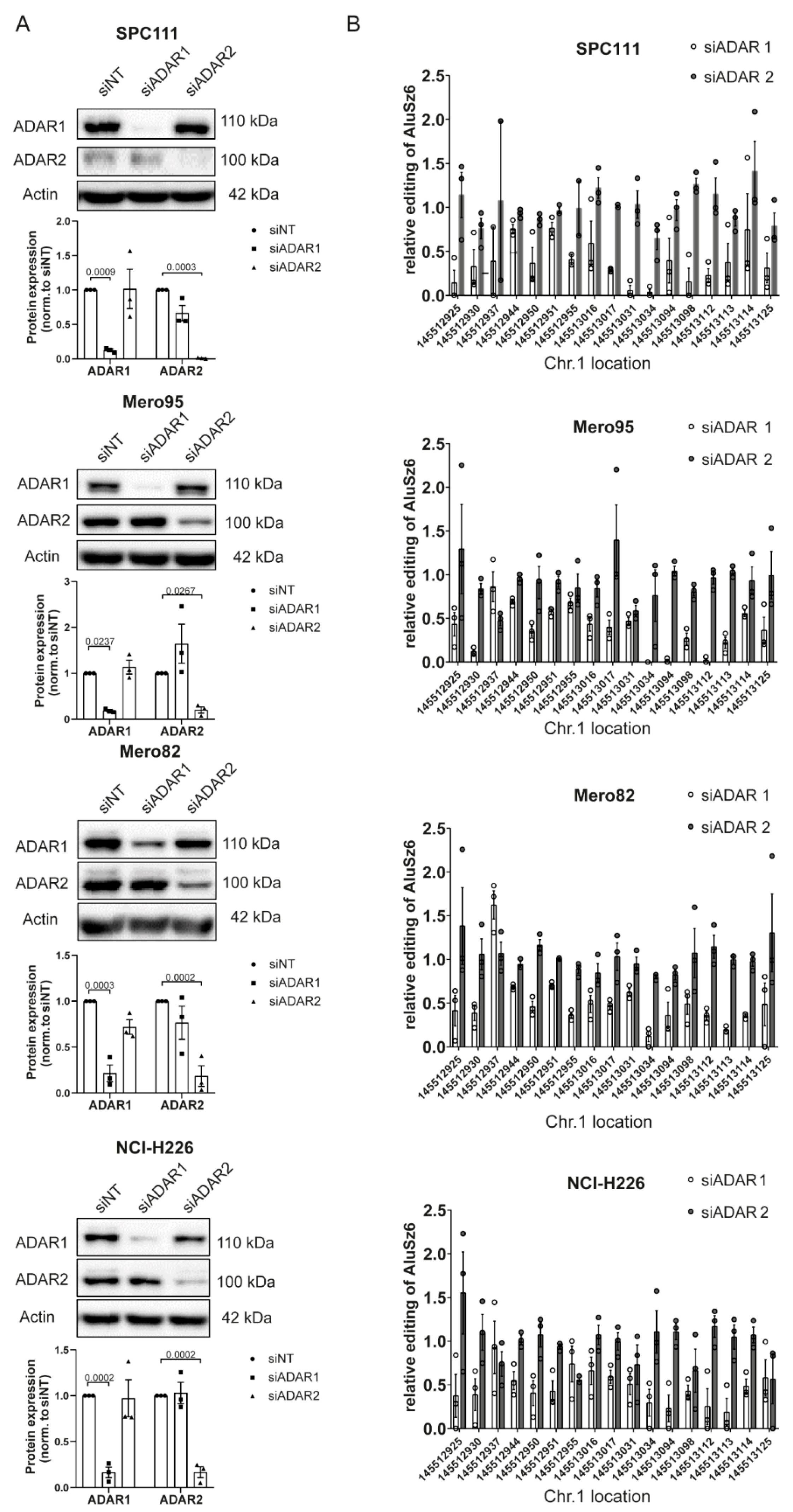
3.3. RBM8A 3′UTR Editing Is Mediated by Both ADAR1 and ADAR2
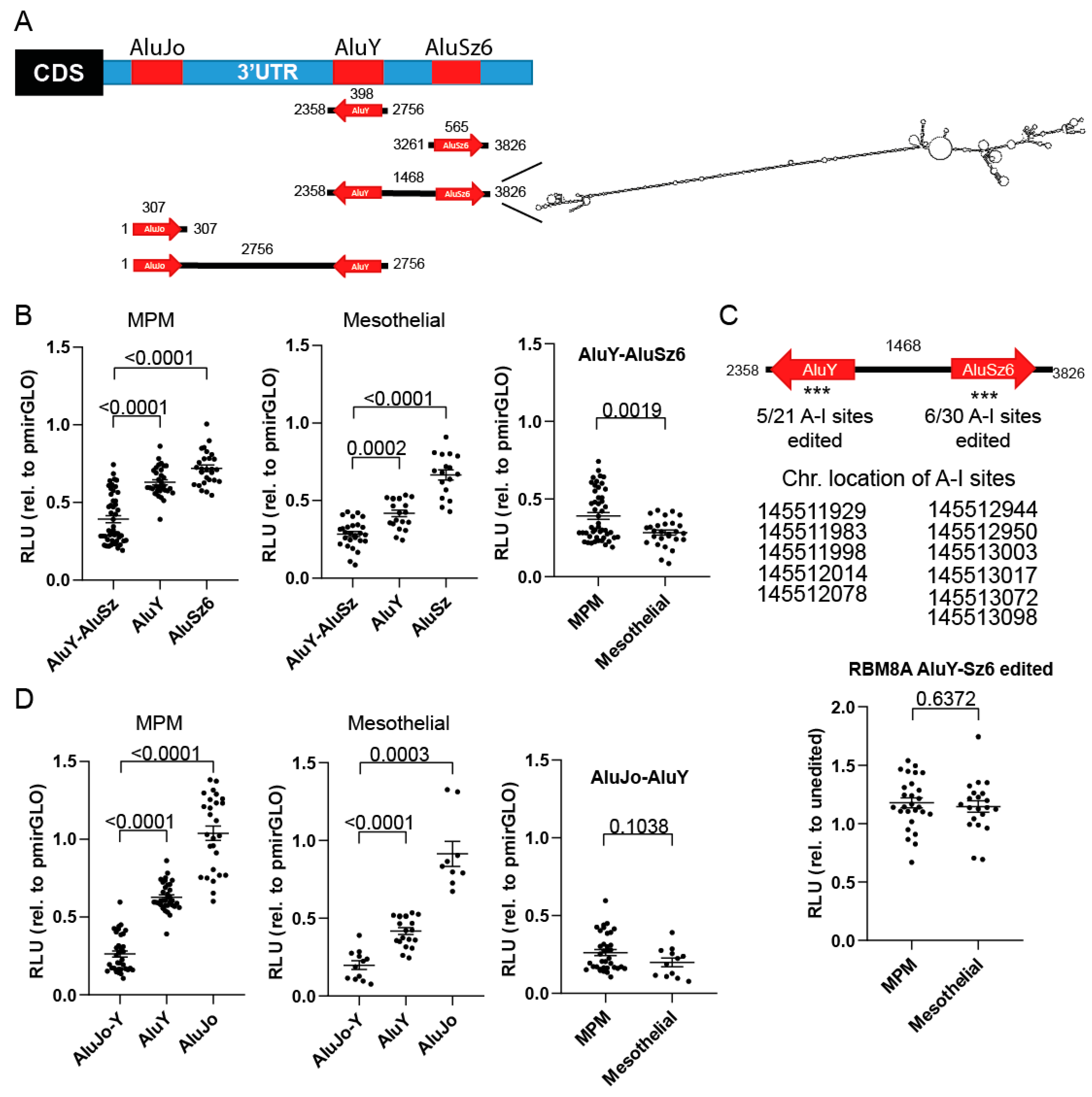
3.4. RBM8A 3′UTR Editing Increases Protein Expression
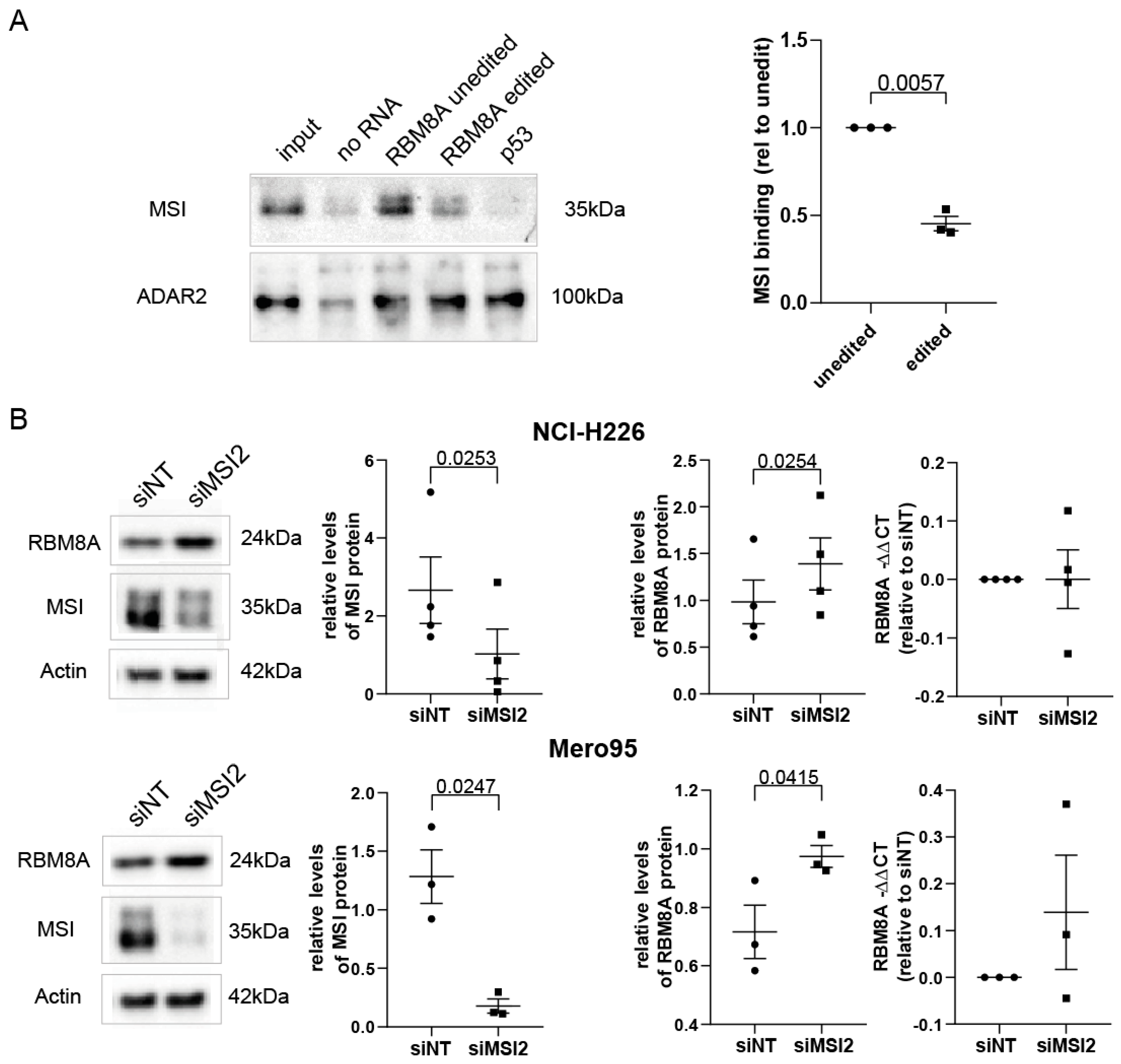
3.5. MSI2 Preferentially Interacts with Unedited RBM8A 3′UTR, and Silencing MSI2 Results in Increased RBM8A Levels
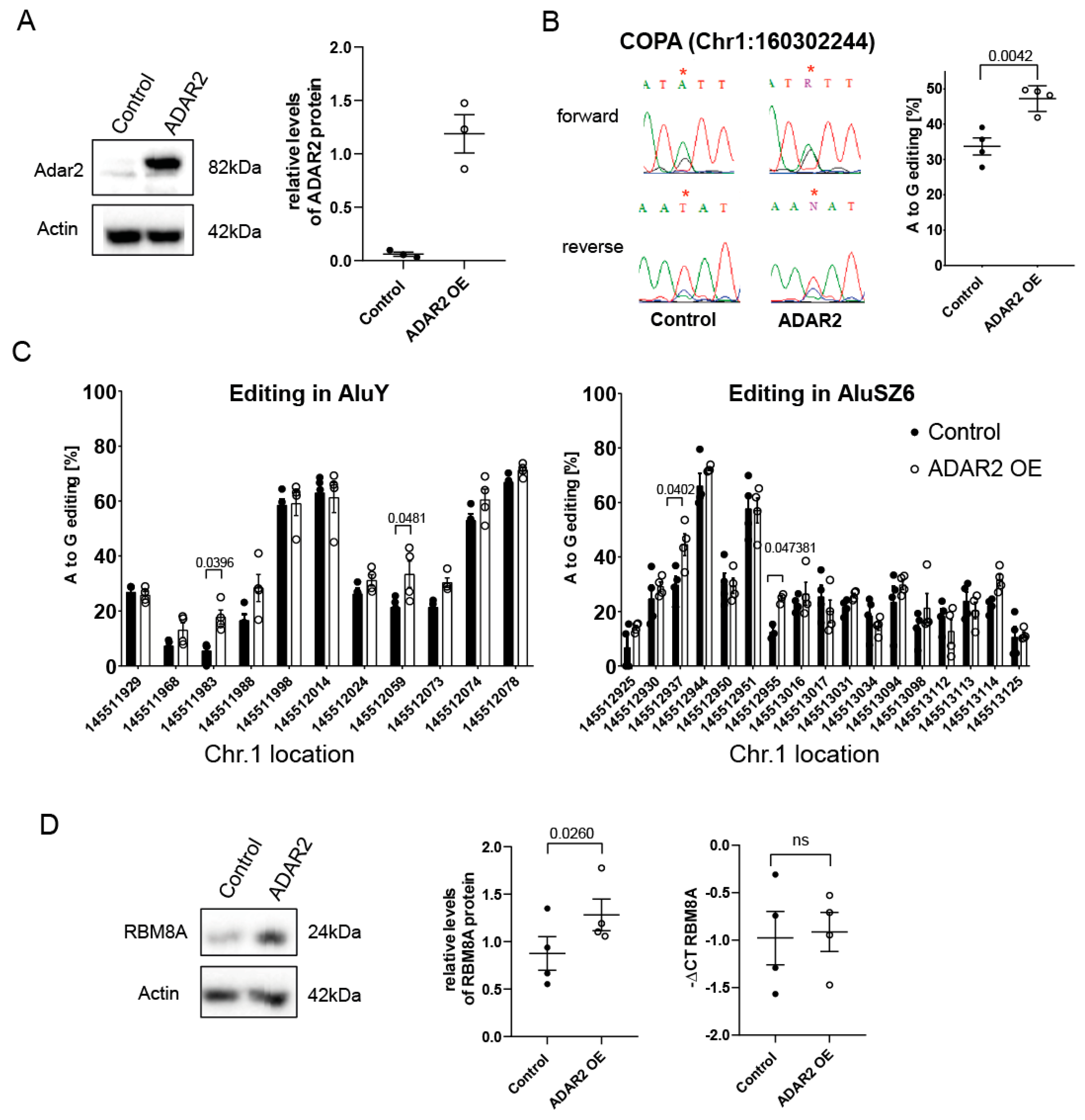
3.6. ADAR-Mediated Editing Increases RBM8A Protein Levels
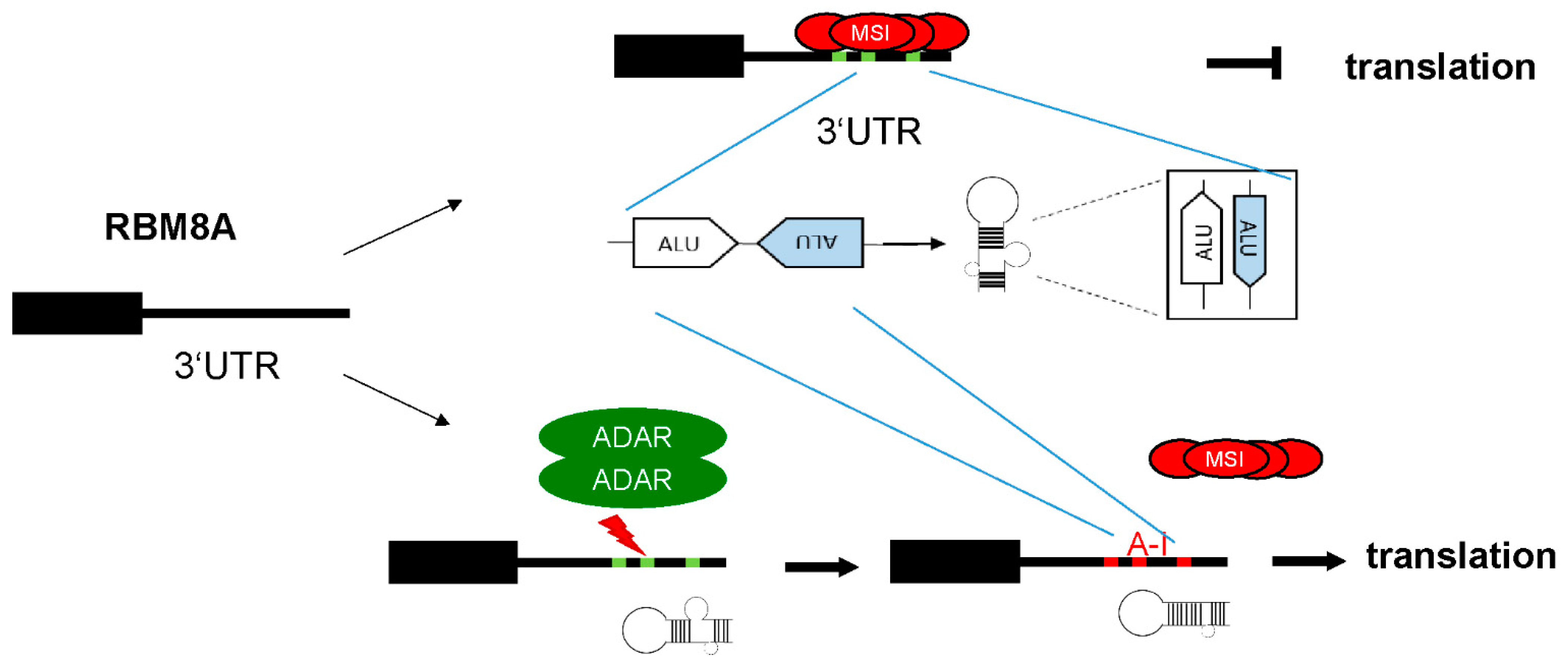
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Felley-Bosco, E.; MacFarlane, M. Asbestos: Modern Insights for Toxicology in the Era of Engineered Nanomaterials. Chem. Res. Toxicol. 2018, 31, 994–1008. [Google Scholar] [CrossRef] [PubMed]
- Carbone, M.; Adusumilli, P.S.; Alexander, H.R., Jr.; Baas, P.; Bardelli, F.; Bononi, A.; Bueno, R.; Felley-Bosco, E.; Galateau-Salle, F.; Jablons, D.; et al. Mesothelioma: Scientific clues for prevention, diagnosis, and therapy. CA Cancer J. Clin. 2019, 69, 402–429. [Google Scholar] [CrossRef] [Green Version]
- Okonska, A.; Buhler, S.; Rao, V.; Ronner, M.; Blijlevens, M.; van der Meulen-Muileman, I.H.; de Menezes, R.X.; Wipplinger, M.; Oehl, K.; Smit, E.F.; et al. Functional Genomic Screen in Mesothelioma Reveals that Loss of Function of BRCA1-Associated Protein 1 Induces Chemoresistance to Ribonucleotide Reductase Inhibition. Mol. Cancer Ther. 2020, 19, 552–563. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.F.; Nowak, N.J.; Shows, T.B.; Aplan, P.D. MAGOH interacts with a novel RNA-binding protein. Genomics 2000, 63, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C. What Are 3′UTRs Doing? Cold Spring Harb. Perspect. Biol. 2019, 11, a034728. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Tian, B.; Manley, J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017, 18, 18–30. [Google Scholar] [CrossRef]
- Woodward, L.A.; Mabin, J.W.; Gangras, P.; Singh, G. The exon junction complex: A lifelong guardian of mRNA fate. Wiley Interdiscip. Rev. RNA 2017, 8, e1411. [Google Scholar] [CrossRef]
- Boehm, V.; Britto-Borges, T.; Steckelberg, A.L.; Singh, K.K.; Gerbracht, J.V.; Gueney, E.; Blazquez, L.; Altmuller, J.; Dieterich, C.; Gehring, N.H. Exon Junction Complexes Suppress Spurious Splice Sites to Safeguard Transcriptome Integrity. Mol. Cell 2018, 72, 482–495.e487. [Google Scholar] [CrossRef] [Green Version]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef]
- Sudo, H.; Tsuji, A.B.; Sugyo, A.; Kohda, M.; Sogawa, C.; Yoshida, C.; Harada, Y.N.; Hino, O.; Saga, T. Knockdown of COPA, identified by loss-of-function screen, induces apoptosis and suppresses tumor growth in mesothelioma mouse model. Genomics 2010, 95, 210–216. [Google Scholar] [CrossRef] [Green Version]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e516. [Google Scholar] [CrossRef] [Green Version]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Frontini, F.; Qi, W.; Hariharan, A.; Ronner, M.; Wipplinger, M.; Blanquart, C.; Rehrauer, H.; Fonteneau, J.F.; Felley-Bosco, E. Endogenous retrovirus expression activates type-I interferon signaling in an experimental mouse model of mesothelioma development. Cancer Lett. 2021, 507, 26–38. [Google Scholar] [CrossRef]
- Rehrauer, H.; Wu, L.; Blum, W.; Pecze, L.; Henzi, T.; Serre-Beinier, V.; Aquino, C.; Vrugt, B.; de Perrot, M.; Schwaller, B.; et al. How asbestos drives the tissue towards tumors: YAP activation, macrophage and mesothelial precursor recruitment, RNA editing, and somatic mutations. Oncogene 2018, 37, 2645–2659. [Google Scholar] [CrossRef] [Green Version]
- Hariharan, A.; Suna, S.; Wipplinger, M.; Felley-Bosco, E. RNA editing in mesothelioma: A look forward. Open Biol. 2020, 10, 200112. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadis, A.; Rich, A.; Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004, 2, e391. [Google Scholar] [CrossRef]
- Erson-Bensan, A.E. RNA-biology ruling cancer progression? Focus on 3′UTRs and splicing. Cancer Metastasis Rev. 2020, 39, 887–901. [Google Scholar] [CrossRef]
- Echeverry, N.; Ziltener, G.; Barbone, D.; Weder, W.; Stahel, R.A.; Broaddus, V.C.; Felley-Bosco, E. Inhibition of autophagy sensitizes malignant pleural mesothelioma cells to dual PI3K/mTOR inhibitors. Cell Death Dis. 2015, 6, e1757. [Google Scholar] [CrossRef] [Green Version]
- Kresoja-Rakic, J.; Felley-Bosco, E. Desthiobiotin-Streptavidin-Affinity Mediated Purification of RNA-Interacting Proteins in Mesothelioma Cells. J. Vis. Exp. 2018, e57516. [Google Scholar] [CrossRef] [Green Version]
- Frei, C.; Opitz, I.; Soltermann, A.; Fischer, B.; Moura, U.; Rehrauer, H.; Weder, W.; Stahel, R.; Felley-Bosco, E. Pleural mesothelioma side populations have a precursor phenotype. Carcinogenesis 2011, 32, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Murigneux, V.; Le Hir, H. Transcriptome-wide modulation of splicing by the exon junction complex. Genome Biol. 2014, 15, 551. [Google Scholar] [CrossRef]
- Schaffer, A.A.; Kopel, E.; Hendel, A.; Picardi, E.; Levanon, E.Y.; Eisenberg, E. The cell line A-to-I RNA editing catalogue. Nucleic Acids Res. 2020, 48, 5849–5858. [Google Scholar] [CrossRef]
- Ramaswami, G.; Li, J.B. RADAR: A rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014, 42, D109–D113. [Google Scholar] [CrossRef] [Green Version]
- Kiran, A.M.; O’Mahony, J.J.; Sanjeev, K.; Baranov, P.V. Darned in 2013: Inclusion of model organisms and linking with Wikipedia. Nucleic Acids Res. 2013, 41, D258–D261. [Google Scholar] [CrossRef]
- Picardi, E.; Manzari, C.; Mastropasqua, F.; Aiello, I.; D’Erchia, A.M.; Pesole, G. Profiling RNA editing in human tissues: Towards the inosinome Atlas. Sci. Rep. 2015, 5, 14941. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Li, D.; Zhang, W.; Guo, M.; Zhan, Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012, 31, 4415–4427. [Google Scholar] [CrossRef] [Green Version]
- Marin-Bejar, O.; Huarte, M. RNA pulldown protocol for in vitro detection and identification of RNA-associated proteins. Methods Mol. Biol. 2015, 1206, 87–95. [Google Scholar] [CrossRef]
- Wang, X.W.; Forrester, K.; Yeh, H.; Feitelson, M.A.; Gu, J.R.; Harris, C.C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. USA 1994, 91, 2230–2234. [Google Scholar] [CrossRef] [Green Version]
- Wang, X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. RNA 2008, 14, 1012–1017. [Google Scholar] [CrossRef] [Green Version]
- Oehl, K.; Vrugt, B.; Wagner, U.; Kirschner, M.B.; Meerang, M.; Weder, W.; Felley-Bosco, E.; Wollscheid, B.; Bankov, K.; Demes, M.C.; et al. Alterations in BAP1 Are Associated with Cisplatin Resistance through Inhibition of Apoptosis in Malignant Pleural Mesothelioma. Clin. Cancer Res. 2021, 2277–2291. [Google Scholar] [CrossRef]
- Singh, A.; Pruett, N.; Hoang, C.D. In vitro experimental models of mesothelioma revisited. Transl. Lung Cancer Res. 2017, 6, 248–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shankavaram, U.T.; Varma, S.; Kane, D.; Sunshine, M.; Chary, K.K.; Reinhold, W.C.; Pommier, Y.; Weinstein, J.N. CellMiner: A relational database and query tool for the NCI-60 cancer cell lines. BMC Genom. 2009, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2019, 48, D682–D688. [Google Scholar] [CrossRef]
- Salicioni, A.M.; Xi, M.; Vanderveer, L.A.; Balsara, B.; Testa, J.R.; Dunbrack, R.L., Jr.; Godwin, A.K. Identification and structural analysis of human RBM8A and RBM8B: Two highly conserved RNA-binding motif proteins that interact with OVCA1, a candidate tumor suppressor. Genomics 2000, 69, 54–62. [Google Scholar] [CrossRef]
- Grosso, S.; Marini, A.; Gyuraszova, K.; Voorde, J.V.; Sfakianos, A.; Garland, G.D.; Tenor, A.R.; Mordue, R.; Chernova, T.; Morone, N.; et al. The pathogenesis of mesothelioma is driven by a dysregulated translatome. Nat. Commun. 2021, 12, 4920. [Google Scholar] [CrossRef] [PubMed]
- Saldi, T.; Riemondy, K.; Erickson, B.; Bentley, D.L. Alternative RNA structures formed during transcription depend on elongation rate and modify RNA processing. Mol. Cell 2021, 81, 1789–1801.e1785. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, M.M.; Thomas, J.M.; Zheng, Y.; Tran, K.; Phelps, K.J.; Scott, A.I.; Havel, J.; Fisher, A.J.; Beal, P.A. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol. 2016, 23, 426–433. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.K.; Sato, S.; Lazinski, D.W. Substrate recognition by ADAR1 and ADAR2. RNA 2001, 7, 846–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukao, A.; Tomohiro, T.; Fujiwara, T. Translation Initiation Regulated by RNA-Binding Protein in Mammals: The Modulation of Translation Initiation Complex by Trans-Acting Factors. Cells 2021, 10, 1711. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.T.; Lu, Y.; Chu, K.L.; Yang, X.; Park, S.M.; Choo, Z.N.; Chin, C.R.; Prieto, C.; Schurer, A.; Barin, E.; et al. HyperTRIBE uncovers increased MUSASHI-2 RNA binding activity and differential regulation in leukemic stem cells. Nat. Commun. 2020, 11, 2026. [Google Scholar] [CrossRef] [PubMed]
- Battelli, C.; Nikopoulos, G.N.; Mitchell, J.G.; Verdi, J.M. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell Neurosci. 2006, 31, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Tokunaga, A.; Yoshida, T.; Hashimoto, M.; Mikoshiba, K.; Weinmaster, G.; Nakafuku, M.; Okano, H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell Biol. 2001, 21, 3888–3900. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, H.; Imai, T.; Imataka, H.; Tsujimoto, M.; Matsumoto, K.; Okano, H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 2008, 181, 639–653. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; An, O.; Ren, X.; Chan, T.H.M.; Tay, D.J.T.; Tang, S.J.; Han, J.; Hong, H.; Ng, V.H.E.; Ke, X.; et al. RNA editing mediates the functional switch of COPA in a novel mechanism of hepatocarcinogenesis. J. Hepatol. 2021, 74, 135–147. [Google Scholar] [CrossRef]
- Moore, S.; Jarvelin, A.I.; Davis, I.; Bond, G.L.; Castello, A. Expanding horizons: New roles for non-canonical RNA-binding proteins in cancer. Curr. Opin. Genet. Dev. 2018, 48, 112–120. [Google Scholar] [CrossRef]
- Blijlevens, M.; van der Meulen-Muileman, I.H.; de Menezes, R.X.; Smit, E.F.; van Beusechem, V.W. High-throughput RNAi screening reveals cancer-selective lethal targets in the RNA spliceosome. Oncogene 2019, 38, 4142–4153. [Google Scholar] [CrossRef]
- Kosti, I.; Jain, N.; Aran, D.; Butte, A.J.; Sirota, M. Cross-tissue Analysis of Gene and Protein Expression in Normal and Cancer Tissues. Sci. Rep. 2016, 6, 24799. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Luna, A.; Rajapakse, V.N.; Koh, C.C.; Wu, Z.; Liu, W.; Sun, Y.; Gao, H.; Menden, M.P.; Xu, C.; et al. Quantitative Proteome Landscape of the NCI-60 Cancer Cell Lines. iScience 2019, 21, 664–680. [Google Scholar] [CrossRef] [Green Version]
- Sinha, A.; Huang, V.; Livingstone, J.; Wang, J.; Fox, N.S.; Kurganovs, N.; Ignatchenko, V.; Fritsch, K.; Donmez, N.; Heisler, L.E.; et al. The Proteogenomic Landscape of Curable Prostate Cancer. Cancer Cell 2019, 35, 414–427.e416. [Google Scholar] [CrossRef] [Green Version]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Wang, J.; Wang, X.; Zhu, J.; Liu, Q.; Shi, Z.; Chambers, M.C.; Zimmerman, L.J.; Shaddox, K.F.; Kim, S.; et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014, 513, 382–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hmeljak, J.; Sanchez-Vega, F.; Hoadley, K.A.; Shih, J.; Stewart, C.; Heiman, D.I.; Tarpey, P.; Danilova, L.; Drill, E.; Gibb, E.A.; et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov. 2018, 8, 1548–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, E.Z.; Jacobson, B.A.; Patel, M.R.; Okon, A.M.; Li, S.; Xiong, K.; Vaidya, A.J.; Bitterman, P.B.; Wagner, C.R.; Kratzke, R.A. Small-molecule inhibition of oncogenic eukaryotic protein translation in mesothelioma cells. Investig. New Drugs 2014, 32, 598–603. [Google Scholar] [CrossRef]
- Grosso, S.; Pesce, E.; Brina, D.; Beugnet, A.; Loreni, F.; Biffo, S. Sensitivity of global translation to mTOR inhibition in REN cells depends on the equilibrium between eIF4E and 4E-BP1. PLoS ONE 2011, 6, e29136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, K.; Shi, Z.; Zhulyn, O.; Denans, N.; Barna, M. Pervasive translational regulation of the cell signalling circuitry underlies mammalian development. Nat. Commun. 2017, 8, 14443. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.; Donehower, L.A.; Cooper, T.A.; Neilson, J.R.; Wheeler, D.A.; Wagner, E.J.; Li, W. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat. Commun. 2014, 5, 5274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Shen, B.; Zhang, D.; Wang, Y.; Tang, Z.; Ni, N.; Jin, X.; Luo, M.; Sun, H.; Gu, P. miR-29a regulates the proliferation and differentiation of retinal progenitors by targeting Rbm8a. Oncotarget 2017, 8, 31993–32008. [Google Scholar] [CrossRef] [Green Version]
- Busacca, S.; Germano, S.; De Cecco, L.; Rinaldi, M.; Comoglio, F.; Favero, F.; Murer, B.; Mutti, L.; Pierotti, M.; Gaudino, G. MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am. J. Respir. Cell Mol. Biol. 2010, 42, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, S.; Galeano, F.; Alon, S.; Raho, S.; Galardi, S.; Polito, V.A.; Presutti, C.; Vincenti, S.; Eisenberg, E.; Locatelli, F.; et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.G.; Park, F.D.; Koechlein, C.S.; Kritzik, M.; Reya, T. Musashi signaling in stem cells and cancer. Annu. Rev. Cell. Dev. Biol. 2015, 31, 249–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, C.G.; Riemondy, K.; Chapnick, D.A.; Bunker, E.; Liu, X.; Kuersten, S.; Yi, R. Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration. Nucleic Acids Res. 2016, 44, 3788–3800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abukar, A.; Wipplinger, M.; Hariharan, A.; Sun, S.; Ronner, M.; Sculco, M.; Okonska, A.; Kresoja-Rakic, J.; Rehrauer, H.; Qi, W.; et al. Double-Stranded RNA Structural Elements Holding the Key to Translational Regulation in Cancer: The Case of Editing in RNA-Binding Motif Protein 8A. Cells 2021, 10, 3543. https://doi.org/10.3390/cells10123543
Abukar A, Wipplinger M, Hariharan A, Sun S, Ronner M, Sculco M, Okonska A, Kresoja-Rakic J, Rehrauer H, Qi W, et al. Double-Stranded RNA Structural Elements Holding the Key to Translational Regulation in Cancer: The Case of Editing in RNA-Binding Motif Protein 8A. Cells. 2021; 10(12):3543. https://doi.org/10.3390/cells10123543
Chicago/Turabian StyleAbukar, Asra, Martin Wipplinger, Ananya Hariharan, Suna Sun, Manuel Ronner, Marika Sculco, Agata Okonska, Jelena Kresoja-Rakic, Hubert Rehrauer, Weihong Qi, and et al. 2021. "Double-Stranded RNA Structural Elements Holding the Key to Translational Regulation in Cancer: The Case of Editing in RNA-Binding Motif Protein 8A" Cells 10, no. 12: 3543. https://doi.org/10.3390/cells10123543
APA StyleAbukar, A., Wipplinger, M., Hariharan, A., Sun, S., Ronner, M., Sculco, M., Okonska, A., Kresoja-Rakic, J., Rehrauer, H., Qi, W., Beusechem, V. W. v., & Felley-Bosco, E. (2021). Double-Stranded RNA Structural Elements Holding the Key to Translational Regulation in Cancer: The Case of Editing in RNA-Binding Motif Protein 8A. Cells, 10(12), 3543. https://doi.org/10.3390/cells10123543






